3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2015; 6(12):1346-1351. doi:10.7150/jca.13361 This issue Cite
Short Research Communication
Identification of Regulatory-RNAs for Alternative Splicing of Ron Proto-Oncogene
School of Life Sciences, Gwangju Institute of Science and Technology, Gwangju 500-712, Korea.
1These authors contributed equally to this manuscript.
Received 2015-7-28; Accepted 2015-9-7; Published 2015-11-1
Abstract
RON receptor tyrosine kinase is a proto-oncogene that induces cell migration and matrix invasion. RONΔ160 protein, which is produced by exclusion of exon 5 and 6, promotes cell migration, matrix invasion and protection from apoptosis. Alternative splicing regulation of exon 5 and 6 is not well understood. In this manuscript, we identified several new RNA regulatory elements for alternative splicing of Ron proto-oncogene. Firstly, we demonstrated that RNA sequences from EcoRI cleavage sites regulate alternative splicing of Ron exon 5 and 6. Secondly, we showed that the ~30 nt RNA at upstream end of exon 4 and the ~33 nt RNA at downstream end of exon 7 also modulate splicing of exon 5 and 6. Thirdly, our results indicate that the RNA sequences of the ends in exon 4 and 7 are required for the regulatory functions of the RNA from restriction enzyme cleavage sites. Our results provide a new insight for regulation of alternative splicing of Ron proto-oncogene.
Keywords: Ron, pre-mRNA splicing, Regulatory-RNA sequence, Ron, exon 5, exon 6
Introduction
Pre-mRNA splicing is required for gene expression in higher eukaryotes. In this process, introns in pre-mRNA are removed and exons are ligated to produce mRNA. Pre-mRNA splicing occurs in a large RNA-protein complex called spliceosome, composed of U snRNP and more than 250 proteins [1]. 5' splice-site, 3' splice-site, branch-point and polypyrimidine tract sequences are required for pre-mRNA splicing. Alternative splicing is a process that can produce various mature mRNAs encoding different proteins with different function or even opposite functions from a single gene. More than 95% of human genes are able to be alternatively produced. Alternative splicing increases protein diversity and is elaborately regulated. The regulation of alternative splicing occurs in two directions. Firstly, RNA sequences in exons or introns play important regulatory roles [2]. Secondly, proteins promote or inhibit alternative exon splicing to regulate alternative splicing [3]. These proteins can function by either contacting or not contacting RNA sequences.
RON receptor tyrosine kinase is a proto-oncogene that induces cell migration and matrix invasion [4]. RON is a produced from 180-kDa precursor through proteolysis. Ron is a heterodimeric protein, composed of a 40-kDa extracellular α chain and 150-kDa transmembrane β chain that has intrinsic protein tyrosine kinase activity [5]. Macrophage-stimulating protein (MSP) activates RON by autophosphorylation and upregulating RON kinase activity. In tumor cells, RON activation induces invasive growth and motility [6]. Alternative splicing of Ron pre-mRNA produces different isoforms to be produced. A well-known isoform is RONΔ165, which is produced by exclusion of exon 11 in alternative splicing [7]. RONΔ165 is not proteolysed, thus a single chain polypeptides are accumulated [7]. RONΔ165 is constantly activated without stimulating. Alternative splicing of exon 11 has been mostly studied by my group and Biamonti's group. It has been demonstrated that SRSF1, SRSF2, hnRNP A1 interact with exon 11 or exon 12 to stimulate exon 11 inclusion/exclusion [8-10]. SRSF1 and hnRNP A1 were also shown that regulate cell invasion through regulating exon 11 splicing [8, 9].
Another Ron isoform is RONΔ160, which is produced by exclusion of exon 5 and 6 that encodes the first IPT domain in the extracellular β chain. RONΔ160 promotes cell migration, matrix invasion and protection from apoptosis [11]. Increased RONΔ160 expression has been demonstrated to induce cellular transformation in vitro and metastasis in vivo [12]. These oncogenic properties have been shown to be caused by abnormal accumulation of ß-catenin and target gene expression such as c-myc and cyclin D [13].
Although biological role of RONΔ160 is very clear, its alternative splicing regulation is not well understood. Here we applied Ron minigene system to demonstrate that the RNA sequences from EcoRI cleavage sequence, RNA sections at the ends of exon 4 and 7 includes regulatory sequences for alternative splicing of exon 5 and 6. In addition, we show that multiple RNA sequences from restriction enzyme cleavage sequences do not play regulatory roles when the RNA sequences at the ends of exon 4 and 7 are absent.
Results and Discussions
RNA sequences from EcoRI cleavage site play regulatory roles for RNA splicing of exon 5 and 6 in Ron pre-mRNA
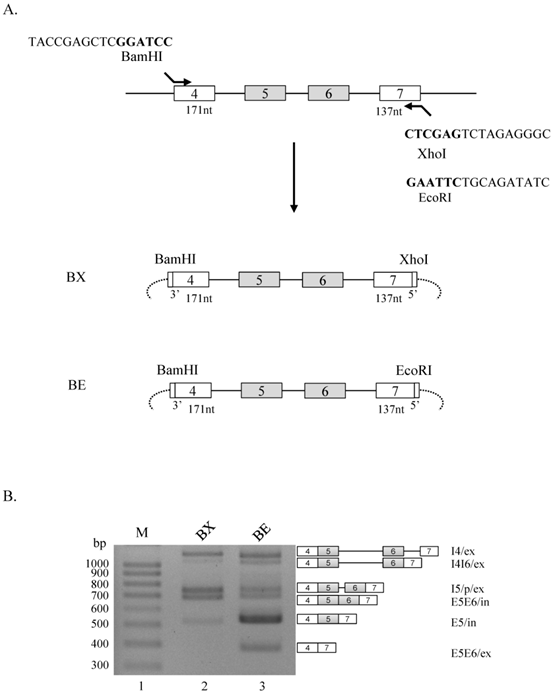
We initiated this experiment to produce Ron minigenes that include exon 4 through exon 7. We decided to include the 3' splice site at upstream end of exon 4, and the 5' splice site at downstream end of exon 7. To clone the genomic DNA into pcDNA 3.1(+) plasmid, we first used a primer that basepairs with the upstream end of exon 4 and also includes a BamHI cleavage sequence at immediate upstream from the 3' splice site of exon 4, and the other primer that basepairs with the downstream end of exon 7 and also contains a XhoI cleavage sequence at immediate downstream from the 5' splice site of exon 7 (BX minigene, figure 1A). To produce another minigene, whereas the same upstream primer that was used in BX minigene was applied, the downstream primer includes EcoRI cleavase sequence instead of XhoI cleavage sequence (BE minigene, figure 1A). BX and BE minigenes have the same contexts except the sequences downstream from 5' splice site of exon 7. To analyze the effects of XhoI and BamHI cleavage sequences on splicing of Ron exon 5 and 6, we extracted RNA from minigene transfected cells and performed RT-PCR using primer pairs that basepair with exon 4 and vector sequences. As shown in figure 1B, in addition to producing exon 5 and 6 included (E5E6/in), exon 5 included (E5/in) isoform, BX minigene also produces only intron 4 spliced (I4/ex), partial cryptic splice site activated (I5/p/ex) [14] and intron 4 and 6 spliced (I4I6/ex) isoforms (lane 2). However, BE minigene demonstrated different splicing pattern. The results in figure 1B show that, in addition to all of the isoforms produced by BX minigene, an isoform in that both exon 5 and 6 excluded is also produced (E5E6/ex, lane 3). Moreover, I5/p/ex and E5E6/in isoforms are reduced. Furthermore, the results also show that E5/in isoform was increased in a significant level as compared with BX minigene. The results in figure 1 indicate that RNA sequences from EcoRI cleavage site play regulatory roles for alternative splicing of exon 5 and 6 in Ron pre-mRNA.
RNA sequences at the end of exon 4 and 7 play regulatory roles in Ron alternative splicing
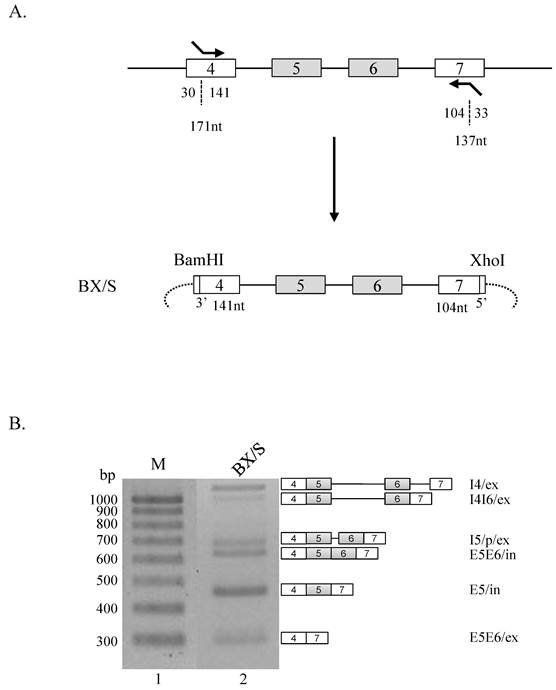
We next asked if intact exon 4 or 7 is required for the regulatory effects of the RNA sequences from restriction enzyme cleavage sites. To address this question, we constructed minigenes in which exon 4 or exon 7 is not intact. In detail, we used primers that basepair with 30 nt downstream from the 5' end of exon 4 and and includes BamHI cleavage sites, and another primer that basepair with 33 nt downstream from the 3' end of exon 7 and contains XhoI cleavage sites (figure 2A). To differentiate this minigene from BX minigene, this shorter minigene constructed was named as BX/S. Strikingly, BX/S minigene (figure 2B, lane 2) produced much different splicing isoforms compared with BX minigene (figure 1B, lane 2). As shown in the figure 2B, E5E6/ex isoform was produced, a significant increase of E5/in isoform, and the decrease of I5/p/ex and E5E6/in isoforms were observed. These results demonstrate that RNA sequences at the end of exon 4 and 7 play regulatory roles in alternative splicing of exon 5 and 6 in Ron pre-mRNA.
RNA sequences from restriction enzyme cleavage sites do not play regulatory roles without the RNA sequences of the ends in exon 4 and 7
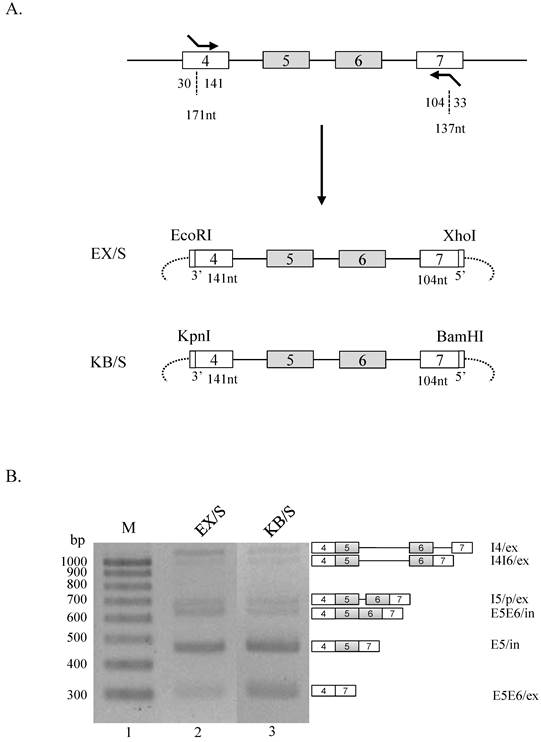
We further asked whether RNA sequences from different restriction enzyme cleavage sites regulate Ron splicing in the shorter exon 4 and 7 contexts. To this end, we produced EX/s and KB/s minigenes, in which EcoRI and XhoI cleavage sequences, or KpnI and BamHI cleavage sequences were applied independently (figure 3A). Using BX/S minigene as a control, we compared splicing event of EX/s and KB/s minigenes. Although an increase of E5E6/ex isoform was detected in KB/S minigene, significant alteration in splicing pattern was not observed (figure 3B). Therefore, we conclude that the restriction enzyme sequences do not play critical regulatory roles when the ends of exon 4 and 7 are absent.
In this manuscript, we identified new RNA regulatory elements for alternative splicing of Ron proto-oncogene. We demonstrate that RNA sequences from EcoRI cleavage sites, ~30 nt RNAs at upstream end of exon 4 and the ~33 nt RNA at downstream end of exon 7 regulate splicing of exon 5 and 6. Our results indicate that the RNA sequences of the ends in exon 4 and 7 play roles in the regulation of RNA sequences from different restriction enzyme sites. The RNA from EcoRI cleavage site promotes splicing of exon 4 and 5, as shown in the increase of E5/in and E5E6/ex isoforms, but inhibits splicing of exon 6. We propose that the RNA sequence from EcoRI cleavage site provides an splicing inhibitory sequences, then nearby exon (exon 6) splicing is inhibited, consequently, distant exon (exon 4 and 5) splicing is increased. The proteins directly contacting the RNA sequence need to be determined. We also found that the RNA sequences at ends of exon 4 and 7 have similar effects as the RNA from EcoRI cleavage sites. How these two RNA sequences combine together to inhibit exon 6 splicing and promote splicing of exon 4 and 5 also needs to be determined. We also show here that, when the ends RNA sequences are absent, RNA sequences from different restriction enzyme sites do not alter alternative splicing in a significant level. Thus our results provide an insight that the ends RNA sequences can be deleted to avoid unexpected effects.
RNA sequences from EcoRI cleavage site play regulatory roles for RNA splicing of exon 5 and 6 in Ron pre-mRNA. (A) Strategy of producing BX and BE minigenes are shown. Exons are shown with boxes, introns are shown with lines and vector sequences are with arcs. Partial primer sequences including restriction enzyme cleavage sites are shown. (B) RT-PCR analysis for alternative splicing of exon 5 and 6 using BX and BE minigenes are shown. Produced isoforms and their names are marked at the right panel.
RNA sequences at the end of exon 4 and 7 play regulatory roles in Ron alternative splicing. (A) Strategy of producing BX/S minigene. Exon 4 and 7 length, and the lengths incorporated into the minigene are shown with number of nucleotides (nt). (B) RT-PCR analysis of BX/S minigene is shown. Isoforms and their names are shown at right panel.
Materials and Methods
Plasmid construction
Ron exon 4-7 genomic DNA was amplified from human genomic DNA using following primer: E4Bam.F and E7Xho.R for BX minigene, E4Bam.F, E7Eco.R for BE minigene. The amplified products were cloned into pcDNA3.1(+) plasmid. BX/S, EX/S and KB/S minigenes were produced using overlapping PCR reaction with BX minigene as a template, and with following primers: BX/S (E4(-30)Bam.F/E7(-33)Xho.R), EX/S (E4(-30)Eco.F/E7(-33)Xho.R), KB/S (E4(-30)Kpn.F/E7(-33)Bam.R). All of the primer sequences are listed in Table 1.
List of primers.
| Name | Sequences |
|---|---|
| Construction | |
| E4Bam.F | 5'- GTTAGGGATCCGTTTTCCAGGTACCTATCCAAG -3' |
| E7Xho.R | 5'- ATTACCTCGAGCGTGCTAGCAGACACTCAGTC -3' |
| E7Eco.R | 5'- ATTTAGAATTCCGTGCTAGCAGACACTCAGTC -3' |
| E4(-30)Bam.F | 5'- AACATGGATCCTGCCGCCACTTCCTGACCTG -3' |
| E7(-33)Xho.R | 5'- CTTACCTCGAGACAGCCCGGCTGGTGCCTAC -3' |
| E4(-30)Eco.F | 5'- AGTATGAATTCTGCCGCCACTTCCTGACCTG -3' |
| E4(-30)Kpn.F | 5'- AACATGGTACCTGCCGCCACTTCCTGACCTG -3' |
| E7(-33)Bam.R | 5'- CTAACGGATCCACAGCCCGGCTGGTGCCTAC -3' |
| RT-PCR | |
| Exon4.F | 5'- GTTTTCCAGGTACCTATCCAAG -3' |
| pcDNA.R | 5'- CTAGAAGGCACAGTCGAGGCT -3' |
RNA sequences from restriction enzyme cleavage sites do not play regulatory roles without the RNA sequences of the ends in exon 4 and 7. (A) Strategy to making EX/S and KB/S is shown. (B) RT-PCR analysis of EX/S and KB/S minigenes are shown.
Cell culture and Transfection
MDA MB 231 cells were grown in RPMI 1640 media supplemented with 10% FBS at 37°C and 5% CO2. Polyethyleneimide (PEI) (Sigma) was used for transfection of Ron minigene plasmids into cells as previously described. Briefly, 0.5 ㎍of minigene was mixed with 1 ug PEI and added to cell. After 4 h transfection, media was changed with fresh media. The cells were incubated for 48 h before total RNA extraction.
RT-PCR
Total RNA was extracted using RiboEx (GeneAll) following the manufacturer's protocol. Reverse transcription was performed using 0.5 ㎍ RNA with oligo (dT) primer and ImProm-IITM reverse transcriptase (Promega). 0.5µl of the reaction mixture was amplified by PCR using G-Taq polymerase (Cosmo Genetech). Spliced products of minigenes were detected with Exon4.F and pcDNA.R primers. All of primer sequences are listed in Table 1.
Acknowledgements
This work was supported by the NRF-2013-R1A1A2062582 grants to Haihong Shen, the NRF-2013-R1A1A2061321 grant to Xuexiu Zheng funded by the National Research Foundation (NRF) of the Korean Ministry of Education, Science, and Technology (MEST).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701-18
2. House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nature structural & molecular biology. 2006;13:937-44
3. Cho S, Moon H, Loh TJ, Oh HK, Williams DR, Liao DJ. et al. PSF contacts exon 7 of SMN2 pre-mRNA to promote exon 7 inclusion. Biochimica et biophysica acta. 2014;1839:517-25
4. Thangasamy A, Rogge J, Ammanamanchi S. Recepteur d'origine nantais tyrosine kinase is a direct target of hypoxia-inducible factor-1alpha-mediated invasion of breast carcinoma cells. The Journal of biological chemistry. 2009;284:14001-10
5. Yoshimura T, Yuhki N, Wang MH, Skeel A, Leonard EJ. Cloning, sequencing, and expression of human macrophage stimulating protein (MSP, MST1) confirms MSP as a member of the family of kringle proteins and locates the MSP gene on chromosome 3. The Journal of biological chemistry. 1993;268:15461-8
6. Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M. et al. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927-33
7. Collesi C, Santoro MM, Gaudino G, Comoglio PM. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Molecular and cellular biology. 1996;16:5518-26
8. Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM. et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Molecular cell. 2005;20:881-90
9. Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C. et al. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic acids research. 2013;41:8665-79
10. Moon H, Cho S, Loh TJ, Oh HK, Jang HN, Zhou J. et al. SRSF2 promotes splicing and transcription of exon 11 included isoform in Ron proto-oncogene. Biochimica et biophysica acta. 2014;1839:1132-40
11. Chen YQ, Zhou YQ, Angeloni D, Kurtz AL, Qiang XZ, Wang MH. Overexpression and activation of the RON receptor tyrosine kinase in a panel of human colorectal carcinoma cell lines. Experimental cell research. 2000;261:229-38
12. Zhou YQ, He C, Chen YQ, Wang D, Wang MH. Altered expression of the RON receptor tyrosine kinase in primary human colorectal adenocarcinomas: generation of different splicing RON variants and their oncogenic potential. Oncogene. 2003;22:186-97
13. Xu XM, Zhou YQ, Wang MH. Mechanisms of cytoplasmic {beta}-catenin accumulation and its involvement in tumorigenic activities mediated by oncogenic splicing variant of the receptor originated from Nantes tyrosine kinase. The Journal of biological chemistry. 2005;280:25087-94
14. Ma Q, Zhang K, Yao HP, Zhou YQ, Padhye S, Wang MH. Inhibition of MSP-RON signaling pathway in cancer cells by a novel soluble form of RON comprising the entire sema sequence. International journal of oncology. 2010;36:1551-61
Author contact
![]() Corresponding author: Phone: 82-62-715-2507, Fax: 82-62-715-2484, E-mail address:haihongshenac.kr
Corresponding author: Phone: 82-62-715-2507, Fax: 82-62-715-2484, E-mail address:haihongshenac.kr

 Global reach, higher impact
Global reach, higher impact