Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1260-1275. doi:10.7150/jca.12659 This issue Cite
Research Paper
Hsa-miR-301a-3p Acts as an Oncogene in Laryngeal Squamous Cell Carcinoma via Target Regulation of Smad4
1. Department of Otolaryngology, Head & Neck Surgery, The First Hospital Affiliated with Shanxi Medical University, Taiyuan, Shanxi, 030001, China
2. Shanxi Key Laboratory of Otolaryngology Head & Neck Cancer, Taiyuan, Shanxi, 030001, China
3. Department of Otolaryngology, Head & Neck Surgery, The First Hospital Affiliated with Liaoning Medical University, Jinzhou, Liaoning, 121001, China
# These authors contributed equally to this work.
Received 2015-5-11; Accepted 2015-8-28; Published 2015-10-20
Abstract
Laryngeal squamous cell carcinoma (LSCC) is the second most common malignant head and neck squamous cell carcinoma. Exploring the molecular indicators of malignant behavior will enhance our knowledge of this type cancer and provide novel options for its prevention, diagnosis, and treatment. MicroRNA might exert regulatory roles as oncogenes or anti-oncogenes. We studied the expression of miR-301a-3p in LSCC tissues and cell lines and conducted a functional analysis of miR-301a-3p to confirm if miR-301a-3p functions as an oncogene in LSCC. We found Smad4 to be one of the potential target genes of miR-301a-3p, and it functioned as a tumor suppressor in LSCC. Hsa-miR-301a-3p participated in the epithelial-mesenchymal transition (EMT) process, which is considered to be linked to the process of LSCC development. Our present findings indicate that miR-301a-3p acts as an oncogene by directly regulating the anti-oncogene Smad4, thereby playing a role in the occurrence and development of LSCC. The present findings are expected to help in the development of novel targets for the prevention and treatment of LSCC.
Keywords: Laryngeal neoplasms, Squamous cell carcinoma, MicroRNAs, Hsa-miR-301a-3p, Smad4
Introduction
Head and neck squamous cell carcinoma (HNSCC) stands sixth among malignant cancers in the world [1]. As the most common type of HNSCC, laryngeal squamous cell carcinoma (LSCC) occupies the second place among HNSCCs and represents about 5.7-7.6% of malignant tumors [2,3], especially in the northern area of China, including Shanxi Province [4]. Although in recent years, considerable research has focused on the diagnosis and treatment of LSCC, the 5-year survival rate of LSCC patients remains medium level, and the mortality is mainly due to tumor recurrence and local lymph node metastasis [5,6]. Thus, understanding the basis of occurrence and development of LSCC is of importance in clinical practice.
MicroRNAs (miRNAs) are endogenous, small (22-25-nucleotide long), noncoding RNAs and have specific expression in different cells and tissues [7,8].They are involved in several biological processes such as development, cell proliferation, differentiation, and apoptosis [9,10]. The dysregulation of miRNAs might play pivotal roles in tumorigenesis in breast cancer [11,12], liver cancer [13,14], gastric cancer [15,16], and colorectal cancer [17,18]. Thus, researching the function and expression of miRNAs will contribute to the understanding of the pathogenesis, diagnosis, treatment, and prognosis of such tumors.
To investigate the underlying miRNAs in human LSCC, we used a miRNA array and identified a candidate biomarker, namely, hsa-miR-301a-3p (miR-301a-3p). miR-301a-3p has been reported in several types of cancer [19-21], but its function and role in LSCC are unclear. Recent studies have demonstrated that EMT plays an important role in invasion and metastasis of cancer. TGF-β/Smad signal pathway is a crucial pathway in EMT progress and Smad4 is characterized as a mediator of TGF-β/Smad signal passway, so Smad4 also takes part in EMT progress [22]. Smad4 was identified as a tumour suppressor gene at 18q21.1, but the study in LSCC had been reported rarely. Smad4 might be a downstream target gene of miR-301a-3p with the assistance of bioinformatics methods. The aims of this study were to investigate the function and role of miR-301a-3p and Smad4 in LSCC and to determine whether miR-301a-3p regulates Smad4 in a targeted manner. We also investigate the role of miR-301a-3p and Smad4 in EMT and evaluated the clinical significance in the development and progression of LSCC. Our findings might provide a new insight into the mechanism of miRNA and its gene regulation in LSCC.
Materials and methods
Ethics Statement
This study was conducted in accordance with the Helsinki declaration. Before surgery, all of patients signed a written, informed consent, acknowledging that they understood their rights and obligations. The study was approved by the Research Ethics Committee at Shanxi Medical University.
Cell lines and clinical samples
Human LSCC cell line Hep-2, HEK-293 cells were purchased from ATCC. Human LSCC cell line Tu-177 or human bronchial epithelial cell line 16HBE was purchased from Shanghai Bioleaf Biotech Company (Shanghai, China). Cells were plated in RPMI-1640 (Tu-177) (HyClone, Logan, UT, USA) or DMEM (Hep-2, HEK-293, 16HBE) (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (HyClone) and 100mg/mL penicillin/streptomycin (HyClone) and were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Laryngeal carcinoma and corresponding adjacent normal margin (ANM) tissues were obtained from 145 patients undergoing surgery at The First Hospital affiliated with Shanxi Medical University, China. According to the clinical data integrity, follow-up information, and paraffin specimen condition, we finally chose 120 patients for our research. The excluded patients comprised 9 patients who died (5 from heart attack or cerebrovascular accident, 2 from falls, and 2 from unknown reasons). The paraffin-embedded LSCC tissues included 120 patients and 120 cases of corresponding ANM tissues which were selected 45 cases in use of IHC detection of Smad4. We also chose 6 pairs of fresh LSCC and corresponding ANM tissues from patients undergoing surgery in our hospital in 2014 for qRT-PCR. The patients undergoing follow-up were treated at our department between 2002 and 2013, and recorded with detailed clinical and follow-up data.
Homo sapiens Smad4 overexpression vector construction
To manufacture a Smad4 overexpressing vector, Smad4 cDNA was amplified with total RNA from HEK-293 cells by reverse transcription-PCR. The pcDNA™3.1 (+) vector was purchased from Invitrogen (Carlsbad, CA, USA). The next steps were performed according to the instructions. The following primers of Smad4 were used: BamH I F: 5'cgcggatccgccaccATGGACAATATGTCTATTACGAATACAC 3'; Smad4 Xho I R: 5'ccgctcgagTCAGTCTAAAGGTTGTGGGTCTGCA 3'. In order to make sure the overexpression of Smad4 in level of transcript and protein, Smad4 was detected with the methods of qRT-PCR and western blot in LSCC cell lines Hep-2 and Tu-177 after transfection.
Cell transfection
Cells were plated in 6-well dishes (2.0 ×105cells per well). A miR-301a-3p inhibitor, a negative control (NC) precursor (RiboBio, Guangzhou, China), Smad4 vector pcDNA™3.1(+)-Smad4 plasmid, and Smad4 NC were transfected at final concentrations of 40nM each by using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions. After transfection, cells were harvested and used for qRT-PCR, western blot analysis, and cell function experiments.
Real-time PCR
Total RNA was extracted from cell lines and tissues using TRIzol (Invitrogen) or the miRNeasy FFPE kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. qRT-PCR was performed using the SYBR® Green PCR kit (Toyobo, Osaka, Japan) and the ABI PRISM® 7500 Sequence Detection System (Applied Biosystems, FosterCity, CA, USA). The primer sequences were as follows (Sangon, Shanghai, China): hsa-miR-301a-3p: forward, 5ʹ-ACACTCCAGCTGGGCAGTGCAATAGTATTGTC-3ʹ and reverse, 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ; Smad4: forward, 5ʹ-TGCTGTTGCAAAGGCTGCTT-3ʹ and reverse, 5ʹ-GCTTCTGGCATAGCTGCATT-3ʹ. U6 or 18S rRNA was used as reference for miRNAs or mRNAs respectively. The primer sequences were as follows: U6: forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; 18S rRNA: forward, 5ʹ-CCTGGATACCGCAGCTAGGA-3ʹ and reverse, 5ʹ-GCGGCGCAATACGAATGCCCC-3ʹ. The qRT-PCR results, recorded as threshold cycle numbers (Ct), were normalized against an internal control. The 2-△△Ct method was used to quantify the relative levels of gene expression [23]. Each sample was analyzed in triplicate. The final measurement result was the average of the values obtained from 3 experiments.
Dual-luciferase reporter assay
To identify whether miR-301a-3p directly regulated Smad4 mRNA expression, the dual-luciferase reporter assay for target validation was applied. HEK-293 cells were transfected with psiCHECK™-2 Vector (Promega, Madison, WI, USA) containing wild-type firefly luciferase with Smad4 or firefly luciferase with mut-Smad4 3ʹ-UcTR and with miR-301a-3p, or negative-control oligonucleotide (RiboBio). After 48h co-transfection, cells were harvested, and luciferase activity was measured with the Dual-Luciferase Reporter Gene Assay Kit (Promega) according to the manufacturer's instructions.
MTS assay
The in vitro cell viability in Hep-2 and Tu-177 cells was evaluated using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega). Briefly, 1×104 cells were seeded in 96-well culture plates. After transfection and culture, 10µL MTS was added to each well, and the cells were incubated for 4h. The optical density was measured at 490nm using a microplate spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Colony-forming assay
To determine colony-forming ability, cells were transfected with miR-301a-3p inhibitor or Smad4 plasmid. Cells were seeded in 96-well plates and incubated for 7 days at 37°C with 5% CO2. After 70% ethanol fixation, the plate was stained with crystal violet for 20min, and cells were counted and photographed.
EDU cell proliferation assay
One of the most sensitive and accurate methods for determining cell division is the EDU assay. Transfected cells were seeded on cover slips. When the confluency rate reached 80-90%, the medium was discarded and 100μL EDU medium (50μM) was added. Incubation was performed at 37°C for 2h. Cells were fixed in 4% paraformaldehyde for 20min. For permeabilization, 0.5% TritonX-100 in phosphate-buffered saline (PBS) was applied for 20min. 100µL 1×Apollo® stain liquid was added per well, and cells were incubated at room temperature away from light for 30min. Then, 100µL DAPI stain liquid was added per well. Next, the cells were washed with PBS and observed under a fluorescence microscope.
Cell migration and invasion assays
For the migration assay, cells transfected with the miR-301a-3p inhibitor or NC were harvested and plated on the upper chamber of a Transwell unit (BD Biosciences, San Jose, CA, USA) in 500µL serum-free RPMI-1640, and 1mL RPMI-1640 containing 10% FBS was added to the bottom chamber. After incubation for 24h at 37°C, cells remaining on the upper chamber were removed by scrubbing, and the migrated cells were fixed with 4% paraformaldehyde, stained with crystal violet for 10min, and photographed. For invasion assays, the upper chamber was covered with Matrigel (BD Biosciences) for 2h. The other steps were similar to the migration assay.
Western blot analysis
The transfected cells were washed and lysed. The proteins were collected by centrifugation and quantified by using the bicinchoninic acid protein assay kit (Thermo Scientific). Aliquots containing 40µg proteins with 2×loading buffer (0.25 mol/L Tris-Cl, pH 6.8, 10% sodium dodecyl sulfate, 0.5% bromophenyl blue, and 50% glycerol) were boiled for 5min, loaded into 10% Tris-HCl polyacrylamide gels (80V, 50min) and transferred electrophoretically to a PVDF membrane (Millipore, Billerica, MA, USA) for immune detection of candidate proteins. The membrane was blocked with 5% skimmed milk at 4°C overnight and then incubated with primary antibodies overnight. Anti-Smad4 antibody (Santa Cruz Biotechnology, Dallas, TX, USA, 1:1000), anti-E-cadherin antibody (CellSignaling Technology, Danvers, MA, USA, 1:1000), anti-N-cadherin antibody (CellSignaling, 1:1000), anti-vimentin antibody (Abcam, Cambridge, MA, USA, 1:800), anti-matrix metalloproteinase2 (MMP2) antibody (Abcam, 1:1000), anti-MMP9 antibody (Abcam, 1:1000), and GAPDH antibody (KangChen Biotech, Shanghai, China, 1:10000) were used for western blot analysis, according to the manufacturer's instructions. The secondary antibodies were goat anti-rabbit IgG at a dilution of 1:20000 (SouthernBiotech, Birmingham, AL, USA). The blots were transferred to X-ray films (Kodak, USA). Bands were analyzed using Image J software for quantification, and normalization was done using GAPDH band intensities.
Flow cytometry assay
Cells were collected by trypsinization, fixed in ice-cold 75% ethanol in PBS, counted, and treated with 20mg/mL RNase. Each sample contained 105-106 cells/mL. Cell death was analyzed using the Annexin-V FITC two-color flow cytometry kit (KeyGen Biotech, Nanjing, China) according to the manufacturer's instructions. Cells were stained with 10µL propidium iodide for 15min to test the cell cycle. Next, the cell suspensions were diluted using binding buffer and analyzed with a BD FACSCalibur Flow Cytometer (BD Biosciences).
Immunohistochemistry
Immunohistochemical staining was performed in 2 different tissue types: LSCC and corresponding ANM tissues. Paraffin-embedded specimens were cut into 4 μm sections and baked at 65°C for 3h. After deparaffinization, rehydration, antigen retrieval, blocking of endogenous peroxidase, and treatment with normal serum, the sections were incubated with anti-Smad4 antibody (Santa Cruz, 1:100) overnight at 4°C. Then, the sections were incubated with the streptavidin-horseradish peroxidase complex. This was followed by DAB staining and hematoxylin staining. Smad4 expression was evaluated semiquantitatively by the proportion of tumor cells with positive staining. Staining intensity was scored as 0, no staining (same as negative controls incubated by PBS); 1, weak intensity; 2, moderate intensity; 3, strong intensity. Positive staining percentage was scored as 1, <10%; 2, 11-50%; 3, 51-80%; and 4, >81%. The final assessment was the mean combined score of staining intensity and positive staining percentage. According to the overall score, specimens were classified into two grades: negative expression (-), 1∼2 points; positive expression (+), 3∼7 points. To determine the expression pattern, at least five fields (×400) and at least 100 cells per field were selected for microscopy randomly. The sections were scored three times independently by two pathologists who were blinded to the clinical parameters, and the final score was the average of the scores by these 2 observers.
Statistical analysis
Statistical analysis was performed using the SPSS 20.0 statistical software package. Mean ± standard deviation values of parameters were obtained from at least 3 separate experiments. The differences between groups were analyzed using Student's t-test. Pearson's correlation was used to analyze the relationship between the expression of miR-301a-3p and Smad4 in LSCC tissues. Survival was analyzed by the Kaplan-Meier method with the log rank test. Multivariate analysis was performed by use of Cox proportional hazards model. The difference was deemed statistically significant at P <0.05.
Results
miR-301a-3p regulates Smad4 expression by interaction with Smad4 3ʹ-UTR
We attempted to investigate expression levels of miR-301a-3p and Smad4 mRNA in human LSCC cell lines Hep-2 and Tu-177 as well as human bronchial epithelial cell line 16HBE, and 6 pairs of LSCC tissues and corresponding ANM tissues by qRT-PCR tentatively (detailed clinical data shown as Supplementary Table 1). As shown in Figure 1A, miR-301a-3p expressions were significantly higher in human LSCC tissues than in the ANM tissues. In Figure 1C, miR-301a-3p expressions were significantly higher in human LSCC cells than in the 16HBE cells. The expression levels of Smad4 mRNA in human LSCC tissues and cancer cell lines were shown in Figure 1B and 1D. As expected, Smad4 expression was significantly lower in human LSCC cells and tissues than in the 16HBE cells and ANM tissues.
To identify the potential target gene of miR-301a-3p, we searched for candidate genes using miRNA databases such as TargetScan [24] and found Smad4 to be a predicted target gene of miR-301a-3p. Moreover we found their binding sites (Figure 1E).We used the dual-luciferase reporter gene system to confirm whether there is a targeted modulation of miR-301a-3p and Smad4. We retrieved the 3ʹ-UTR sequence and constructed wild-type and mutant-type vectors. psi-CHECK2-Smad4-3ʹ-UTR and miR-301a-3p co-transfected in HEK-293 cells. The results showed that miR-301a-3p was able to significantly repress luciferase activity of wild-type Smad4-3ʹ-UTR, while the mutant type abrogated this effect (Figure 1F). Therefore, the luciferase assays revealed Smad4 to be a direct target of miR-301a-3p.
Anti-miR-301a-3p can inhibit growth and proliferation in LSCC cell lines
To investigate the function of miR-301a-3p in LSCC, we used the “loss-of-function” with miR-301a-3p inhibitor, which was transfected into human LSCC cell lines and compared with the negative control (NC) and blank groups. The MTS assay showed that the miR-301a-3p inhibitor reduced the cell proliferation ability of Hep-2 and Tu-177 cells to a greater extent compared with the NC and the blank (Figure 2A, 2B). After Hep-2 and Tu-177 cells were transfected with the miR-301a-3p inhibitor, the EDU assay revealed that duplication activity was inhibited. The cells were counted under a fluorescent microscope, and the proliferation rate of the miR-301a-3p inhibitor group was found to be significantly lower than that of the NC and blank groups (Figure 2C). Then, we observed the effect of miR-301a-3p inhibitor on colony formation in LSCC cell lines. The colony-formation rates decreased after transfection with miR-301a-3p inhibitor (Figure 2D).
Anti-miR-301a-3p can induce apoptosis and arrest the cell cycle at the G0/G1 phase in LSCC cell lines
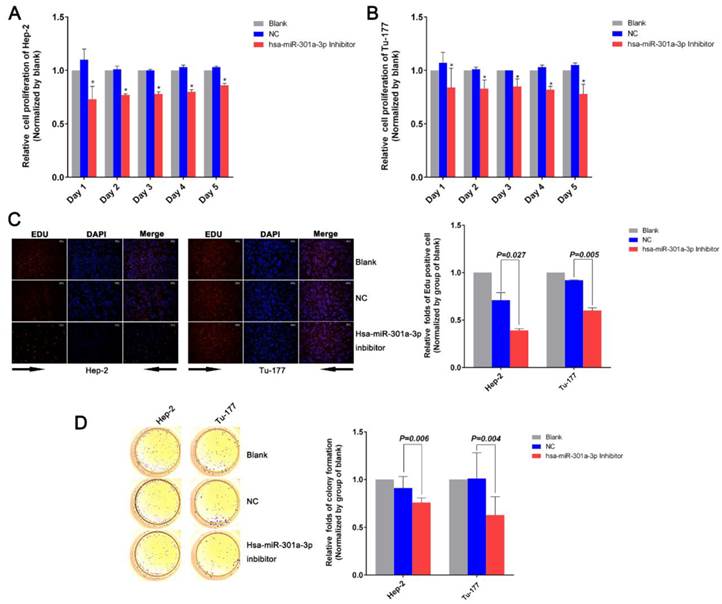
After treatment with the miR-301a-3p inhibitor and miR-301a-3p inhibitor NC, the changes in cells apoptosis rate and cell cycle were assessed by flow cytometry (FCM). In Hep-2 cells transfected with the miR-301a-3p inhibitor, FCM showed that the cell cycle was arrested at the G0/G1 phase, suggested the suppression of the cell proliferation (Figure 3A). The same phenomenon was also demonstrated in the Tu-177 cells (Figure 3B). Cells in which apoptosis occurred at early stages of miR-301a-3p inhibitor-transfected cells were more than the NC cells and blank cells (Figure 3C). Therefore, we can infer that LSCC cell apoptosis might be occurring through cell cycle arrest at the G0/G1 phase.
miR-301a-3p regulates Smad4 expression by interaction with Smad4 3ʹ-UTR. (A) miR-301a-3p expression levels in human LSCC and corresponding ANM tissues were detected by qRT-PCR. (B) Smad4 expression levels in human LSCC and corresponding ANM tissues were detected by qRT-PCR. (C) miR-301a-3p expression levels in human LSCC cell lines and human bronchial epithelial cell line 16HBE were detected by qRT-PCR. (D) Smad4 expression levels in human LSCC cell lines and human bronchial epithelial cell line 16HBE were detected by qRT-PCR. (E) The binding sites of miR-301a-3p and Smad4 were found using miRNA databases Targetscan. (F) The dual-luciferase reporter gene system was used to address targeted modulation of miR-301a-3p and Smad4. miR-301a-3p could significantly repress luciferase activity of wild-type Smad4-3ʹ-UTR when transfected with the Smad4-3ʹ-UTR plasmid. (ANM: Adjacent Normal Margin. All experiments were performed in triple.)
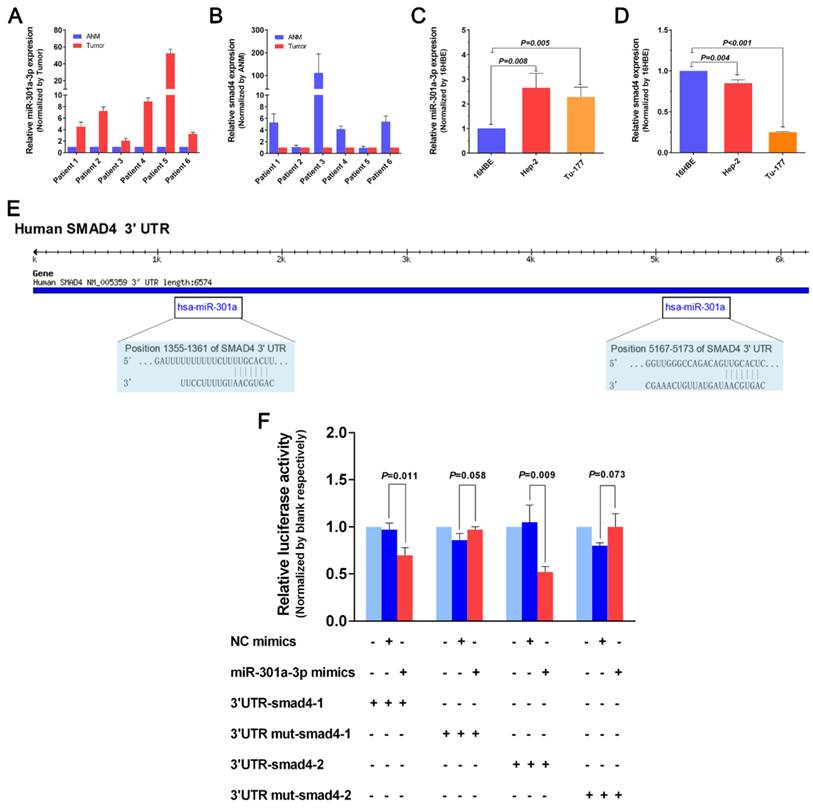
Anti-miR-301a-3p can inhibit cell growth and proliferation. (A) Cell viability in Hep-2 was analyzed by the MTS assay. (B) Cell viability in Tu-177 was analyzed by the MTS assay. (C) After transfection, cell proliferation ability was measured by the EDU assay. (D) Colony-formation ability was determined by the colony-forming assay. (All experiments were performed in triple.)
Smad4 overexpression can also lead to a series of cellular phenomena as anti-miR-301a-3p
When we overexpressed Smad4 using a Smad4 vector pcDNA3.1™(+)-Smad4 plasmid to investigate its function, Smad4 mRNA and protein expression level were increased in Smad4 group compared with the vector group (Normalized by blank group) in Hep-2 and Tu-177 cells (Figure 4A, 4B).
MTS assay results showed that cell proliferation ability in Hep-2 or Tu-177 was lower in the Smad4 group than in the blank and vector groups (Figure 4C, 4D). The EDU assay showed that increased expression of Smad4 could inhibit cell proliferation and relative quantity of LSCC cell lines Hep-2 and Tu-177 (Figure 4E). The colony-formation rates decreased after transfection with Smad4 (Figure 4F). The cell cycle was arrested at the G0/G1 phase (Figure 5A, 5B). Similarly, the early apoptotic cells in Smad4-transfected cells were more than those in the blank and vector cells (Figure 5C).
Anti-miR-301a-3p and overexpressing Smad4 can inhibit migration and invasion, and reverse epithelial-mesenchymal transition in LSCC cell lines
The migration and invasion of cancer cells were analyzed by the Transwell assay. As shown in Figure 6A, miR-301a-3p inhibitor-transfected cells showed less migration through membranes than NC. The results of the invasion assay also indicated that inhibition of miR-301a-3p expression significantly reduced human LSCC cell invasion ability in both LSCC cell lines (Figure 6C). Further migration and invasion experiments indicated that an increase in Smad4 expression resulted in a remarkable decrease in migratory and invasive potential (Figure 6B, D).
Western blot analysis was used to explore the relationship of miR-301a-3p and Smad4 and the genes involved in epithelial-mesenchymal transition (EMT). The level of E-cadherin was found to increase, while the levels of N-cadherin, vimentin, MMP2, and MMP9 decreased (Figure 7).
Anti-miR-301a-3p can induce apoptosis in LSCC cells and arrest the cell cycle at the G0/G1 phase. (A, B) Cell cycle of Hep-2 and Tu-177 cells after transfection with miR-301a-3p inhibitor or NC. (C) Flow cytometry-based analysis of apoptosis in Hep-2 and Tu-177 cells transfected with miR-301a-3p inhibitor or NC. (All experiments were performed in triple.)
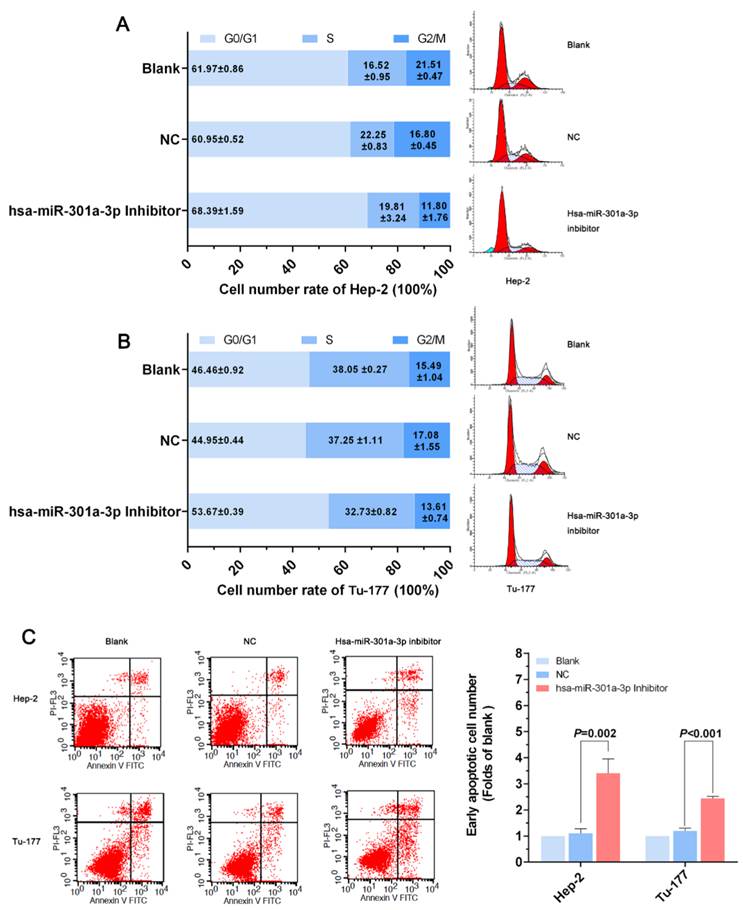
Overexpression of Smad4 can also inhibit cell growth and proliferation. (A) Smad4 mRNA expression level was detected by qRT-PCR after transfection. (B) Smad4 protein expression level was detected by western blot analysis after transfection. (C, D) Cell viability was analyzed by MTS assay. (E) Cell proliferation ability was measured by the EDU assay after transfection. (F) Colony-formation ability was detected by the colony-forming assay. (All experiments were performed in triple.)
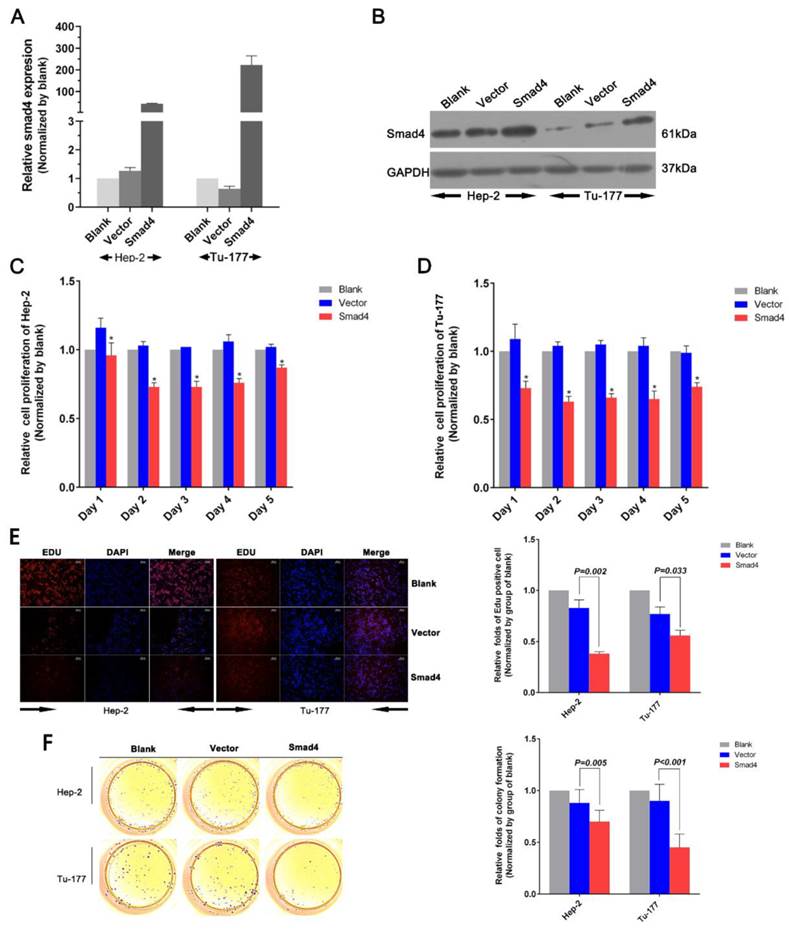
Overexpression of Smad4 can induce apoptosis in LSCC cells and arrest the cell cycle at the G0/G1 phase. (A, B) Flow cytometry analysis of the cell cycle of Hep-2 cells and Tu-177 cells after transfection with Smad4 or vector. (C) Flow cytometry analysis of apoptosis in Hep-2 cells and Tu-177 cells transfected with Smad4 or vector. (All experiments were performed in triple.)
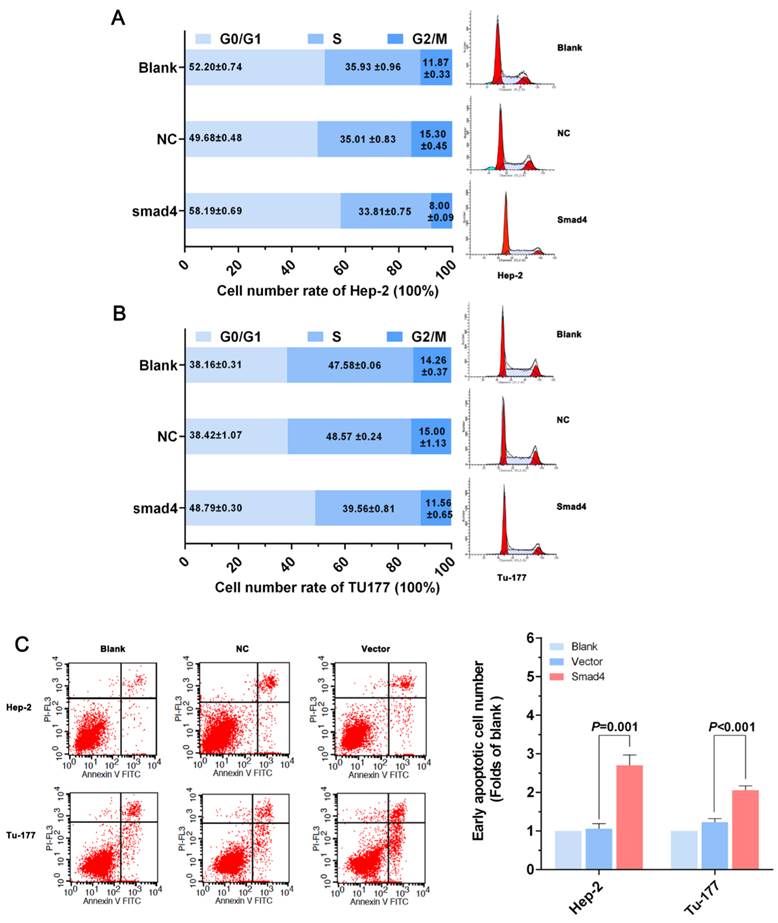
Anti-miR-301a-3p and overexpression of Smad4 can inhibit cell migration and invasion. (A, C) Migration and invasion assays were performed using Transwell chambers after cells were transfected with miR-301a-3p inhibitor or NC. (B, D) Migration and invasion assays were performed using Transwell chambers after cells were transfected with Smad4 or vector. (All experiments were performed in triple.)
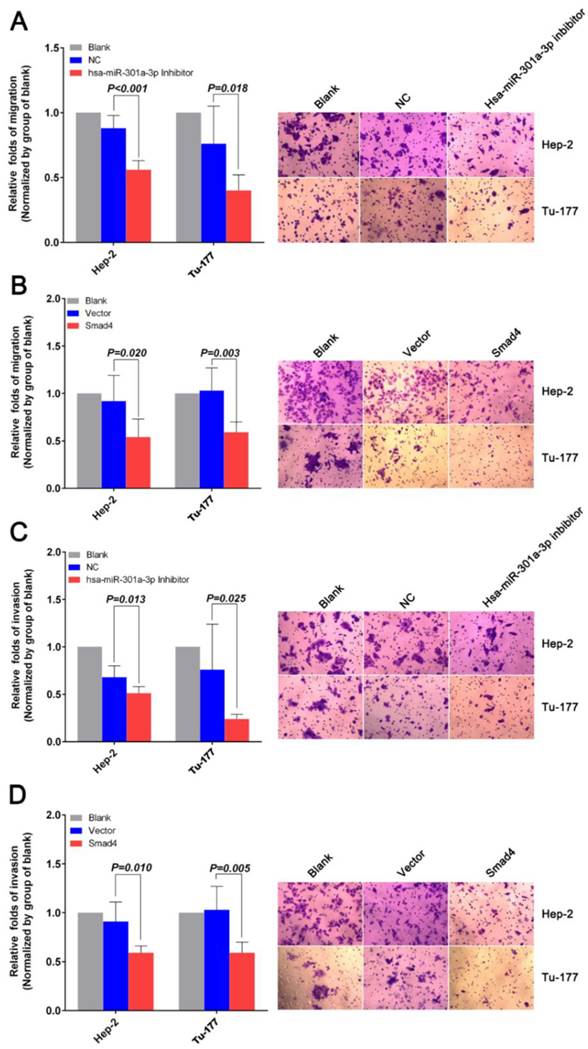
miR-301a-3p and Smad4 play a part in EMT in human LSCC. The expression levels of EMT related factors in Hep-2 and Tu-177 cells were examined by western blot analysis after transfection. (A) Hsa-miR-301a-3p inhibitor, (B) Smad4 overexpression. (All experiments were performed in triple.)
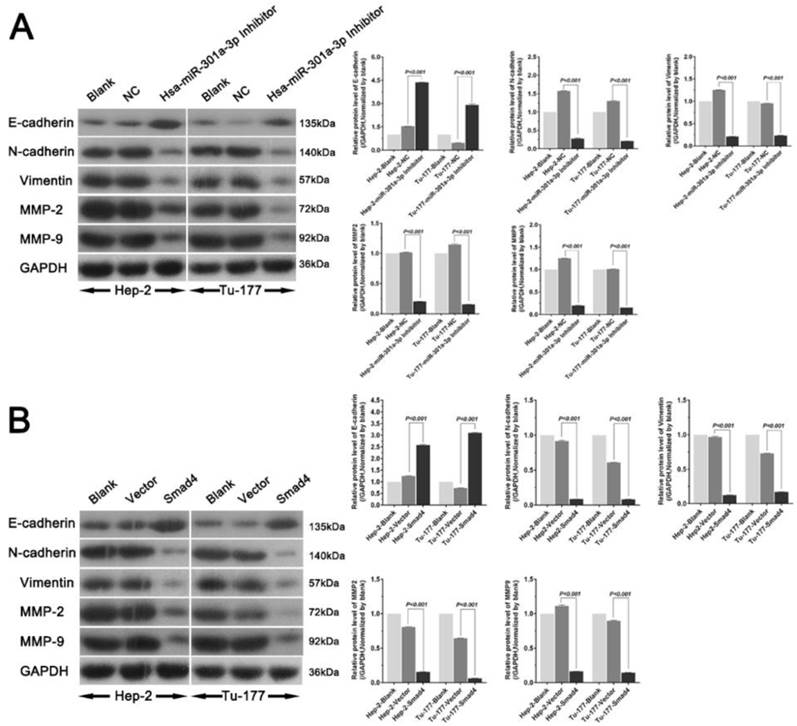
Association of miR-301a-3p and Smad4 expression with malignant clinicopathological characteristics in LSCC patients
A summary of study population and clinicopathological variables is in Supplementary Table 2. The above findings indicate that miR-301a-3p and Smad4 act as oncogene and tumor suppressor gene, respectively, at the cellular level. qRT-PCR results from LSCC tissues and corresponding ANM tissues showed the expression of miR-301a-3p was higher in LSCC samples than in the normal samples (Figure 8A). The average of miR-301a-3p relative expression was 2.957±2.067, and the median was 2.464. Low expression was defined as ≤2.464 (60 out of 120 patients), and high expression was defined as >2.464 (the remaining 60 patients). By investigating the relationship between the expression of miR-301a-3p and the clinicopathological parameters in the LSCC patients, miR-301a-3p expression was found to be associated with primary tumor, while no relationship with age, sex, primary sites, histologic differentiation ,cervical lymph node metastasis or clinical stage was found (Table 1).
Immunohistochemistry studies of pathological sections showed that the level of Smad4 expression was lower in LSCC tissues than that in adjacent healthy tissues. According to the scoring criteria, negative and positive staining were detected in 70.83% (85/120) and 29.17% (35/120) tumor samples, respectively, while 71.11% (32/45) samples of adjacent normal tissues yielded positive staining (P<0.01). Negative Smad4 expression was observed in moderately or poorly differentiated LSCC specimens. Under microscopic observation, the positive expression of Smad4 was mainly localized in the cytoplasm and occasionally in the nuclei of cells of carcinoma and adjacent tissue (Figure 8B-I). The data of the association between Smad4 expression and the clinicopathological characters in 120 LSCC patients are presented in Table 1. There were significant correlations between Smad4 expression and histologic differentiation, as well as cervical lymph node metastasis. However, there were no significant associations with age, sex, primary sites, primary tumor and clinical stage. Spearman's correlation analysis performed to compare the correlation of Smad4 and miR-301a-3p in 120 samples showed them to be significantly inversely correlated (R=-0.458, p<0.001) (Table 2). Thus, miR-301a-3p and Smad4 might serve as potential prognostic markers predicting patient survival.
Association of miR-301a-3p and Smad4 expression with clinicopathological features in LSCC patients
| Clinicopathological features | Case | miR-301a-3p | χ2 | p | Smad4 | χ2 | p | ||
|---|---|---|---|---|---|---|---|---|---|
| High | Low | + | - | ||||||
| Age | |||||||||
| ≤61 years | 55 | 26 | 29 | 0.302 | 0.583 | 17 | 38 | 0.149 | 0.699 |
| >61 years | 65 | 34 | 31 | 18 | 47 | ||||
| Sex | |||||||||
| Male | 108 | 53 | 55 | 0.370 | 0.543 | 29 | 79 | 2.801 | 0.094 |
| Female | 12 | 7 | 5 | 6 | 6 | ||||
| Primary sites | |||||||||
| Supraglottic | 59 | 31 | 28 | 0.18※ | 15 | 44 | 1.401 | 0.496 | |
| Glottic | 57 | 29 | 28 | 18 | 39 | ||||
| Subglottic | 4 | 0 | 4 | 2 | 2 | ||||
| Histologic differentiation | |||||||||
| Good | 40 | 15 | 25 | 3.750 | 0.053 | 19 | 21 | 9.761 | 0.002 |
| Moderate to Poor | 80 | 45 | 35 | 16 | 64 | ||||
| Primary tumor | |||||||||
| T1+T2 | 73 | 42 | 31 | 4.232 | 0.040 | 17 | 56 | 3.118 | 0.077 |
| T3+T4 | 47 | 18 | 29 | 18 | 29 | ||||
| Cervical lymph node metastasis | |||||||||
| N0 | 96 | 48 | 48 | 0.208 | 0.648 | 32 | 64 | 4.034 | 0.045 |
| N+ | 24 | 12 | 12 | 3 | 21 | ||||
| Clinical stage | |||||||||
| I+II | 65 | 32 | 33 | 0.034 | 0.855 | 21 | 44 | 0.677 | 0.411 |
| III+IV | 55 | 28 | 27 | 14 | 41 | ||||
※Fisher's exact test
Correlation of Smad4 and miR-301a-3p expression in 120 samples of LSCC
| Smad4 IHC | miR-301a-3p relative expression | Total Case | Correlation coefficient (Spearman's test) | p | |
|---|---|---|---|---|---|
| High | Low | ||||
| + | 5 | 30 | 35 | -0.458 | <0.001 |
| _ | 55 | 30 | 85 | ||
| Total | 60 | 60 | 120 | ||
High expression of miR-301a-3p and low expression of Smad4 indicate poor prognosis of LSCC
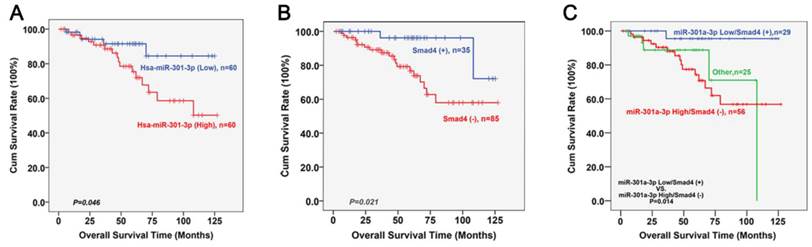
The follow-up was continued until December 2013. Fifteen patients were lost to follow-up. During the follow-up, 21 patients died (10, from in situ recurrence; 8, cervical lymph node metastases; 2, metastasis to the lung; and 1, metastasis to the liver). Kaplan-Meier survival analyses of LSCC patients who had undergone surgery based on the high or low expression of miR-301a-3p and Smad4 are shown as figure 9A and 9B. Low expression levels of miR-301a-3p or positive expression levels of Smad4 was correlated with better survival (P=0.046, P=0.021, respectively; log rank test). Further division was made based on the expression status of miR-301a-3p combined with Smad4. In figure 9C, high expression of miR-301a-3p but negative expression of Smad4 were found to be associated with poor survival (P=0.014). The Cox regression analysis was performed with enter method, and it was indicated that T stage (OR=2.861, P=0.011, 95%CI=1.270∼6.449) and Smad4 (OR=0.133, P=0.011, 95%CI=0.028∼0.634) was independent prognostic factors. Notably, when the status of miR-301a-3pHigh/Smad4- was entered into Cox model, it was a significantly independent predictor of LSCC survival (OR=2.039, P=0.046, 95%CI=1.012∼4.106).
miR-301a-3p and Smad4 are potential prognostic markers in human LSCC. (A) The expression of miR-301a-3p was examined in human LSCC and adjacent normal paraffin samples (ANM) by qRT-PCR (All experiments were performed in triple). (B-I) Representative immunohistochemistry images of Smad4 in LSCC (Magnification, 400×). (B) Histologic differentiation good (Positive expression, +), (C) Histologic differentiation poor (Negative expression,-) , (D) Cervical lymph node metastasis N0 (Positive expression, +), (E) Cervical lymph node metastasis N+ (Negative expression,-), (F) Hsa-miR-301a-3p low (Positive expression, +), (G) Hsa-miR-301a-3p high (Negative expression,-), (H) ANM (Positive expression, +), (I) ANM (Negative expression,-).
Discussion
Laryngeal Squamous Cell Carcinoma (LSCC) is an aggressive type of HNSCC [25]. Despite surgical resection-based comprehensive treatment being widely adopted, the overall 5-year survival rateis around 50%, which suggests that satisfactory improvements have not been made in this direction [26]. Therefore, in-depth understanding of the molecular mechanisms of LSCC development and progression are of great clinical significance. In the current scenario, miRNAs have come to be the focal point in molecular oncology studies.
miRNAs are a special kind of endogenous, non-coding RNA involved in cell proliferation, apoptosis, and metastases, among other such phenomena[27, 28]. Dysregulated miRNAs may play an important role by acting as oncogenes or anti-oncogenes [29]. miR-301a-3p has been reported to be upregulated in several malignant tumors, including pancreatic cancer[30], ovarian cancer[31], breast cancer[32], and hepatocellular cancer[33]. However, relatively little is known about the role of miR-301a-3p in LSCC. First, we used cell lines and a small number of fresh tissues (6 pairs) tentatively to test the expressions of miR-301a-3p and Smad4 by qRT-PCR. The expression of miR-301a-3p was significantly higher in LSCC cell lines Hep-2 and Tu-177 and fresh LSCC tissues than in 16HBE cells and corresponding ANM tissues. However, the expression of Smad4 in LSCC cell lines and tissues were lower than the control group. These results reminded the possibility of regulation between miR-301a-3p and Smad4. Therefore, in follow-up experiments, we designed a dual-luciferase assay to investigate possibility of targeted modulation between miR-301a-3p and Smad4, and we tested the expressions of miR-301a-3p and Smad4 in 120 clinical paraffin specimens by qRT-PCR or immunohistochemistry to study the correlation between miR-301a-3p and Smad4 expression and pathologic parameters. Observably, qRT-PCR results showed that miR-301a-3p was also higher in LSCC paraffin samples than in the normal paraffin samples and the role of miR-301a-3p expression was closely related to primary tumor. This is consistent with findings by Ma et al., wherein miR-301a-3p was markedly upregulated in pro-metastatic breast cancer cell lines and primary tumor samples from patients with distant metastases [34]. We next suppressed miR-301a-3p by transfection with a miR-301a-3p inhibitor in LSCC cell lines and performed functional experimental studies. Cancer cell growth, proliferation, migration, and invasion were suppressed by the repression of miR-301a-3p; apoptosis was enhanced; and the cell cycle was arrested at the G0/G1 phase. The survival curve indicated that the upregulation of miR-301a-3p was significantly associated with poorer patient survival. These results reinforce the opinions of several researchers that miR-301a-3p functions as an oncogene and results in the occurrence and development of LSCC.
To explore miR-301a-3p downstream target genes, we used a series of algorithms and identified Smad4 as a potential target gene. The possibility of targeted modulation was confirmed by a dual-luciferase assay, and miR-301a-3p and Smad4 were found to be significantly inversely correlated. Smad4 is a critical regulatory protein in TGF-β/BMP signaling pathways [35, 36]. Deletion or degradation of Smad4 in tumors can specifically inhibit the tumor suppressor effect of TGF-β [37]. Smad4 was first found in pancreatic cancer [38] and was regarded as a tumor-suppressor gene in several cancers such as esophageal cancer [39], colorectal cancer [40], and prostate cancer [41]. Hao et al. found that the mRNA level of Smad4 was markedly inhibited in pancreatic cancer tissues compared with the normal tissues [42], consistent with previous reports of Smad4 downregulation in pancreatic adenocarcinomas [43, 44]. In line with these findings, our qRT-PCR analysis showed that the expression of Smad4 in LSCC cell lines and tissues were lower than the control group. Liu et al. investigated Smad4 expression in breast carcinoma samples to evaluate the association between Smad4 and outcome in breast cancer [45].Our immunohistochemistry analysis further supported their findings of the role of Smad4 expression in histologic differentiation and cervical lymph node metastasis. Patients with low Smad4 expression tended to have shorter overall survival compared with those with higher Smad4 expression. Therefore, Smad4 overexpression in LSCC cell lines can enhance apoptosis and inhibit cancer cell growth, proliferation, migration, and invasion. As indicated earlier, the cell cycle was blocked at the G0/G1 phase, suggesting that Smad4 acts as a tumor suppressor, thereby facilitating LSCC carcinogenesis and progression.
Cell migration is a complex, multi-step process that plays an important role in cancer progression. Cell invasion is related to, and encompasses, cell migration. However, cells do more than just migrate. EMT plays a critical role in cancer progression and metastasis, and cancer cells can acquire the capacity to spread and invade other tissues when they undergo EMT[46]. Smad4 participates in the EMT when induced by the TGF-β signaling pathway [47]. We inhibited miR-301a-3p or overexpressed Smad4 in cell lines and detected the expression of key molecular markers in EMT. Epithelial marker E-cadherin increased while mesenchymal marker N-cadherin decreased, implying that EMT might have been interrupted by transfection. Moreover, the expression of mesenchymal marker vimentin decreased. Recent research has found that tumor-associated MMPs can stimulate processes associated with EMT [48]. We also confirmed this viewpoint. The level of MMP2 and MMP9 decreased when the cells were transfected. This illustrates that dysregulation of miR-301a-3p and Smad4 affect EMT in LSCC.
Kaplan-Meier survival analyses of miR-301a-3p and Smad4 in LSCC patients. (A) Kaplan-Meier curves for LSCC patients with high and low expression of miR-301a-3p. (B) Kaplan-Meier curves for LSCC patients with positive and negative expression of Smad4. (C) Kaplan-Meier curves for LSCC patients with different expression status of miR-301a-3p combined with Smad4.
To sum up, we transfected miR-301a-3p inhibitor into LSCC cells in vitro and found that suppressing the expression of miR-301a-3p could block LSCC cell growth, promote apoptosis, and inhibit invasion or metastasis. By directly regulating its target gene Smad4, miR-301a-3p acts as an oncogene to play an important biological role. miR-301a-3p/Smad4 may represent a novel pathway to target in relation with the occurrence and development of LSCC. For instance, testing miR-301a-3p and Smad4 in the blood or biopsied tissue of patients might facilitate early detection of LSCC. Hence, the present findings enhance current knowledge for the treatment and evaluation of LSCC.
Abbreviations
LSCC: laryngeal squamous cell carcinoma; HNSCC: Head and neck squamous cell carcinoma; miRNA/miR: microRNA; NC: negative control; UTR: untranslated region; qRT-PCR: reverse transcription and quantitative polymerase chain reaction; ANM: adjacent normal margin; FBS: fetal bovine serum; FFPE: formalin fixed paraffin embedded; PBS: phosphate-buffered saline; MMP: matrix metalloproteinase; SPSS: Statistical product and service solutions.
Supplementary Material
Supplementary Tables 1- 2.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No.81172584) and Shanxi Undergraduate Education Innovation Project (2015BY32).
Author contributions
Yan Lu, Jian Kang, Binquan Wang conceived and designed the experiments. Yan Lu, Wei Gao, Chunming Zhang, Shuxin Wen performed the experiments. Yan Lu, Hui Huangfu, Wei Gao, Jian Kang analysed the results. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Black RJ, Bray F, Ferlay J. et al. Cancer incidence and mortality in the European Union: cancer registry data and estimates of national incidence for 1990. Eur J Cancer. 1997;33:1075-1107
2. Chu EA, Kim YJ. Laryngeal cancer: diagnosis and preoperative work-up. Otolaryngol Clin North Am. 2008;41:673-95
3. Dequanter D, Lothaire P, Zouaoui K. et al. Epidemiology and clinical characteristics of larynx and hypopharynx carcinoma: a comparative study in the Hainaut and review of the literature. Acta Chir Belg. 2012;112:423-5
4. Gao W, Zhang CM, Feng Y. et al. Fascin-1, ezrin and paxillin contribute to the malignant progression and are predictors of clinical prognosis in laryngeal squamous cell carcinoma. PLoS One. 2012;7:e50710
5. Ganly I, Patel SG, Matsuo J. et al. Predictors of outcome for advanced-stage supraglottic laryngeal cancer. Head Neck. 2009;31:1489-95
6. Hermans R. Staging of laryngeal and hypopharyngeal cancer: value of imaging studies. Eur Radiol. 2006;16:2386-400
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
8. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205
9. Sun W, Julie Li YS, Huang HD. et al. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng. 2010;12:1-27
10. Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215-39
11. Iorio MV, Ferracin M, Liu CG. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-70
12. Singh B, Ronghe AM, Chatterjee A. et al. MicroRNA-93 regulates NRF2 expression and is associated with breast carcinogenesis. Carcinogenesis. 2013;34:1165-72
13. Gooderham NJ, Koufaris C. Using microRNA profiles to predict and evaluate hepatic carcinogenic potential. Toxicol Lett. 2014;228:127-32
14. Tsai WC, Hsu SD, Hsu CS. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-97
15. Zabaleta J. MicroRNA: A Bridge from H. pylori Infection to Gastritis and Gastric Cancer Development. Front Genet. 2012;3:294
16. Gorur A, Balci Fidanci S, Dogruer Unal N. et al. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep. 2013;40:2091-96
17. Mosakhani N, Sarhadi VK, Borze I. et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer. 2012;51:1-9
18. Drusco A, Nuovo GJ, Zanesi N. et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One. 2014;9:e96670
19. Shi W, Gerster K, Alajez NM. et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926-37
20. Xu XD, He XJ, Tao HQ. et al. Abnormal expression of miR-301a in gastric cancer associated with progression and poor prognosis. J Surg Oncol. 2013;108:197-202
21. Zhou P, Jiang W, Wu L. et al. miR-301a is a candidate oncogene that targets the homeobox gene Gax in human hepatocellular carcinoma. Dig Dis Sci. 2012;57:1171-80
22. Valcourt U, Kowanetz M, Niimi H. et al. TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;4:1987-2002
23. Patel N, Tahara SM, Malik P. et al. Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem J. 2011;434:473-82
24. Peterson SM, Thompson JA, Ufkin ML. et al. Common features of microRNA target prediction tools. Front Genet. 2014;5:23
25. Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5:311-6
26. Wang W, Lin P, Han C. et al. Vasculogenic mimicry contributes to lymph node metastasis of laryngeal squamous cell carcinoma. J Exp Clin Cancer Res. 2010;29:60
27. Lagos-Quintana M, Rauhut R, Lendeckel W. et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-8
28. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20
29. Zhang B, Pan X, Cobb GP. et al. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12
30. Lu Z, Li Y, Takwi A. et al. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57-67
31. Cui J, Eldredge JB, Xu Y. et al. MicroRNA expression and regulation in human ovarian carcinoma cells by luteinizing hormone. PLoS One. 2011;6:e21730
32. Shi W, Gerster K, Alajez NM. et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926-37
33. Jiang J, Gusev Y, Aderca I. et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419-27
34. Ma F, Zhang J, Zhong L. et al. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene. 2014;535:191-7
35. Freeman TJ, Smith JJ, Chen X. et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology. 2012;142:562-71 e2
36. Yang G, Yang X. Smad4-mediated TGF-β signaling in tumorigenesis. Int J Biol Sci. 2010;6:1-8
37. Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108-25
38. Hahn SA, Schutte M, Hoque AT. et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350-3
39. Fukuchi M, Masuda N, Miyazaki T. et al. Decreased Smad4 expression in the transforming growth factor-beta signaling pathway during progression of esophageal squamous cell carcinoma. Cancer. 2002;95:737-43
40. Losi L, Bouzourene H, Benhattar J. Loss of Smad4 expression predicts liver metastasis in human colorectal cancer. Oncol Rep. 2007;17:1095-9
41. Ding Z, Wu CJ, Chu GC. et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269-73
42. Hao J, Zhang S, Zhou Y. et al. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2011;585:207-13
43. Wilentz RE, Iacobuzio-Donahue CA, Argani P. et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002-6
44. Wilentz RE, Su GH, Dai JL. et al. Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol. 2000;156:37-43
45. Liu NN, Xi Y, Callaghan MU. et al. SMAD4 is a potential prognostic marker in human breast carcinomas. Tumour Biol. 2014;35:641-50
46. Iwatsuki M, Mimori K, Yokobori T. et al. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293-9
47. Takano S, Kanai F, Jazag A. et al. Smad4 is essential for down-regulation of E-cadherin induced by TGF-beta in pancreatic cancer cell line PANC-1. J Biochem. 2007;141:345-51
48. Orlichenko LS, Radisky DC. Matrix metalloproteinases stimulate epithelial-mesenchymal transition during tumor development. Clin Exp Metastasis. 2008;25:593-600
Author contact
![]() Corresponding author: Dr. BinquanWang, Department of Otolaryngology, Head & Neck Surgery, The First Hospital Affiliated with Shanxi Medical University, 85 Jiefang Road South, Taiyuan, China, Tel: (+86-0351)4639218, email: wbq_xyorg
Corresponding author: Dr. BinquanWang, Department of Otolaryngology, Head & Neck Surgery, The First Hospital Affiliated with Shanxi Medical University, 85 Jiefang Road South, Taiyuan, China, Tel: (+86-0351)4639218, email: wbq_xyorg

 Global reach, higher impact
Global reach, higher impact