Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(2):192-199. doi:10.7150/jca.13414 This issue Cite
Research Paper
Effects of Lovastatin on MDA-MB-231 Breast Cancer Cells: An Antibody Microarray Analysis
1. National Engineering Laboratory for Rice and Byproduct Deep Processing, Central South University of Forestry and Technology, Changsha, Hunan 410004, China;
2. Medical College, Hunan Normal University, Changsha, Hunan 410013, China;
3. College of Biology, Hunan University, Changsha, Hunan 410082, China.
4. Department of Urology, University of Michigan, Ann Arbor, MI 48109, USA
* These authors contributed equally to this work.
Received 2015-8-1; Accepted 2015-10-27; Published 2016-1-1
Abstract
Despite the tremendous improvement in cancer therapeutics, treatment of late-stage breast cancer remains a challenge for both basic scientists and clinicians. Lovastatin, a natural product derived from Aspergillus terreus or Monascus ruber, has been widely used as cholesterol-lowing drug in the clinic. It also has anti-cancer properties through poorly defined molecular mechanisms. In the present study, we employed a novel antibody microarray technology to investigate the molecular mechanisms through which lovastatin inhibits breast cancer. We found that lovastatin up-regulated 17 proteins and down-regulated 20 proteins in MDA-MB-231 breast cancer cells. These included proteins that modulate apoptosis, cell proliferation, differentiation, signal transduction, epithelial-to-mesenchymal transition and tumor metastasis. Modulation of these pathways may mediate, in part, the inhibitory activity of lovastatin on breast cancer.
Keywords: Natural products, Lovastatin, Breast cancer, Antibody microarray, Hypoxia
Introduction
In spite of the tremendous advancement of modern cancer diagnostics and therapeutics, breast cancer remains the most common form of cancer in women and one of the leading causes of cancer-related mortality worldwide especially in highly-developed countries [1]. As in most other countries, breast cancer cases in China account for 12.2% of all newly diagnosed breast cancers and about 10% of all deaths from breast cancer worldwide [2]. Although the existing treatment modalities are effective in treating early breast cancer, they have limited usefulness in treating metastatic breast cancer and in particular the triple-negative (ER-, PR-, HER2-) breast cancers. Although these patients are chemosensitive at the initial stage of treatment, they relapse frequently and have an overall survival of only 18-24 months. Currently, a few combination therapy protocols have been adopted for breast cancer but toxicity problems frequently arise that compromise the final effectiveness of these combinations. Consequently, new combinations of current drugs as well as new drugs with novel targets, especially for triple-negative breast cancer, are urgently needed and represent an area of active investigation.
Natural products include small-molecule secondary metabolites that contribute to organism survival. They have historically been a mainstay source of anti-cancer drugs or adjuvants, but in the late 20th century they lost favor because of the emergence of targeted therapies, which rely on antibodies or small synthetic molecules identified by high throughput screening. Although targeted therapies have greatly improved the treatment of a few cancers, the benefit has remained limited for many solid tumors, leading to renewed interest in natural products [3]. A recent analysis of the strategies used in the discovery of new medicines showed that 36% of the first-in-class small-molecules approved by US FDA between 1999 and 2008 were natural products or natural products derivatives [4]. Natural products such as curcumin, resveratrol and sulfarophane are gaining acceptance as adjuvant anti-cancer agents with minimal or no side effects [5].
Lovastatin, also called Monacolin K, a natural product derived from Monascus or Aspergillus-fermented rice, is a member of the statin class of drugs known as 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase inhibitors. Statins have been shown to have pleiotropic cardiovascular and anti-atherosclerotic effects, including cholesterol-lowering and anti-oxidant activities, immunomodulation, regulation of inflammatory response, etc. In contrast to hydrophilic statins such as pravastatin, lipophilic statins such as lovastatin can directly cross cell membranes [6]. Besides its lipid-regulating effects, numerous in vitro and preclinical studies have shown that lovastatin has anti-proliferative, anti-angiogenic, anti-metastatic and pro-apoptotic properties in many types of cancer cells including breast cancer [7, 8]. When the cytostatic activity of lovastatin was compared among a variety of cell lines, the IC50s were in the range of 0.3-2.0 μM [9]. This makes lovastatin a promising drug to be administered as a cytostatic agent in conjunction with other cytotoxic agents. Indeed, lovastatin has been shown to interact synergistically with chemotherapeutic agents such as 5-fluorouracil [10], cisplatin [11] and 1-β-D-arabinofuranosylcytosine [12] in different kinds of cancer cells in in vitro studies. In animal models, lovastatin potentiated the anti-tumor effects of doxorubicin [13] and TNF-α [14].
Antibody-based microarray is a novel innovative technology for the study of non-fractionated complex proteomes that enables the researchers to better understand the pathogenesis of diseases. Because of its high sensitivity and increased specificity, this strategy provides a more powerful tool for the identification of proteome-wide changes of differentially expressed proteins over other proteomic methods such as LC-MS/MS and NMR analysis [15]. In this study, antibody microarrays were employed to analyze the proteome of lovastatin-treated and control MDA-MB-231 cells which were cultured under hypoxia. Quantitative real-time RT-PCR and Western blot analysis were used to validate the differential expression of mRNA or proteins. The proteins that were up- or down-regulated by lovastatin were grouped according to their biological functions and their potential roles in mediating lovastatin's anti-cancer effects discussed.
Materials and Methods
Cell culture and treatments
MDA-MB-231and MDA-MB-468 human breast cancer cells were cultured routinely in DMEM supplemented with 10% FBS in a humidified incubator at 37°C with 5% CO2 according to the standard culture procedure. The cells were tested negative for mycoplasma before experiments. For treatment, the cells were seeded in culture dishes or plates (about 1.5 x 104 cells/cm2) and allowed to grow overnight before treatment. The next day, lovastatin was added to the cells at various concentrations and the cells were cultured under normoxia (21% O2) or hypoxia (1% O2) for the desired period of time. Vehicle alone was added to the culture medium serving as the untreated control. Hypoxic environment, which was used to mimic the in vivo tumor microenvironment, was generated in a Forma 3131 Hypoxic Incubator (Thermo Fisher Scientific, Waltham, MA, USA) through injecting N2 gas into the chamber.
Cell proliferation assay
The cells were seeded into 96-well plates at 1 - 2 x 103 cells/100 μl/well, treated with different concentrations of lovastatin for 48 h under hypoxia. During the last 1 - 4 h of incubation, 20 μl of CellTiter Blue cell viability assay reagent (Promega, Madison, WI, USA) was added to each well. The production of the fluorescent dye resorufin was directly measured using a BioTek (Waltham, MA, USA) Synergy 2 microplate reader with fluorescence recorded at 560/590nm.
Cell apoptosis study
The cells were seeded into 35-mm dishes at 2 x 105 cells/dish and treated with different concentrations of lovastatin or vehicle for 48 h under hypoxia. The cells were stained with an Annexin V/PI double staining kit (Seven Sea Bio, Shanghai, China) according to the manufacturer's instructions. Apoptosis was detected using a BD FACSCanto II Flow Cytometer (Becton Dickenson, Mountain View, CA, USA) and analyzed using the CellQuest Pro software.
Cell lysate preparation
Whole cell lysates were prepared from cultured cells using 1X cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA) with 1X protease inhibitor cocktail (Complete Mini, Roche, Mannheim, Germany) and 1 mM phenylmethanesulfonyl fluoride (Sigma, St Louis, MO, USA) added. Briefly, after washing with cold PBS, 1X cell lysis buffer was added and the cells scraped off using a plastic cell lifter and collected into a pre-chilled 1.5-ml tube. The cells were allowed to lyse on ice for 30 min and centrifuged at 14,000 x g for 10 min at 4°C. The supernatant (whole cell lysate) was collected, snap-frozen in liquid nitrogen, and stored at -80°C. Nuclear lysates were extracted according to our recently published method [16].
Antibody microarray analysis
Antibody Microarray (ASB600) was obtained from Full Moon BioSystems (Sunnyvale, CA, USA). Each glass slide contains 656 highly specific and well-characterized antibodies in duplicate. These antibodies were raised against cellular proteins that play important roles in diverse biological functions such as cell cycle regulation, cell differentiation, apoptosis, stress response and DNA damage repair, cancer metastasis, and signal transduction. Proteins (whole cell lysates) were extracted as described above, biotinylated and then hybridized to the microarray and detected with fluorescent-labeled strepatavidin using Antibody Microarray Detection Kit (Spring Bioscience, Pleasanton, CA, USA) according to the manufacturer's protocol. A change of around 2-fold was used as a cut-off standard to evaluate the differential expression of proteins between lovastatin-treated and control cells.
Western blot analysis
Nuclear lysates were separated on 10% PAGE denaturing gels followed by incubation with the anti-HIF-1α or anti-Histone H3 antibody, the HRP-conjugated secondary antibody, and ECL development according to our standard protocol [17].
RNA extraction and real-time reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from treated or control cells with the RNA Isolator reagent (Vazyme, Nanjing, China). One μg of total RNA was used as the template for reverse transcription using HiScript 1st Strand cDNA Synthesis Kit (Vazyme). Standard quantitative PCR reactions were conducted using SuperMix qPCR reagent (Vazyme) in a CFX Connect Real-Time PCR Detection System (Bio-Rad) using the previously described conditions. PCR primers used in the present study were summarized in Table 1. GAPDH was used as a normalization control for gene expression.
Statistical and bioinformatic analyses
For proliferation assay and quantitative RT-PCR analysis of gene expression, the data from 4 - 6 replicate samples from at least two independent experiments were expressed as mean ± SEM and analyzed by one-way ANOVA followed by Student's t test (unpaired) to determine the statistical significance. P < 0.05 was considered significant. GO enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool.
Results
Lovastatin's anti-cancer effects in breast cancer cells
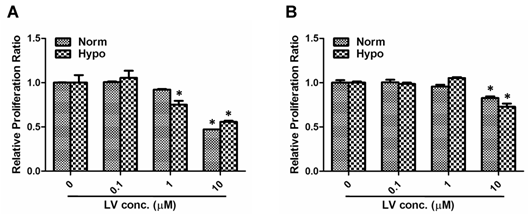
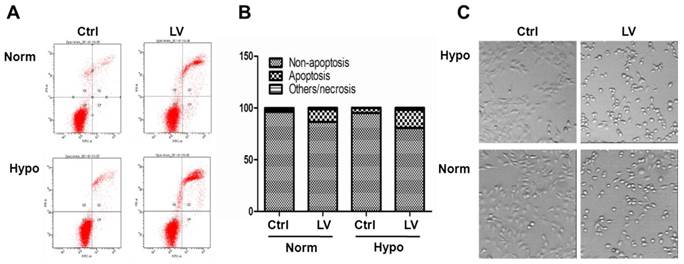
We have chosen MDA-MB-231 and MDA-MB-468 as representative cell lines of triple-negative breast cancer phenotype [18]. Lovastatin, when used at a concentration range between 0.1 and 10 μM, dose-dependently inhibited proliferation of MDA-MB-231 cells (Figure 1A) or MDA-MB-468 cells (Figure 1B) under both normoxia and hypoxia. LV-induced inhibition of cell proliferation was more prominent in MDA-MB-231 cells than in MDA-MB-468 cells. Furthermore, lovastatin induced apoptosis in MDA-MB-231 cells under normoxia and hypoxia (Figures 2A & 2B). Cell images taken at the end of the 48-h treatment period also showed characteristic changes of cell apoptosis including shrinkage and rounding of the cells in LV-treated group compared with the control group (Figure 2C).
List of PCR Primers Used in This Study
| No | Name | GenBank Accession # | Forward Sequence | Reverse Sequence | Product Size |
|---|---|---|---|---|---|
| 1 | DR3 | U72763.1 | AGATGTTCTGGGTCCAGGTG | GCTGTCCAAGGGTGACAGAT | 176 |
| 2 | Keratin 19 | AF202321.1 | CACACTTATACCCTTGCAGCC | GAGGATAGGGAGAGGGGGTT | 104 |
| 3 | p57Kip2, V1 | NM_000076.2 | TGCACGAGAAGGTACACTGG | ACCAGATGTGGGAGATGGAG | 265 |
| 4 | p57Kip2, V2 | U22398.1 | CTGATCTCCGATTTCTTCGC | AAAAACAAAACCGAACGCTG | 279 |
| 5 | Histone H1a | NM_005325.3 | GTCTGAAACAGTGCCTCCCG | TGCAGCCTTAGCAGGTTTCT | 91 |
| 6 | Histone H1b | NM_005322.2 | TTTCTTGCCACCATGTCGGA | TTGCCTTCTTCTTAGCCGGG | 85 |
| 7 | MHC I (HLA-A) | Z46633.1 | GCGGCTACTACAACCAGAGC | CCAGGTAGGCTCTCAACTGC | 237 |
| 8 | NuMA, V1 | NM_006185.3 | GCTCCTGTGCCTTCTACCTG | TCTCAGCTAGCTCCAGCTCC | 259 |
| 9 | NuMA, V2 | NM_001286561.1 | GCTCCTGTGCCTTCTACCTG | TCTCAGCTAGCTCCAGCTCC | 259 |
| 10 | TGM2 | NM_004613.2 | CCTCGTGGAGCCAGTTATCA | GTCGCTCTCGAAGTTCACCA | 316 |
| 11 | HIF-1α | NM_001530.3 | GCCAGATCTCGGCGAAGTAA | CCAGTTAGTTCAAACAGCATCCA | 165 |
| 12 | GAPDH | NM_002046.5 | CACCACCAACTGCTTAGC | TTCACCACCTTCTTGATGTC | 333 |
Lovastatin induces growth inhibition in breast cancer cells. MDA-MB-231 (A) or MDA-MB-468 (B) cells seeded in 96-well plates were treated with different concentrations of lovastatin (LV) and cultured under normoxia (21% O2) or hypoxia (1% O2) for 48 h. Cell proliferation was analyzed by measuring fluorescence at 560/590nm after the addition of the CellTiter Blue cell viability assay reagent. * P < 0.05 compared with the control.
Lovastatin induces apoptosis in breast cancer cells. (A) MDA-MB-231 cells seeded in 35-mm dishes were treated with lovastatin (LV, 30 μM) or vehicle and cultured under normoxia or hypoxia for 48 h. The cells were harvested, washed twice with PBS, and resuspended in 1X binding buffer. FITC-labeled Annexin V and propidium iodide were added and incubated for 15 min at room temperature in the dark. Fluorescence was detected using the BD FACSCanto II Flow Cytometer. (B) A bar graph summarizes the percentage distribution of cells in each apoptosis-related portion from one representative experiment. (C) Photomicrographic images of cells 24 h after treatment with either LV or vehicle control (Ctrl). Original magnification: x200.
Differentially expressed proteins in MDA-MB-231 cells treated with lovastatin
To analyze the molecular mechanisms underlying lovastatin's anti-cancer effects in breast cancer cells, MDA-MB-231 cells collected from triplicate dishes were pooled together and subjected to antibody microarray analysis using an antibody microarray. Thirty-seven proteins were identified as significantly changed upon treatment with lovastatin that included 17 up-regulated and 20 down-regulated proteins (Table 2 and Supplementary Figure 1). These altered proteins were further grouped into different pathways using GO enrichment analysis (Table 3). These pathways included proteins that are involved in apoptosis, proliferation, differentiation, signal transduction, EMT and metastasis.
Cell apoptosis
Antibody microarray analysis indicated that several proteins involved in apoptosis regulation were altered when exposed to lovastatin. The up-regulated proteins included death receptor 3 (DR3), tumor necrosis factor receptor type 1-associated death domain protein (TRADD), glutamate-cysteine ligase, Caspase 7, Stat1, Raf1, Mek6, Vitamin D3 receptor and amyloid beta; while the expression of silencer of death domain (SODD), poly [ADP-ribose] polymerase (PARP), apoptosis-inducing factor (AIF), the chromatin DNA-binding protein p63, transglutaminase II (TGM2), cyclic AMP-responsive element-binding protein (CREB) were down-regulated. Among the apoptosis-related proteins up-regulated by lovastatin, DR3 was by far the most significantly changed protein, with an 18.5 fold change.
Cell proliferation and differentiation markers
Inhibition of cell proliferation was another effect exerted by lovastatin in MDA-MB-231 cells. Among the proteins related to cell proliferation, the cyclin-dependent kinase inhibitor p57Kip2, the dual-specificity phosphatase CDC14A and the co-chaperone protein CDC37 were up-regulated; whereas cyclin-dependent kinase CDK4, chromatin DNA-binding protein Histone H1 and nuclear mitotic apparatus protein NuMA were down-regulated by lovastatin treatment. Several cell differentiation markers including keratin, leukocyte surface antigen CD53, the endoplasmic reticulum calcium ATPase II, and the thyroid transcription factor TTF-1 were up-regulated by lovastatin treatment. At the same time, lovastatin treatment led to down-regulation of 5 differentiation markers, i.e., CD16, CD1a, MHC I (HLA-A), B-cell linker protein and CD29 (integrin β1).
Signal transduction
The signal transducers Raf1, amyloid β, Mek6, Stat1, myelin-oligodendrocyte glycoprotein and Vitamin D3 receptor were up-regulated while CREB and γ glutamyl transferase down-regulated by lovastatin treatment.
EMT and metastasis
Among the proteins that are involved in epithelial-to-mesenchymal transition (EMT), including TGM2, hypoxia-inducible factor 1α (HIF-1α) and Histone H1, were down-regulated by lovastatin. Proteins that are involved in tumor metastasis including the intercellular adhesion molecule CD50 (ICAM3), the target of the anti-proliferative antibody CD81, the vascular endothelial glycoprotein CD105 (Endoglin) and the matrix metalloproteinase MMP-7 (matrilysin) were down-regulated.
Validation of the antibody microarray data
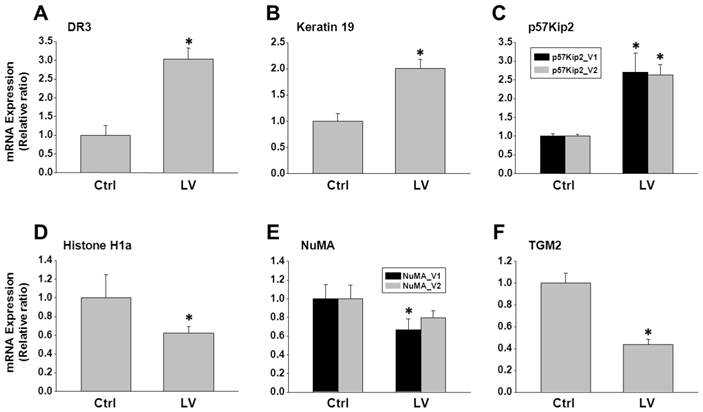
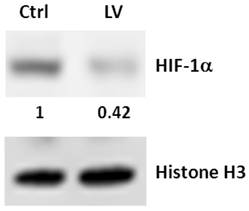
To validate the antibody microarray data, MDA-MB-231 cells were treated with lovastatin or vehicle and subjected to real-time RT-PCR analysis. We choose 3 up-regulated genes (DR3, keratin 19 and p57Kip2) and 5 down-regulated genes (Histone H1, MHC I (HLA-A), NuMA, TGM2 and HIF-1α) for validation. Up-regulation of DR3 (Figure 3A), keratin 19 (Figure 3B) and p57Kip2 (Figure 3C) and down-regulation of Histone H1a (Figure 3D), NuMA (Figure 3E) and TGM2 (Figure 3F) were confirmed by real-time PCR analysis of RNA extracted from lovastatin-treated MDA-MB-231 cells compared with the control cells. Expression of HIF-1α mRNA was not altered by lovastatin and, therefore, we validated the protein expression of HIF-1α using Western blot analysis. We found that nuclear HIF-1α protein level was down-regulated in MDA-MB-231 cells treated with lovastatin compared with vehicle control (Figure 4). These data were consistent with HIF-1α's role as part of a transcription factor, which is mainly subjected to post-translational regulation of the protein stability through proteasome-mediated degradation [19].
List of proteins up- or down-regulated by LV in MDA-MB-231 cells
| Group | Protein Name | GenBank Accession # | Fold Change |
|---|---|---|---|
| Up-regulated | Death Receptor 3 (DR3) | U72763.1 | 18.48 |
| Keratin, Pan | AF202321.1 | 9.00 | |
| Vitamin D3 Receptor | AB002168.2 | 7.33 | |
| TRADD | NM_003789.3 | 6.00 | |
| Glutamate-cysteine Ligase | AB262176.1 | 5.50 | |
| Raf1 | NM_002880.3 | 4.60 | |
| Stat1 | GU211347.1 | 3.43 | |
| Caspase 7 | NM_001227.4 | 3.33 | |
| CDC14A Phosphatase | AF122013.1 | 3.03 | |
| Amyloid Beta | NM_000484.3 | 2.97 | |
| CD53 | NM_001040033.1 | 2.50 | |
| Thyroid Transcription Factor 1 (TTF-1) | U33749.1 | 2.15 | |
| CDC37 | NM_007065.3 | 2.09 | |
| ER Ca+2 ATPase II | NM_001681.3 | 2.00 | |
| Mek6 | U49732.1 | 2.00 | |
| Myelin-oligodendrocyte Glycoprotein | NM_206809.3 | 2.00 | |
| p57Kip2 | U22398.1 | 1.87 | |
| Down-regulated | p63 | BC039815.1 | 0.03 |
| CD16 | BC036723.1 | 0.11 | |
| CD50 | NM_002162.3 | 0.13 | |
| CD1a | NM_001763.2 | 0.15 | |
| Silencer of Death Domain (SODD) | NM_004874.3 | 0.20 | |
| MHC I (HLA-A) | Z46633.1 | 0.22 | |
| CD105 | BC014271.2 | 0.24 | |
| PARP | NM_001618.3 | 0.25 | |
| CREB | AY347527.1 | 0.25 | |
| B-cell Linker Protein | NM_013314.3 | 0.26 | |
| CD29 | BC020057.1 | 0.31 | |
| Gamma Glutamyl Transferase | BC128239.1 | 0.32 | |
| Cyclin-Dependent Kinase 4 (CDK4) | NM_000075.3 | 0.41 | |
| MMP-7 | NM_002423.3 | 0.41 | |
| CD81 | BT019507.1 | 0.43 | |
| Apoptosis-Inducing Factor (AIF) | AF100928.1 | 0.44 | |
| Histone H1 | NM_005325.3 | 0.50 | |
| HIF-1α | NM_001530.3 | 0.54 | |
| NuMA | NM_006185.3 | 0.56 | |
| Transglutaminase II (TGM2) | NM_004613.2 | 0.56 |
GO enrichment analysis of proteins regulated by LV in MDA-MB-231 cells
| Category | Count* | % | P Value | Genes |
|---|---|---|---|---|
| Apoptosis | 15(9+6) | 40.5 | 3.41E-06 | DR3, TRADD, Glutamate-cysteine ligase, Caspase 7, Stat1, Raf1, Mek6, Vitamin D3 receptor, Amyloid beta; SODD, PARP, AIF, p63, TGM2, CREB |
| Differentiation | 9(4+5) | 24.3 | 2.77E-06 | Keratin, CD53, ER Ca+2 ATPase II, TTF-1; CD16, CD1a, MHC (HLA-A), B-cell linker protein, CD29 |
| Proliferation | 6(3+3) | 16.2 | 2.77E-06 | p57Kip2, CDC14A, CDC37; CDK4, NuMA, Histone H1 |
| Cell signaling | 8(6+2) | 21.6 | 1.31E-05 | Raf1, Amyloid beta, Mek6, Stat1, myelin-oligodendrocyte glycoprotein, Vitamin D3 receptor; CREB, Gamma glutamyl transferase |
| EMT & metastasis | 7(0+7) | 18.9 | 3.76E-05 | TGM2, HIF-1a, Histone H1, CD50, CD81, CD105, MMP-7 |
*Total count (up- + down-regulated)
Analysis of genes selected from the antibody microarray data by RT-PCR. Total RNA extracted from MDA-MB-231 cells treated with lovastatin or vehicle under hypoxia for 24 h was reverse-transcribed into cDNA. Real-time PCR was performed using primers specific for each of the genes with SYBR Green as the fluorescent dye. A: DR3; B: Keratin 19; C: p57Kip2; D: Histone H1a; E: NuMA; F: TGM2. * P < 0.05.
Western blot analysis of the effect of lovastatin on HIF-1α expression in MDA-MB-231 cells. Whole cell lysates from MDA-MB-231 cells treated with lovastatin or vehicle under hypoxia for 24 h were separated by SDS-PAGE and electro-blotted to PVDF membranes, which were probed with an antibody to human HIF-1α or Histone H3. Numbers beneath the HIF-1α blot represent signal density quantification after normalization to the Histone H3 signal.
Discussion
Although the beneficial effects of statins has been documented in breast cancer prevention and treatment [20], their use in triple-negative breast cancer has gained little recognition until recently [21, 22]. Results from our laboratory (data not shown) and others [21, 23] have indicated that lovastatin preferentially suppressed proliferation and induced apoptosis in triple-negative breast cancer cells. These observations may have significant clinical implications in that lovastatin may be used as an ideal natural product to treat a subset of clinically challenging breast cancer patients. To this end, we deemed it necessary to further elucidate the molecular mechanisms underlying the anti-cancer effects of lovastatin. Although a LC-MS/MS-based proteomic strategy has been used by Klawitter et al to identify the proteomic changes induced by lovastatin in MDA-MB-231 cells [15], antibody microarray analysis represents a novel strategy with high sensitivity and increased specificity. Furthermore, we used a hypoxic culture environment that more faithfully mimics the actual growth conditions of tumor cells in vivo.
Our antibody microarray analysis revealed that 17 proteins were up-regulated and 20 down-regulated by lovastatin in the highly malignant MDA-MB-231 cells. GO enrichment analysis further grouped these differentially regulated proteins into different pathways. Notably, the apoptotic pathway stands out as a prominent pathway among these lovastatin-induced signaling pathways. Statins including lovastatin have been shown to induce apoptosis in a myriad of cell types including breast cancer cells [24]. It was therefore not surprising that a variety of apoptosis-related proteins were regulated by lovastatin in MDA-MB-231 cells. In this group of proteins, DR3 was the one with the highest protein change ratio. Lovastatin-induced up-regulation of DR3 was accompanied by the increased levels of its downstream intracellular partners, i.e., TRADD and Caspase 7, and the decreased level of the substrate PARP.
Death receptors (DRs) are members of the tumor necrosis factor receptor (TNFR) superfamily. When DRs bind to their ligands, death domains recruit the intracellular adaptor protein FADD which results in the activation of Caspases as well as an increase of Bax and decrease of BCl-2 to induce apoptosis [25]. Activation of DR3 triggers the Caspase cascade through the interaction with TRADD in a variety of cancer types including human breast carcinoma [26]. Stimulation of DR expression is implicated in the induction of apoptosis which is related to the elimination of chemoresistance in cancer cells [27]. Considering the fact that the efflux protein P-glycoprotein (P-gp) is a direct target for statin interactions [28], our results suggest that the natural compound lovastatin may serve the role of reducing or overcoming chemoresistance in breast cancer. Furthermore, DR3 serves as a functional receptor for vascular endothelial growth inhibitor [29] and targeting of DR3 by ribozyme impaired the in vitro migration of breast cancer cells [30]. These observations suggest that DR3 plays a negative regulatory role in breast carcinogenesis and up-regulation of DR3 and its downstream partners may be one of the molecular modulators to slow down the development of breast cancer.
Another important finding of our antibody microarray analysis was the involvement of the EMT and tumor metastasis pathways. Specifically, HIF-1α, Histone H1 and TGM2 comprise the backbone of the EMT signaling network. TGM2 is the most abundantly expressed member of the transglutaminase family. It is distributed in various parts of a cell, including the extracellular matrix, plasma membrane, the cytosol and the nucleus. Nuclear TGM2 especially impacts on the regulation of gene expression via post-translational mechanisms including interaction with transcriptional factors and related proteins such as HIF-1, Sp1, E2F1 and histones [31]. Histone proteins, including H1, have been shown to be phosphorylated by nuclear TGM2 in vitro [32], suggesting an important role for TGM2 in the regulation of chromatin function and gene expression.
It has recently been shown that TGM2 expression in mammary epithelial cells is associated with activation of nuclear transcription factors such as NF-κB and HIF-1α. The increased stability and activation of HIF-1 in cancer cells contribute to altered glycolytic metabolism, invasion, metastases, treatment resistance and disease relapse [33]. Additionally, TGM2 expression is able to induce the developmentally regulated program of EMT and confer cancer stem cell traits in mammary epithelial cells [34]. These observations imply that TGM2 plays a crucial role in promoting the aggressive phenotypes of breast cancer including increased drug resistance and tumor metastasis. In our microarray study, we found that HIF-1α, Histone H1, and TGM2 were all down-regulated by lovastatin. It is possible that lovastatin down-regulates the expression of EMT-related genes in MDA-MB-231 cells through the modulation of TGM2- HIF-1α-Histone H1 cascade. This possibility needs to be investigated in further in vitro and in vivo studies.
In summary, in the present study, we demonstrated that lovastatin exerts anti-cancer effects in the triple-negative breast cancer cell line MDA-MB-231. We further used an antibody microarray analysis to elucidate the molecular mechanisms underlying lovastatin's anti-cancer effects in MDA-MB-231 cells under a hypoxic culture environment. Among the lovastatin-regulated proteins (17 up and 20 down), the prominent pathways included the apoptosis pathway and the EMT-metastasis pathway among others. Modulation of these pathways may explain, in part, the anti-breast cancer effects of lovastatin. These findings provide an experimental basis for further elucidation of the molecular targets of lovastatin.
Supplementary Material
Supplementary Figure 1.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Project 81472496), the S&T Plan Key Project of Hunan (2013WK4006), the Grain-oil Process and Quality Control 2011 Collaborative and Innovative Grant from Hunan Province, the Special Fund for Agro-scientific Research in the Public Interest of China (201303071-2-1), the Key Project of Department of Education of Hunan (14A089), and the Research Project of the S&T Bureau of Changsha (k1403024-31).
Abbreviations
AIF: Apoptosis-inducing factor
CREB: cyclic AMP-responsive element-binding protein
DR3: Death receptor 3
EMT: Epithelial-to-mesenchymal transition
ER: Estrogen receptor
GO: Gene ontology
HER2: Human epidermal growth factor receptor 2
HIF-1: Hypoxia-inducible factor 1
LV: Lovastatin
PARP: Poly [ADP-ribose] polymerase
PR: Progesterone receptor
RT-PCR: Reverse transcription-polymerase chain reaction
SODD: Silencer of death domain
TGM2: Transglutaminase II
TRADD: Tumor necrosis factor receptor 1-associated death domain protein
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD. et al. Breast cancer in China. Lancet Oncol. 2014;15:e279-89
3. Basmadjian C, Zhao Q, Bentouhami E, Djehal A, Nebigil CG, Johnson RA. et al. Cancer wars: natural products strike back. Front Chem. 2014;2:20
4. Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507-19
5. Vira D, Basak SK, Veena MS, Wang MB, Batra RK, Srivatsan ES. Cancer stem cells, microRNAs, and therapeutic strategies including natural products. Cancer Metastasis Rev. 2012;31:733-51
6. Xu H, Yang YJ, Yang T, Qian HY. Statins and stem cell modulation. Ageing Res Rev. 2013;12:1-7
7. Chan KK, Oza AM, Siu LL. The statins as anticancer agents. Clin Cancer Res. 2003;9:10-9
8. Yang T, Liu J, Luo F, Lin Q, Rosol TJ, Deng X. Anticancer properties of Monascus metabolites. Anticancer Drugs. 2014;25:735-44
9. Prasanna P, Thibault A, Liu L, Samid D. Lipid metabolism as a target for brain cancer therapy: synergistic activity of lovastatin and sodium phenylacetate against human glioma cells. J Neurochem. 1996;66:710-6
10. Agarwal B, Bhendwal S, Halmos B, Moss SF, Ramey WG, Holt PR. Lovastatin augments apoptosis induced by chemotherapeutic agents in colon cancer cells. Clin Cancer Res. 1999;5:2223-9
11. Feleszko W, Zagozdzon R, Golab J, Jakobisiak M. Potentiated antitumour effects of cisplatin and lovastatin against MmB16 melanoma in mice. Eur J Cancer. 1998;34:406-11
12. Holstein SA, Hohl RJ. Interaction of cytosine arabinoside and lovastatin in human leukemia cells. Leuk Res. 2001;25:651-60
13. Feleszko W, Mlynarczuk I, Balkowiec-Iskra EZ, Czajka A, Switaj T, Stoklosa T. et al. Lovastatin potentiates antitumor activity and attenuates cardiotoxicity of doxorubicin in three tumor models in mice. Clin Cancer Res. 2000;6:2044-52
14. Feleszko W, Balkowiec EZ, Sieberth E, Marczak M, Dabrowska A, Giermasz A. et al. Lovastatin and tumor necrosis factor-alpha exhibit potentiated antitumor effects against Ha-ras-transformed murine tumor via inhibition of tumor-induced angiogenesis. Int J Cancer. 1999;81:560-7
15. Klawitter J, Shokati T, Moll V, Christians U. Effects of lovastatin on breast cancer cells: a proteo-metabonomic study. Breast Cancer Res. 2010;12:R16
16. Li H, He G, Yao H, Song L, Zeng L, Peng X. et al. TGF-beta Induces Degradation of PTHrP Through Ubiquitin-Proteasome System in Hepatocellular Carcinoma. J Cancer. 2015;6:511-8
17. Deng X, He G, Levine A, Cao Y, Mullins C. Adenovirus-mediated expression of TIMP-1 and TIMP-2 in bone inhibits osteolytic degradation by human prostate cancer. Int J Cancer. 2008;122:209-18
18. Camirand A, Fadhil I, Luco AL, Ochietti B, Kremer RB. Enhancement of taxol, doxorubicin and zoledronate anti-proliferation action on triple-negative breast cancer cells by a PTHrP blocking monoclonal antibody. Am J Cancer Res. 2013;3:500-8
19. Tang CM, Yu J. Hypoxia-inducible factor-1 as a therapeutic target in cancer. J Gastroenterol Hepatol. 2013;28:401-5
20. Kwan ML, Habel LA, Flick ED, Quesenberry CP, Caan B. Post-diagnosis statin use and breast cancer recurrence in a prospective cohort study of early stage breast cancer survivors. Breast Cancer Res Treat. 2008;109:573-9
21. Campbell MJ, Esserman LJ, Zhou Y, Shoemaker M, Lobo M, Borman E. et al. Breast cancer growth prevention by statins. Cancer Res. 2006;66:8707-14
22. Kumar AS, Benz CC, Shim V, Minami CA, Moore DH, Esserman LJ. Estrogen receptor-negative breast cancer is less likely to arise among lipophilic statin users. Cancer Epidemiol Biomarkers Prev. 2008;17:1028-33
23. Dimitroulakos J, Ye LY, Benzaquen M, Moore MJ, Kamel-Reid S, Freedman MH. et al. Differential sensitivity of various pediatric cancers and squamous cell carcinomas to lovastatin-induced apoptosis: therapeutic implications. Clin Cancer Res. 2001;7:158-67
24. Sassano A, Platanias LC. Statins in tumor suppression. Cancer Lett. 2008;260:11-9
25. Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228-37
26. van der Weide K, de Jonge-Peeters SD, Kuipers F, de Vries EG, Vellenga E. Combining simvastatin with the farnesyltransferase inhibitor tipifarnib results in an enhanced cytotoxic effect in a subset of primary CD34+ acute myeloid leukemia samples. Clin Cancer Res. 2009;15:3076-83
27. Choi KE, Hwang CJ, Gu SM, Park MH, Kim JH, Park JH. et al. Cancer cell growth inhibitory effect of bee venom via increase of death receptor 3 expression and inactivation of NF-kappa B in NSCLC cells. Toxins (Basel). 2014;6:2210-28
28. Wang E, Casciano CN, Clement RP, Johnson WW. HMG-CoA reductase inhibitors (statins) characterized as direct inhibitors of P-glycoprotein. Pharm Res. 2001;18:800-6
29. Zhang N, Sanders AJ, Ye L, Jiang WG. Vascular endothelial growth inhibitor in human cancer (Review). Int J Mol Med. 2009;24:3-8
30. Ge Z, Sanders AJ, Ye L, Mansel RE, Jiang WG. Expression of death receptor-3 in human breast cancer and its functional effects on breast cancer cells in vitro. Oncol Rep. 2013;29:1356-64
31. Kuo TF, Tatsukawa H, Kojima S. New insights into the functions and localization of nuclear transglutaminase 2. FEBS J. 2011;278:4756-67
32. Mishra S, Saleh A, Espino PS, Davie JR, Murphy LJ. Phosphorylation of histones by tissue transglutaminase. J Biol Chem. 2006;281:5532-8
33. Mimeault M, Batra SK. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J Cell Mol Med. 2013;17:30-54
34. Agnihotri N, Kumar S, Mehta K. Tissue transglutaminase as a central mediator in inflammation-induced progression of breast cancer. Breast Cancer Res. 2013;15:202
Author contact
![]() Corresponding author: Xiyun Deng, E-mail: dengxiyunmededu.cn; Tel: +86-731-88912426
Corresponding author: Xiyun Deng, E-mail: dengxiyunmededu.cn; Tel: +86-731-88912426

 Global reach, higher impact
Global reach, higher impact