3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(2):153-159. doi:10.7150/jca.13748 This issue Cite
Review
DEC1 and DEC2 Crosstalk between Circadian Rhythm and Tumor Progression
1. Department of Pathology, Wakayama Medical University School of Medicine, Wakayama 641-8509, Japan
2. Department of Biochemistry, Nihon University School of Dentistry at Matsudo, Chiba 271-8587, Japan
Received 2015-9-4; Accepted 2015-10-18; Published 2016-1-1
Abstract
Clock genes, major regulators of circadian rhythm, are involved in tumor progression. We have shown that clock genes basic helix-loop-helix (BHLH) transcription factors, differentiated embryonic chondrocyte gene 1 (DEC1/BHLHE40/Sharp2/Stra13) and DEC2 (BHLHE41/Sharp1) play important roles in circadian rhythm, cell proliferation, apoptosis, hypoxia response, various stresses, and epithelial-to-mesenchymal transition (EMT) of tumor cells. Various stresses, such as exposure to transforming growth factor-beta (TGF-β), hypoxia, cytokines, serum-free, and anti-tumor drugs affect DEC1 and DEC2 expression. An increased or decreased expression of DEC1 and DEC2 regulated tumor progression. However, DEC1 and DEC2 have opposite effects in tumor progression, where the reason behind remains unclear. We found that DEC2 has circadian expression in implanted mouse sarcoma cells, suggesting that DEC2 regulates tumor progression under circadian rhythm. In addition to that, we showed that DEC1 and DEC2 regulate target genes via positive or negative feedback system in tumor progression. We propose that DEC1 and DEC2 act as an accelerator or a brake in tumor progression. In this review, we summarize current progress of knowledge in the function of DEC1 and DEC2 genes in tumor progression.
Keywords: DEC1, DEC2, clock gene, circadian rhythm, immunohistochemistry, tumor progression
The significance of circadian rhythm
All living beings from bacteria to human have circadian rhythm, which is dominantly regulated by clock genes [1, 2]. The molecular mechanism of circadian rhythm depends on negative feed-back system by clock genes. CLOCK and BMAL1 which are clock products bind to E-boxes in the promoters of DEC1, DEC2, PER1 and CRY1 which produce DEC1, DEC2, PER1, CRY1 and CRY2 proteins [3-6]. These proteins are modified by ubiquitination, phosphorylation, and SUMOylation for eventual degradation [7-11]. The remaining proteins either make PER and CRY or DEC1 and DEC2 dimers and suppress the CLOCK/BMAL1 transactivation [3-6, 12, 13]. This negative feed-back system plays important roles in 24-hour rhythmic regulation for circadian rhythm in suprachiasmatic nucleus (SCN), peripheral tissues, or mesenchymal stem cells (MSCs) [14-16]. The disturbance of circadian rhythm may induce metabolic syndrome, diabetes, Alzheimer's disease, depression, sleep disorder, or cancer [17-25]. Night work may increase cancer risk, possibly via suppression of melatonin release [17]. Night time workers have 1.76-fold higher risk for prostate cancer, 2.09-fold higher risk for lung cancer, and 1.74-fold higher risk for colon cancer compared to individuals who have never worked at night [17]. Excessive daytime sleeplessness in children is associated with a high incidence of 60% of cancer and 80% of children with neoplasm in the central nervous system and sleep disorder is account for 40% of the entire group of children with cancer [24]. He Y et al. have shown that mutations in the DEC2 gene are involved in human short sleep phenotype and that DEC2 regulates sleep length in mammals [25]. Therefore, regular life-style may stabilize negative feed-back system and therefore maintain the circadian rhythm. On the other hand, irregular life style may disrupt the negative feed-back system and circadian rhythm and consequently induce various diseases and carcinogenesis.
Previously we reported DEC2, vascular endothelial growth factor (VEGF) and cytochrome P450 (CYP) 2D6 have circadian expression in implanted sarcoma and HepG2 cells [26, 27]. Also, other researchers have shown that BMAL1, PER1, PER2, CRY1, CRY2, VEGF, CYP3A4, retinoid related orphan receptor (ROR)α1 and peroxisome proliferator-activated receptor (PPAR)γ have circadian expression in sarcoma, HepG2, MCF-7 and Panc-1 cells [28-31]. Additionally, we have proven that DEC2 suppressed VEGF expression in sarcoma and oral cancer cells or CYP2D6 expression in hepatocellular carcinoma cells [26, 27]: Our studies have shown that clock genes regulate their target genes under circadian rhythm in tumor cells. In addition to the clock genes, c-Myc, Wee1, cyclin D1, p53, ARF and SNAIL also shows circadian expression [16, 32-36]. Our recent report showed that MSCs have circadian rhythm [16]. MSCs interact with tumor cells, promoting tumor progression [37, 38]. We may consider that the circadian rhythm of tumor is not only limited to tumor cells but also MSCs. As described above, some tumor cells have circadian rhythm. Thus, invasion, angiogenesis, EMT and resistance to apoptosis may regulated by circadian rhythm. The evidence showed that major regulators of tumor malignancy such as c-Myc, VEGF, p53, cyclin D1, and SNAIL were directly regulated by DEC1, DEC2 and PER1 [26, 39-43]. In addition to that, the expression of DEC1 and PER1 were associated with tumor malignancy [39, 44, 45]. Therefore, we speculate that tumor cells may undergo malignancy under circadian rhythm. This speculation may link to chronotherapy.
Chronotherapy is useful for treatment of anti-tumor drugs in terms of its minimum cytotoxicity and effectiveness in tumor suppression, depending on the timing of administration [46]. The evidence revealed that the timing of administration of 5-fluorouracil (FU) affected the degree of the cytotoxicity in cancer patients [46, 47]. It was observed that the administration of treatment at 4:00 a.m. resulted in minimum cytotoxicity in cancer patients. In addition to that, CLOCK/BMAL1 modulates sensitivity to drug-induced toxicity, and DEC1 and DEC2 are downstream factors of CLOCK/BMAL1 [4, 48]. These facts imply that tumor cells have circadian rhythm like normal cells, suggesting the need to know the persistent trend of clock genes in tumor cells. It is certain that clock genes play an important role not only in normal cells but also tumor cells. These results indicate that tumor progression is closely associated with circadian rhythm.
Molecular structures of DEC1 and DEC2 and their significance in the mutants
Human DEC1 has 412 amino acid residues with bHLH and Orange domains, whereas human DEC2 has 482 amino acid residues with alanine and glycine-rich regions in the C-terminus [49]. DEC2 has high (97%) and moderate (57%) similarities to the bHLH region and the Orange domain of DEC1, respectively [50]. The molecular structures of DEC1 and DEC2 are similar to Hairy/E (Sp1) /HES proteins, though DEC1 and DEC2 do not possess WRPR motif, which is necessary for interaction with corepressor Groucho in the C-terminus [50-52]. DEC1 and DEC2 are expressed in a variety of developing and adult tissues as well as MSCs [16, 49, 53-55]. Generally, DEC1 binds to E-boxes to suppress the target genes via histone deacetylase (HDAC) [4, 55-57]. We demonstrated for the first time that DEC1 deletion mutants of the basic region and a point mutation of the basic region R65A have dominant negative effects on CLOCK/BMAL1 transactivation [4]. Also, we and other groups have shown that a DEC1 deletion mutant of the basic region does not bind to E-boxes and that STAT-dependent cis-elements do not suppress the target genes [4, 55, 57]. Interestingly, the DEC1 deletion mutant of the basic region suppressed metastasis of breast cancer cells, whereas DEC1 overexpression induced metastasis in these cells [58]. Moreover, DEC1 upregulates an apoptosis-related factor survivin through the basic region, whereas a point mutation of DEC1 basic region has little effect on the survivin activity [59].
DEC2 suppresses the target genes through E-boxes. We have shown that a DEC2 point mutation R57A in the basic region had little effect on CLOCK/BMAL2 transactivation [60, 61]: However, there are few articles about DEC2 mutation. Our suggestion is to further explore the effect of mutations of DEC1 and DEC2 on circadian rhythm. If these mutations affect the circadian expression of DEC1 and DEC2, they may suppress the circadian expression of DEC1 and DEC2 in tumor cells. Further studies are needed to clarify the significance of these mutations in tumor progression, involving circadian rhythm. In view of all these findings, these studies suggest that the basic region of DEC1 and DEC2 may be a key domain to regulate tumor progression. However, functions of DEC1 and DEC2 often differ in tumor progression. DEC1 has pro-apoptotic effects, and DEC2 has anti-apoptotic effects in human breast cancer cells [41]. The differences in constructs of C-terminus between DEC1 and DEC2 may affect the differential functions in tumor progression.
Immunohistochemical detection of DEC1 and DEC2 in tumor tissues
We have shown that DEC1 is highly expressed in nuclei and cytoplasm of tumor cells compared with adjacent non-tumor cells, which are in line with other studies [39, 40, 62-71]. Previous immunohistochemical analysis showed that DEC1 was localized both in the nuclei and cytoplasm of tumor cells [39, 40, 62-71]. The significance of DEC1 localization in cytoplasm is that. DEC1 regulates apoptosis-related factors via mitochondrial pathway, and regulates phosphorylation of SMAD3 via TGF-β receptor I in EMT [39, 41, 42]. However, we speculate that DEC1 regulates target genes that are not only in nuclei but also in cytoplasm. In immunohistochemistry, DEC1 expression is significantly associated with tumor grade, one-year recurrence, the age of patients, tumor emboli, depth of invasion, lymph node metastasis and pTNM, poor histological differentiation, malignancy and survival rate in invasive breast ductal carcinoma, oral squamous cell carcinoma, esophageal squamous cell carcinoma, hepatocellular carcinoma, or pancreatic ductal adenocarcinoma (Table 1) [44, 64-67]. In addition to that, DEC1 overexpression was positively associated with hypoxia inducible factor 1 (HIF-1) α and Ki-67 in gastric cancer, invasive breast ductal carcinoma and non-small cell lung cancer, whereas it was negatively associated with claudin-1 expression in invasive breast ductal carcinoma [62-64, 68, 69]. Generally, tumor cells are exposed to various stresses such as hypoxia and abnormalities of cytokines and metabolism [72]. Hypoxia treatment immediately induced DEC1 in human oral cancer cells [26]. The reason why DEC1 is highly expressed in tumor cells may associate with various stress of tumor environment. DEC1 expression levels are also decreased in tumor cells compared with adjacent non-tumor cells of non-small cell lung carcinoma (adenocarcinoma and squamous cell carcinoma) [70, 71]. As described above, many reports have shown that immunohistochemical analysis of DEC1 and DEC2 in epithelial tumors, but a few group have reported about the mesenchymal tumors [73]. Future studies are needed for further clarification.
We have shown that DEC2 is subtly expressed in tumor cells compared with adjacent non-tumor pancreatic duct cells in human pancreatic cancer, whereas another group showed that DEC2 is highly expressed in tumor cells of endometrial carcinoma compared with the non-tumor cells (Table 2) [74, 75]. More discrepancies were shown in some reports on PER1 expression in surgical resections. For example, PER1 is highly expressed in tumor cells of oral squamous cell carcinoma and pancreatic cancer compared with adjacent non-tumor cells [45, 76]. However, Chen et al. showed that PER1 is subtly expressed in tumor cells compared with adjacent non-tumor cells in oral squamous cell carcinoma [77]. Taken together, these observations suggest that the expression of clock genes in tumor tissues varies in different laboratories. As clock genes have shown circadian expression in various tissues, we fixed the sampling time for immunohistochemistry as described previously [76]. It remains unclear these discrepancies of immunohitochemical data. Generally, inpatients are exposed to various stress such as sleep disorder and chemotherapy. DEC1, DEC2 and PER1 were affected by various stress such as paclitaxel and cisplatin [42, 45, 78]. Almost articles of DEC1 and DEC2 using inpatient samples have not been mentioned the possibilities of these stress may affect the expression of DEC1 and DEC2. Therefore, there are, at least, two possibilities of these discrepancies. At first, clock genes showed circadian expression in various tumor cells. Therefore, only one point preparation for immunohistochemistry may not enough for clock genes. Secondary, researchers should consider that the expression of clock genes are affected by various stress described as above. Future studies needs to require some points and same time preparation for immunohistochemistry, and divide the data with or without chemotherapy.
Immunohistochemical detection of DEC1 in human tumor tissues
| Tumor type | tumor cells | non-tumor cells | references |
|---|---|---|---|
| Pancreatic ductal carcinoma | strong | weak | [39, 66] |
| Hepatocellular carcinoma | |||
| Well differentiated type | strong | weak | [65] |
| Poorly differentiated type | weak | weak | [65] |
| Invasive breast ductal carcinoma | strong | weak | [63, 64,71] |
| Non-small cell lung cancer | weak | strong | [62, 70] |
| Gastric cancer | strong | weak | [68, 69] |
| Oral squamous cell carcinoma | strong | weak | [40, 44] |
| Esophageal squamous cell carcinoma | strong | weak | [67] |
Immunohistochemical detection of DEC2 in human tumor tissues
| Tumor type | tumor cells | non-tumor cells | references |
|---|---|---|---|
| Pancreatic ductal carcinoma | weak | strong | [74] |
| Endometrial carcinoma | strong | weak | [75] |
Factors upstream and downstream of DEC1 and DEC2
In our previous study, there are many factors up and downstream of DEC1 (Figure 1). CLOCK/BMAL1 induces DEC1 transcription by binding to E-box [3, 4, 12]. We have shown that hypoxia, irradiation, paclitaxel, tumor necrosis factor (TNF)-α, and TGF-β induce DEC1 expression in human oral cancer cells HSC-2 and HSC-3, breast cancer cells MCF-7, and pancreatic cancer cells Panc-1 [39-42]. Moreover, IL-1β or liver X receptors (LXR) induce DEC1 expression in human primary gingival cells, Hepa1c cells, and NIH3T3 cells [79, 80]. Cisplatin treatment decreases DEC1 expression in HSC-3 and CA9-22 cells [78]. Several studies have shown that SMAD3, CD28, sunitinib, RORα, and etoposide induce DEC1 expression in Rat-1, human primary T-cells, renal cell carcinoma, Hepa1c1c7 cells, and NIH3T3 cells [81-85]. It has been proven that HIF-1α induced by hypoxia binds to hypoxia response element in the DEC1 promoter [86].
Up and downstream factors of DEC1.
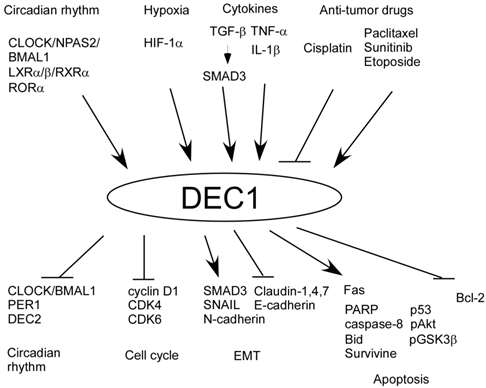
Also, we have shown that there are various factors upstream of DEC2 (Figure 2). For example, TGF-β and polyinosinic -polycytidylic acid (Poly IC) induces DEC2 expression in human pancreatic cancer cells BxPC-3 and in human mesangial cells respectively [74, 87]. Additionally, we have shown that there are many factors downstream of DEC1 and DEC2. DEC1 negatively regulates DEC2, PER1, cyclin D1, CDK4, CDK6, CLOCK/BMAL1/2, claudins 1, 4, 7, E-cadherin, and Bcl-2, whereas DEC1 positively regulates phosphorylated SMAD3 (pSMAD3), SNAIL, N-cadherin, pAkt, pGSK3β, p53, caspase-8, poly (ADP-ribose) polymerase (PARP), Fas, and Bid. On the other hand, DEC2 negatively regulates DEC1, CLOCK/BMAL1/2, PER1, VEGF, SLUG, CCL5, retinoic acid-inducible gene-I (RIG-I), IFN-β, Bim, caspase-8, PARP, p53, Bax, muscle creatine kinase, CYP2D6, and Twist1, whereas DEC2 positively regulates cyclin D1 [3, 4, 26, 27, 39-42, 60, 61, 74, 78-80, 87, 88]. In particular, TGF-β is closely associated with circadian rhythm and it resets clock via DEC1 in rat-1 cells [81]. In addition to that, we showed SMAD3 has circadian expression [16]. Therefore, it is possible that TGF-β may crosstalk between circadian rhythm and tumor progression via SMAD3 and DEC1, and that the circadian rhythm of DEC1 and DEC2 may be regulated by SMAD3. However, the significance of clock reset by TGF-β in tumor progression still remains unclear. It would be of interest to analyze the circadian expression of DEC1 and DEC2 using a tumor injection model in Smad3 knockout mice for understanding crosstalk between TGF-β and circadian rhythm.
Biological functions of DEC1 and DEC2 in normal and tumor cells
In normal cells, DEC1 promotes differentiation of chondrocytes and neurons, whereas DEC1 inhibits mesodermal and adipogenic differentiation and cell proliferation [49, 53, 56, 89]. DEC2 inhibits muscle and adipogenic differentiation and inflammation [87, 90, 91]. DEC1 and DEC2 are expressed in various peripheral tissues, showing circadian expression in SCN [3, 16, 49]. These biological functions of DEC1 and DEC2 may be regulated under circadian rhythm.
Interestingly, abnormalities of DEC1 and DEC2 cause various phenomena in mice and human. A loss of DEC1 induces an autoimmune disorder in mice [92]. DEC1 knockdown or overexpression in MSCs affects the circadian phase of DEC2 and Per1 [93]. A loss of both DEC1 and DEC2 induces sleep disorder and psychiatric diseases in mice [94]. Meanwhile, DEC2 has been found to regulate sleep length in human [25]. In conclusion, DEC1 and DEC2 mediate crosstalk between circadian rhythm and physiological actions.
It seems that endogenous expression levels of DEC1 and DEC2 are low in normal cells [26, 59, 84, 87, 90]. DEC1 and DEC2 knockout mice can survive without any obvious morphologic phenotype except for splenomegaly [92, 95]. However, DEC1 and DEC2 are induced by various stresses, and the increased expression of DEC1 and DEC2 was associated with cell survival [41, 42, 56, 58]. In addition to that, their expression is associated with tumor progression [26, 39, 41, 74]. Therefore, DEC1 and DEC2 may play more important roles in cells under stress than in those without stress. Analysis using tumor cells with a stress inducer may be useful for further understanding of their functions.
Up and downstream factors of DEC2.
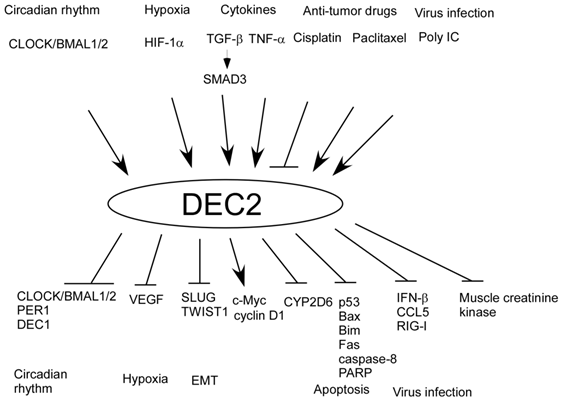
Several papers have reported that DEC1 induces apoptosis, EMT and senescence in tumor cells [39, 41, 67, 85, 96]. On the contrary, DEC2 inhibits apoptosis, EMT and metastasis, showing circadian expression in sarcoma and hepatocellular carcinoma cells [26, 27, 41, 74, 97]. Given that DEC1 and DEC2 play an important role in tumor progression under circadian rhythm, it would be possible that they can be used as target genes for chronotherapy.
However, as mentioned above, the function of clock genes in tumor cells is still controversial [43, 77, 98]. PER1 has anti-apoptotic effects on human oral cancer, pancreatic cancer and hepatocellular carcinoma cells, whereas PER1 has pro-apoptotic effects on human colon cancer and prostate cancer cells [43, 45, 76, 98]. BMAL1 overexpression induces apoptosis by oxaliplatin, inhibiting the colony formation of human colon cancer cells. Additionally, BMAL1 siRNA induces apoptosis and inhibits colony formation of human malignant mesothelioma cells [99, 100]. Li Y et al. have shown that DEC1 has anti-apoptotic effects, regulating survivin under serum-starvation, whereas DEC1 has pro-apoptotic effects, involving DNA damage [59, 85]. It has been found that only medium change affects the expression of PER1, PER2, and BMAL1 in culture cells, upregulating or downregulating their expression in a time-dependent manner [101]. Taking these observations into consideration suggests that DEC1 and DEC2 have pro and anti-apoptotic effects, depending on the time of medium change and sample preparation. As described earlier, differences in C-terminal regions also may affect these functions. Since DEC1 and DEC2 regulate each other by binding their E-boxes, it would be of interest to examine whether combination treatment of DEC1 and DEC2 (overexpression or knockdown) influences tumor progression.
Conclusion
DEC1 and DEC2 suppress CLOCK/BMAL1/2 transactivation most strongly among clock genes [3, 5, 12, 61]. As described above, CLOCK/BMAL1 modulates drug toxicity. Therefore, DEC1 and DEC2 may regulate the toxicity of anti-tumor drugs under circadian rhythm via CLOCK/BMAL1. As described earlier, DEC2 regulates VEGF expression as well as the circadian rhythm by negative feed-back system [26]. In addition to that, DEC1 and DEC2 regulate EMT by positive and negative feed-back system in tumor cells [39, 74]. Thus, DEC1 and DEC2 act as an “accelerator” and a “brake” in tumor progression.
All the available findings to date, this review points towards the fact that DEC1 and DEC2 are the most important roles in crosstalk between circadian rhythm and tumor progression, and attractive target genes for chemotherapy. Further studies are needed to elucidate how DEC1 and DEC2 regulate tumor progression under circadian rhythm.
Acknowledgements
We would like to thank Shenli Hew from the Department of Clinical Research Center, Wakayama Medical University, for proofreading and editing the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369-77 Review
2. Albrecht U, Eichele G. The mammalian circadian clock. Curr Opin Genet Dev. 2003;13:271-7 Review
3. Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M. et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841-4
4. Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, Honma S. et al. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur J Biochem. 2004;271:4409-19
5. Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N. et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J. 2004;382:43-50
6. Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X. et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193-205
7. Keesler GA, Camacho F, Guo Y, Virshup D, Mondadori C, Yao Z. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport. 2000;11:951-5
8. Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855-67
9. Miyazaki K, Nagase T, Mesaki M, Narukawa J, Ohara O, Ishida N. Phosphorylation of clock protein PER1 regulates its circadian degradation in normal human fibroblasts. Biochem J. 2004;380:95-103
10. Wang Y, Rao VK, Kok WK, Roy DN, Sethi S, Ling BMT. et al. SUMO modification of Stra13 is required for repression of cyclin D1 expression and cellular growth arrest. PLoS One. 2012:7
11. Wang Y, Shankar SR, Kher D, Ling BMT, Taneja R. Sumoylation of the basic helix-loop-helix transcription factor sharp-1 regulates recruitment of the histone methyltransferase G9a and function in myogenesis. J Biol Chem. 2013;288:17654-62
12. Kawamoto T, Noshiro M, Sato F, Maemura K, Takeda N, Nagai R. et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem Biophys Res Commun. 2004;313:117-24
13. Griffin EA, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768-71
14. Jin X, Shearman LP, Weaver DR, Zylka MJ, De Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57-68
15. Noshiro M, Furukawa M, Honma S, Kawamoto T, Hamada T, Honma K. et al. Tissue-specific disruption of rhythmic expression of Dec1 and Dec2 in clock mutant mice. J Biol Rhythms. 2005;20:404-18
16. Sato F, Sato H, Jin D, Bhawal UK, Wu Y, Noshiro M. et al. Smad3 and Snail show circadian expression in human gingival fibroblasts, human mesenchymal stem cell, and in mouse liver. Biochem Biophys Res Commun. 2012;419:441-6
17. Parent MÉ, El-Zein M, Rousseau MC, Pintos J, Siemiatycki J. Night work and the risk of cancer among men. Am J Epidemiol. 2012;176:751-9
18. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E. et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043-5
19. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y. et al. High-Fat Diet Disrupts Behavioral and Molecular Circadian Rhythms in Mice. Cell Metab. 2007;6:414-21
20. Thome J, Coogan AN, Woods AG, Darie CC, Häßler F. CLOCK Genes and Circadian Rhythmicity in Alzheimer Disease. J Aging Res. 2011;2011:383091. Review
21. Bhatwadekar AD, Yan Y, Qi X, Thinschmidt JS, Neu MB, Li Calzi S. et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes. 2013;62:273-82
22. Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP. et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950-5
23. Anisimov VN, Vinogradova IA, Panchenko AV, Popovich IG, Zabezhinski MA. Light-at-night-induced circadian disruption, cancer and aging. Curr Aging Sci. 2012;5:170-7
24. Rosen G, Brand SR. Sleep in children with cancer: Case review of 70 children evaluated in a comprehensive pediatric sleep center. Support Care Cancer. 2011;19:985-94
25. He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL. et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866-70
26. Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Imaizumi T, Imanaka T. et al. Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular endothelial growth factor expression. Genes Cells. 2008;13:131-44
27. Matsunaga N, Inoue M, Kusunose N, Kakimoto K, Hamamura K, Hanada Y. et al. Time-dependent interaction between differentiated embryo chondrocyte-2 and CCAAT/enhancer-binding protein α underlies the circadian expression of CYP2D6 in serum-shocked HepG2 cells. Mol Pharmacol. 2012;81:739-47
28. Takiguchi T, Tomita M, Matsunaga N, Nakagawa H, Koyanagi S, Ohdo S. Molecular basis for rhythmic expression of CYP3A4 in serum-shocked HepG2 cells. Pharmacogenet Genomics. 2007;17:1047-56
29. Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S. et al. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63:7277-83
30. Frasch Xiang, S Mao, L Duplessis, T Yuan, L Dauchy, R et al. Oscillation of Clock and Clock Controlled Genes Induced by Serum Shock in Human Breast Epithelial and Breast Cancer Cells: Regulation by Melatonin. Breast Cancer Basic Clin Res. 2012:137
31. Pazienza V, Tavano F, Francavilla M, Fontana A, Pellegrini F, Benegiamo G. et al. Time-Qualified Patterns of Variation of PPARgamma, DNMT1, and DNMT3B Expression in Pancreatic Cancer Cell Lines. PPAR Res. 2012;2012:890875
32. Horiguchi M, Koyanagi S, Hamdan AM, Kakimoto K, Matsunaga N, Yamashita C. et al. Rhythmic control of the ARF-MDM2 pathway by ATF4 underlies circadian accumulation of p53 in malignant cells. Cancer Res. 2013;73:2639-49
33. Mao L, Dauchy RT, Blask DE, Slakey LM, Xiang S, Yuan L. et al. Circadian gating of epithelial-to-mesenchymal transition in breast cancer cells via melatonin-regulation of GSK3β. Mol Endocrinol. 2012;26:1808-20
34. Soták M, Polidarová L, Ergang P, Sumová A, Pácha J. An association between clock genes and clock-controlled cell cycle genes in murine colorectal tumors. Int J Cancer. 2013;132:1032-41
35. Gu X, Xing L, Shi G, Liu Z, Wang X, Qu Z. et al. The circadian mutation PER2S662G is linked to cell cycle progression and tumorigenesis. Cell Death Differ. 2012;19:397-405
36. Iurisci I, Filipski E, Reinhardt J, Bach S, Gianella-Borradori A, Iacobelli S. et al. Improved tumor control through circadian clock induction by seliciclib, a cyclin-dependent kinase inhibitor. Cancer Res. 2006;66:10720-8
37. Spaeth EL, Dembinski JL, Sasser AK, Watson K, Klopp A, Hall B. et al. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One. 2009:4
38. Liu Y, Han ZP, Zhang SS, Jing YY, Bu XX, Wang CY. et al. Effects of inflammatory factors on mesenchymal stem cells and their role in the promotion of tumor angiogenesis in colon cancer. J Biol Chem. 2011;286:25007-15
39. Wu Y, Sato F, Yamada T, Bhawal UK, Kawamoto T, Fujimoto K. et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int J Oncol. 2012;41:1337-46
40. Bhawal UK, Sato F, Arakawa Y, Fujimoto K, Kawamoto T, Tanimoto K. et al. Basic helix-loop-helix transcription factor DEC1 negatively regulates cyclin D1. J Pathol. 2011;224:420-9
41. Liu Y, Sato F, Kawamoto T, Fujimoto K, Morohashi S, Akasaka H. et al. Anti-apoptotic effect of the basic helix-loop-helix (bHLH) transcription factor DEC2 in human breast cancer cells. Genes to Cells. 2010;15:315-25
42. Wu Y, Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M. et al. Basic helix-loop-helix transcription factors DEC1 and DEC2 regulate the paclitaxel-induced apoptotic pathway of MCF-7 humanbreast cancer cells. Int J Mol Med. 2011;27:491-5
43. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. MolCell. 2006;22:375-82
44. You J, Lin L, Liu Q, Zhu T, Xia K, Su T. The correlation between the expression of differentiated embryo-chondrocyte expressed gene l and oral squamous cell carcinoma. Eur J Med Res. 2014;19:21
45. Sato F, Wu Y, Bhawal UK, Liu Y, Imaizumi T, Morohashi S. et al. PERIOD1 (PER1) has anti-apoptotic effects, and PER3 has pro-apoptotic effects during cisplatin (CDDP) treatment in human gingival cancer CA9-22 cells. Eur J Cancer. 2011;47:1747-58
46. Innominato PF, Lévi FA, Bjarnason GA. Chronotherapy and the molecular clock: Clinical implications in oncology. Adv Drug Deliv Rev. 2010;62:979-1001
47. Altinok A, Lévi F, Goldbeter A. Identifying mechanisms of chronotolerance and chronoefficacy for the anticancer drugs 5-fluorouracil and oxaliplatin by computational modeling. Eur J Pharm Sci. 2009;36:20-38
48. Antoch MP, Kondratov RV. Circadian Clock Genes as Modulators of Sensitivity to Genotoxic Stress. Encycl. Stress. 2010:496-9
49. Shen M, Kawamoto T, Yan W, Nakamasu K, Tamagami M, Koyano Y. et al. Molecular characterization of the novel basic helix-loop-helix protein DEC1 expressed in differentiated human embryo chondrocytes. Biochem Biophys Res Commun. 1997;236:294-8
50. Fujimoto K, Shen M, Noshiro M, Matsubara K, Shingu S, Honda K. et al. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem Biophys Res Commun. 2001;280:164-71
51. Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620-34
52. Ishibashi M, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of HES-2, a mammalian helix-loop-helix factor structurally related to Drosophila hairy and Enhancer of split. Eur J Biochem. 1993;215:645-52
53. Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dollé P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052-65
54. Rossner MJ, Dörr J, Gass P, Schwab MH, Nave KA. SHARPs: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci. 1997;10:460-75
55. St-Pierre B, Flock G, Zacksenhaus E, Egan SE. Stra13 homodimers repress transcription through class B E-box elements. J Biol Chem. 2002;277:46544-51
56. Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A. 2000;97:4058-63
57. Ivanova A V, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J Mol Biol. 2004;340:641-53
58. Ehata S, Hanyu A, Hayashi M, Aburatani H, Kato Y, Fujime M. et al. Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 2007;67:9694-703
59. Li Y, Xie M, Yang J, Yang D, Deng R, Wan Y. et al. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene. 2006;25:3296-306
60. Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, Sato F. et al. Transcriptional repression by the basic helix-loop-helix protein Dec2: Multiple mechanisms through E-box elements. Int J Mol Med. 2007;19:925-32
61. Kondo J, Sato F, Fujimoto K, Kusumi T, Imanaka T, Kawamoto T. et al. 57Arg in the bHLH transcription factor DEC2 is essential for the suppression of CLOCK/BMAL2-mediated transactivation. Int J Mol Med. 2006;17:1053-6
62. Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Wykoff CC, Gatter KC. et al. DEC1 (STRA13) protein expression relates to hypoxia-inducible factor 1-alpha and carbonic anhydrase-9 overexpression in non-small cell lung cancer. J Pathol. 2003;200:222-8
63. Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC. et al. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer. 2004;91:954-8
64. Liu Y, Miao Y, Wang J, Lin X, Wang L, Xu HT. et al. DEC1 is positively associated with the malignant phenotype of invasive breast cancers and negatively correlated with the expression of claudin-1. Int J Mol Med. 2013;31:855-60
65. Shi XH, Zheng Y, Sun Q, Cui J, Liu QH, Fei Q. et al. DEC1 nuclear expression: A marker of differentiation grade in hepatocellular carcinoma. World J Gastroenterol. 2011;17:2037-43
66. Wang W, Reiser-Erkan C, Michalski CW, Raggi MC, Quan L, Yupei Z. et al. Hypoxia inducible BHLHB2 is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Biochem Biophys Res Commun. 2010;401:422-8
67. Xu Q, Ma P, Hu C, Chen L, Xue L, Wang Z. et al. Overexpression of the DEC1 protein induces senescence in vitro and is related to better survival in esophageal squamous cell carcinoma. PLoS One. 2012:7
68. Jia Y-F, Xiao D-J, Ma X-L, Song Y-Y, Hu R, Kong Y. et al. Differentiated embryonic chondrocyte-expressed gene 1 is associated with hypoxia-inducible factor 1α and Ki67 in human gastric cancer. Diagn Pathol. 2013;8:37
69. Zheng Y, Jia Y, Wang Y, Wang M, Li B, Shi X. et al. The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in gastric cancer. OMICS. 2009;13:301-6
70. Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y. et al. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumor Biol. 2013;34:1641-50
71. Turley H, Wykoff CC, Troup S, Watson PH, Gatter KC, Harris AL. The hypoxia-regulated transcription factor DEC1(Stra13, SHARP-2) and its expression in human tissues and tumours. J Pathol. 2004;203:808-13
72. Jiang J, Tang YL, Liang XH. EMT: A new vision of hypoxia promoting cancer progression. Cancer Biol Ther. 2011;11:714-23
73. Hu T, He N, Yang Y, Yin C, Sang N, Yang Q. DEC2 expression is positively correlated with HIF-1 activation and the invasiveness of human osteosarcomas. J Exp Clin Cancer Res. 2015;34:22
74. Sato F, Kawamura H, Wu Y, Sato H, Jin D, Bhawal UK. et al. The basic helix-loop-helix transcription factor DEC2 inhibits TGF-beta-induced tumor progression in human pancreatic cancer BxPC-3 cells. Int J Mol Med. 2012;30:495-501
75. Yunokawa M, Tanimoto K, Nakamura H, Nagai N, Kudo Y, Kawamoto T. et al. Differential regulation of DEC2 among hypoxia-inducible genes in endometrial carcinomas. Oncol Rep. 2007;17:871-8
76. Sato F, Nagata C, Liu Y, Suzuki T, Kondo J, Morohashi S. et al. PERIOD1 is an anti-apoptotic factor in human pancreatic and hepatic cancer cells. J Biochem. 2009;146:833-8
77. Chen R, Yang K, Zhao N, Zhao D, Chen D, Zhao CR. et al. Abnormal expression of PER1 circadian-clock gene in oral squamous cell carcinoma. Onco Targets Ther. 2012;5:403-7
78. Wu Y, Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M. et al. BHLH transcription factor DEC2 regulates pro-apoptotic factor Bim in human oral cancer HSC-3 cells. Biomed Res. 2012;33:75-82
79. Noshiro M, Usui E, Kawamoto T, Sato F, Nakashima A, Ueshima T. et al. Liver X receptors (LXRalpha and LXRbeta) are potent regulators for hepatic Dec1 expression. Genes Cells. 2009;14:29-40
80. Bhawal UK, Ito Y, Tanimoto K, Sato F, Fujimoto K, Kawamoto T. et al. IL-1β-mediated up-regulation of DEC1 in human gingiva cells via the Akt pathway. J Cell Biochem. 2012;113:3246-53
81. Kon N, Hirota T, Kawamoto T, Kato Y, Tsubota T, Fukada Y. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat Cell Biol. 2008;10:1463-9
82. Martínez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J. et al. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J Exp Med. 2013;210:1603-19
83. Zhu Y, Xu L, Zhang J, Hu X, Liu Y, Yin H. et al. Sunitinib induces cellular senescence via p53/Dec1 activation in renal cell carcinoma cells. Cancer Sci. 2013;104:1052-61
84. Ozaki N, Noshiro M, Kawamoto T, Nakashima A, Honda K, Fukuzaki-Dohi U. et al. Regulation of basic helix-loop-helix transcription factors Dec1 and Dec2 by RORalpha and their roles in adipogenesis. Genes Cells. 2012;17:109-21
85. Thin TH, Li L, Chung T-K, Sun H, Taneja R. Stra13 is induced by genotoxic stress and regulates ionizing-radiation-induced apoptosis. EMBO Rep. 2007;8:401-7
86. Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277:47014-21
87. Imaizumi T, Sato F, Tanaka H, Matsumiya T, Yoshida H, Yashiro-Aizawa T. et al. Basic-helix-loop-helix transcription factor DEC2 constitutes negative feedback loop in IFN-beta-mediated inflammatory responses in human mesangial cells. Immunol Lett. 2011;136:37-43
88. Suzuki M, Sato F, Bhawal UK. The basic helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates Twist1 through an E-box element. Biochem Biophys Res Commun. 2014;455:390-5
89. Iwata T, Kawamoto T, Sasabe E, Miyazaki K, Fujimoto K, Noshiro M. et al. Effects of overexpression of basic helix-loop-helix transcription factor Dec1 on osteogenic and adipogenic differentiation of mesenchymal stem cells. Eur J Cell Biol. 2006;85:423-31
90. Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem. 2004;279:52643-52
91. Gulbagci NT, Li L, Ling B, Gopinadhan S, Walsh M, Rossner M. et al. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep. 2009;10:79-86
92. Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol. 2001;2:1040-7
93. Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T. et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol. 2008;28:4080-92
94. Baier PC, Brzózka MM, Shahmoradi A, Reinecke L, Kroos C, Wichert SP. et al. Mice Lacking the Circadian Modulators SHARP1 and SHARP2 Display Altered Sleep and Mixed State Endophenotypes of Psychiatric Disorders. PLoS One. 2014;9:e110310
95. Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J. et al. Disturbed clockwork resetting in sharp-1 and sharp-2 single and double mutant mice. PLoS One. 2008:3
96. Peng Y, Liu W, Xiong J, Gui HY, Feng XM, Chen RN. et al. Down regulation of differentiated embryonic chondrocytes 1 (DEC1) is involved in 8-methoxypsoralen-induced apoptosis in HepG2 cells. Toxicology. 2012;301:58-65
97. Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E. et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012
98. Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J. et al. A role for the clock gene Per1 in prostate cancer. Cancer Res. 2009;69:7619-25
99. Zeng ZL, Luo HY, Yang J, Wu WJ, Chen DL, Huang P. et al. Overexpression of the circadian clock gene bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin Cancer Res. 2014;20:1042-52
100. Elshazley M, Sato M, Hase T, Yamashita R, Yoshida K, Toyokuni S. et al. The circadian clock gene BMAL1 is a novel therapeutic target for malignant pleural mesothelioma. Int J Cancer. 2012;131:2820-31
101. Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured rat-1 fibroblasts. J Biol Chem. 2002;277:44244-51
Author contact
![]() Corresponding author: Fuyuki Sato, D.D.S, PhD. Department of Pathology, Wakayama Medical University School of Medicine, Wakayama 641-8509, Japan. Tel: +81 734 41 0634; Fax: +81 734 46 3781, E-mail address: fsatoDEC1DEC2co.jp
Corresponding author: Fuyuki Sato, D.D.S, PhD. Department of Pathology, Wakayama Medical University School of Medicine, Wakayama 641-8509, Japan. Tel: +81 734 41 0634; Fax: +81 734 46 3781, E-mail address: fsatoDEC1DEC2co.jp

 Global reach, higher impact
Global reach, higher impact