3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(2):207-213. doi:10.7150/jca.13732 This issue Cite
Research Paper
Expression of TMEM207 in Colorectal Cancer: Relation between TMEM207 and Intelectin-1
1. Department of Surgical Oncology, Gifu University Graduate School of Medicine, Gifu, Japan
2. Department of Pathology and Translational Research, Gifu University Graduate School of Medicine, Gifu, Japan
Received 2015-9-3; Accepted 2015-11-1; Published 2016-1-1
Abstract
Recent research advances highlighted an intestinal goblet cell-produced lectin, intelectin-1 (also known as omentin-1), as a tumor suppressor. One study indicated that downregulation of intelectin-1 may be related to the unfavorable prognosis among patients with colorectal carcinoma at an advanced stage. The present study was aimed at analyzing the expression of a hitherto uncharacterized transmembrane protein TMEM207 in colorectal carcinoma, and we found that the TMEM207 function is linked to intelectin-1 processing. With specific antibodies, TMEM207 immunoreactivity was detected in 38 of 216 colorectal cancer tissue samples. TMEM207 immunoreactivity correlated inversely with lymph node metastatic status (p < 0.01). TMEM207 expression significantly correlated with the mucinous phenotype of colorectal carcinoma. A coimmunoprecipitation assay revealed an interaction between intelectin-1 and TMEM207 in colorectal cancer cells. A proximal ligation assay indicated that intelectin-1 and TMEM207 were colocalized to the cytoplasm of the colorectal cancer cells. A small-interfering-RNA-mediated knockdown of TMEM207 increased polyubiquitination and proteasome degradation of intelectin-1 in cultured colorectal cancer cells and decreased intelectin-1 secretion. These findings indicate that a loss of TMEM207 expression leads to insufficient intelectin-1 production thus promoting colorectal carcinogenesis.
Keywords: Colorectal cancer, mucinous carcinoma, lymph node metastasis, intelectin-1, TMEM207
Introduction
TMEM207 was identified by researchers in a comprehensive project called the Secreted Protein Discovery Initiative, which was aimed at finding novel secretory and transmembrane proteins [1]. A bioinformatic analysis suggested that TMEM207 may be localized to the endoplasmic reticulum (ER; with reliability of 44.4%) [2]. In line with this bioinformatic hypothesis, immunohistochemical staining revealed that TMEM207 is located in the cytoplasm of intestinal goblet cells. Subsequently, TMEM207 was found to be expressed in signet-ring cell carcinoma and mucinous carcinoma, but little or no expression was detected in other gastric adenocarcinomas [3]. Transgenic mice in which murine TMEM207 is ectopically expressed in cutaneous hair follicle bulge cells spontaneously develop a cutaneous appendage tumor, often of the apocrine secretory phenotype [4]. Very recently, TMEM207 expression was shown to be significantly downregulated in clear cell renal cell carcinoma [2]. These findings indicate that TMEM207 may be a member of a transmembrane protein family participating in tumorigenesis just as TMEM16a (also known as anoctamin-1, ANO1, or DOG1) [5, 6], TMEM25 [7], TMEM45 [8], TMEM88 [9], and other proteins. Nonetheless, the precise pathophysiological role of TMEM207 remains largely unclear.
Human intelectin-1 recognizes multiple glycan epitopes on microbes [10] but does not bind known human glycan epitopes, thus participating in intestinal microbial surveillance [11]. Intelectin-1 has pleiotropic pathophysiological properties. It is also known as omentin-1, which is well characterized as an adipokine and believed to promote insulin-stimulated glucose uptake; in fact, intelectin-1 expression is negatively correlated with obesity (reviewed in [12]). Because recent advances uncovered the tumor suppressor function of intelectin-1 [13-16], a loss of intelectin-1 may be involved in obesity-related carcinogenesis [18].
In this study, we assessed TMEM207 expression in colorectal cancer and unexpectedly identified a previously unreported relation between intelectin-1 and TMEM207 in colorectal cancer cells. Our results suggest that TMEM207 may be important for proper processing of intelectin-1, and a loss of TMEM207 may promote colorectal carcinogenesis.
Materials and methods
Antibodies and cells
A rabbit antibody to human recombinant TMEM207 (amino acid residues 77 to 130) was purchased from Sigma-Aldrich (St. Louis, MO). A conventional rabbit antibody to the synthetic peptide VNYNDQHPNGW (amino acid residues 40 to 50 of TMEM207) was prepared in our laboratory. The detailed procedure for the preparation of this antibody, which was purified by affinity chromatography with the immunogenic peptide, was described previously [3]. A murine monoclonal antibody to human intelectin-1 (clone 3G9) [18] was purchased from Immuno-Biological Laboratories Co., Ltd. (Gunma, Japan). A rabbit anti-GAPDH antibody was purchased from Sigma-Aldrich. A murine monoclonal antiubiquitin antibody was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
Human SW480 colon cancer cells [19] and RCM-1 rectal adenocarcinoma cells [20], the latter of which were from partially exhibited mucinous carcinoma features, were obtained from American Type Culture Collection (Manassas, VA) and Japan Health Science Research Resources Bank (Osaka, Japan), respectively. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco Life Technologies, Grand Island, NY) containing 10% of heat-inactivated fetal bovine serum (FBS) and 10 μg/mL gentamycin (Gibco Life Technologies). The cells were passaged in our laboratory not longer than 6 months after revival.
Immunohistochemical staining
Immunohistochemical detection of TMEM207 was performed by means of tissue microarrays (US Biomax, Rockville, MD). All tissue samples were collected according to the ethical standards and the Health Insurance Portability and Accountability Act (USA). Written informed consent was obtained from all donors. Archival pathological tissue samples of mucinous colon cancers were also analyzed by a proximal ligation assay. The study protocol was approved by the Institutional Review Board of the Gifu University Graduate School of Medicine.
Details of the procedure for immunohistochemical staining were described previously [21]. Briefly, antigen retrieval in deparaffinized tissue slices was performed with 0.25% trypsin for 5 min at 37°C. After incubation for 30 min in 10% normal goat serum, the slides were incubated with various antibodies overnight at 4°C. We used the ImmPRESS Polymerized Reporter Enzyme Staining System (Vector Laboratories Inc., Burlingame, CA).
Immunoprecipitation
SW480 or RCM-1 cells were solubilized by means of the CelLytic M Cell Lysis Reagent (Sigma-Aldrich) containing a proteinase inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). The soluble fraction of the cell lysates was isolated by centrifugation, incubated with the murine anti-intelectin-1 antibody for 2 h at 4°C, and mixed with protein G-Sepharose beads (Sigma-Aldrich) with subsequent incubation for 30 min at 4°C. After that, the beads were washed 4 times with the CelLytic M Cell Lysis Reagent containing the proteinase inhibitor cocktail. Thereafter, we eluted the immunocomplexes by boiling the samples for 3 min in sodium dodecyl sulfate (SDS) sample buffer containing 10 mM dithiothreitol (DTT). Next, the samples were subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE) followed by western immunoblotting with antibodies as described previously [3].
Western blotting
This procedure was performed according to the previously described method [3] with the modification proposed by Towbin et al. [22]. The samples were analyzed by electrophoresis in SDS-containing polyacrylamide gels under reducing conditions. The separated proteins were transferred onto polyvinylidene difluoride membranes (Millipore Co., Bedford, MA) and probed with various antibodies. Immunoreactivity was assessed by means of the Western Blotting Detection Kit (Promega, Madison, WI).
Proximal ligation assay in situ
For this assay [23], we used the Duolink In Situ Detection Reagents Brightfield Kit (Sigma-Aldrich). After antigen retrieval with 0.25% trypsin for 5 min at 37°C, the tissue slices were incubated with blocking buffer for 30 min at room temperature followed by incubation with 1 μg/mL rabbit anti-TMEM207 and mouse anti-intelectin-1 antibodies overnight at 4°C. As an antibody control, some slides were incubated with intact anti-intelectin-1 and the anti-TMEM207 antibody, which was prebound with the immunization peptide. Then, the tissue samples were reacted with secondary antibodies, which were conjugated with unique DNA fragments that served as a template for hybridization of added oligonucleotides. After that, the oligonucleotides were ligated to form a circular template. This template was amplified with complementary labeled oligonucleotide probes. These procedures, including ligation and rolling circle amplification, are described in the manufacturer's manual for the kit. The interaction signals were detected with horseradish peroxidase and 3,3′-diaminobenzidine.
Small interfering RNA (siRNA)-mediated gene silencing in colorectal cancer cells
The detailed procedure was described previously [3]. In this study, we used Ambion Silencer Select siRNAs—s43652 and s43653 (Austin, TX)—to silence the TMEM207 gene. A green fluorescent protein (GFP) siRNA duplex with the target sequence 5′-CGGCAAGCUGACCCUGAAGUUCAU-3′ was used as a nonsilencing control. The siRNA was transfected into cells with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). At 72 h after the transfection, the cells were used for subsequent experiments.
Enzyme-linked immunosorbent assay (ELISA)
Cells (105/well) were cultured in 24-well plates in the Opti-MEM medium (Invitrogen). After 48 h, the culture supernatants were collected, and intelectin-1 concentration was measured by a sandwich ELISA (Immuno-Biological Laboratories Co., Ltd., Gunma, Japan). The assay was performed in triplicate and repeated twice. Statistical analysis involved Student's t test for unpaired observations. Differences with p < 0.05 were considered statistically significant.
Results
Expression of TMEM207 is inversely correlated with nodal metastasis of colorectal carcinoma
We started the experiments by examining TMEM207 expression in tissue microarrays containing 216 samples of colorectal adenocarcinomas. Representative results of immunohistochemical staining are shown in Figure 1. Immunoreactivity of the anti-TMEM207 antibody was observed in 38 of the 216 samples. As shown in Figure 1B-D, TMEM207 immunoreactivity was observed in cytoplasm of cancer cells in the positive cases. Strong TMEM207 immunoreactivity was detected in many cases of colonic mucinous carcinoma (Figure 1D).
Expression of TMEM207 according to tissue microarray analysis. The images show representative immunohistochemical staining with an affinity-purified rabbit antibody to a TMEM207 peptide. The scale bar represents 100 µm in panels A and B and 50 µm in panels C and D. A: No significant immunoreactivity with TMEM207 was observed in adenocarcinoma components of a colon cancer sample associated with lymph node metastasis. The asterisk indicates lymphatic-vessel invasion by cancer cells. B: TMEM207 immunoreactivity was observed in carcinoma cells of a colon adenocarcinoma sample not associated with lymph node metastasis. C: TMEM207 immunoreactivity was detected in the cytoplasm of carcinoma cells in another case of colon adenocarcinoma without lymph node metastasis. D: Strong TMEM207 immunoreactivity was detected in a case of colonic mucinous carcinoma (it had mucinous-carcinoma features).
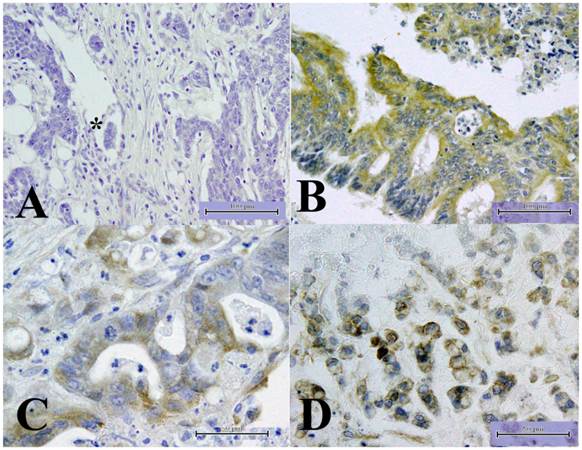
Notably, only 2 of 38 TMEM207-positive colorectal adenocarcinomas involved lymph node metastasis, whereas as many as 54 of 178 TMEM207-negative cancers were associated with lymph node metastasis. TMEM207 expression correlated with lymph node metastasis inversely (χ2 test, p < 0.01).
Expression of TMEM207 in mucinous carcinoma of the colon. The images show representative immunohistochemical staining of archival pathological tissue samples with an affinity-purified rabbit antibody to a TMEM207 peptide. A: Significant immunoreactivity with TMEM207 was observed in mucinous carcinoma components (left-hand side), but little or no immunoreactivity was observed in nonmucinous components (right-hand side) in the same tissue samples. The arrow indicates TMEM207 immunoreactivity in cancer cells adjusted to a mucinous lake. Immunoreactivity in individual cells of each component is shown in panels B and C. B: TMEM207 immunoreactivity was observed in mucinous carcinoma cells. C: Little or no TMEM207 immunoreactivity was observed in nonmucinous carcinoma cells. D: Strong TMEM207 immunoreactivity was detected in the tumor tissue sample from another case of colonic mucinous carcinoma, which exhibited mucinous-carcinoma features with few non-mucinous-carcinoma components. E: Few nonmucinous carcinoma cells with goblet cell features exhibited TMEM207 immunoreactivity.
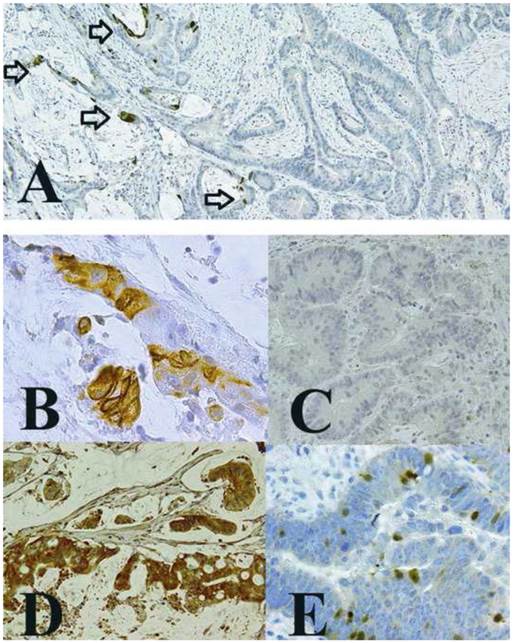
TMEM207 expression is related with the mucinous phenotype of colorectal cancer
Immunohistochemical staining by means of the tissue microarray also revealed that 18 of 28 mucinous carcinomas were immunoreactive with the anti-TMEM207 antibody, in contrast to only 10 of 178 nonmucinous carcinomas. TMEM207 expression significantly correlated with the mucinous phenotype of colorectal carcinoma (χ2 test, p < 0.01).
We then confirmed the TMEM207 immunoreactivity in mucinous carcinoma cells using archival pathological tissue samples harboring both mucinous and nonmucinous components. The cancer cells with a mucinous component showed strong immunoreactivity with the anti-TMEM207 antibody, unlike nonmucinous cancer cells (little or no staining), even in the same tissue samples (Figure 2A-C). TMEM207 immunoreactivity was found in cancer cells adjusted to a mucinous lake. In general, strong TMEM207 immunoreactivity was observed in mucinous carcinoma cells, which floated in a mucinous lake (Figure 2D). TMEM207 immunoreactivity was also detected in cancer cells that contained goblet cell-like cytoplasmic mucin (Figure 2E). We mainly used the conventional rabbit antibody to the synthetic peptide VNYNDQHPNGW (amino acid residues 40 to 50 of TMEM207). Prebinding of the antibody with the immunization peptide significantly diminished the immunoreactivity, confirming specificity of the immunostaining procedure. When we used a commercially available rabbit antibody to human recombinant TMEM207 (amino acid residues 77 to 130; Sigma-Aldrich), we obtained similar results. We concluded that TMEM207 expression is related to the mucinous phenotype of colorectal cancer.
TMEM207 binds to intelectin-1 and they are colocalized to the cytoplasm of the colorectal cancer cells
Recent studies revealed that intelectin-1 is expressed in mucous-producing colorectal cancer cells. The expression pattern of intelectin-1 reported by Washimi et al. [18] indicated a similar distribution of TMEM207 and intelectin-1, thus prompting us to explore the relation of TMEM207 and intelectin-1. To elucidate this relation, we performed coimmunoprecipitation and proximal ligation assays.
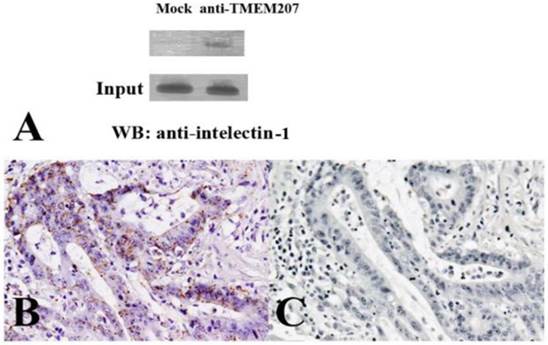
The immunoprecipitation followed by western immunoblotting revealed that TMEM207 bound to intelectin-1 (Figure 3A). The proximal ligation assay in situ yielded positive spots, which result from the events of interaction of the proteins TMEM207 and intelectin-1 in the cytoplasm of mucinous carcinoma cells (Figure 3B). Prebinding of the anti-TMEM207 antibody to the immunization peptide strongly reduced the signals (Figure 3C) and confirmed the close localization of TMEM207 and intelectin-1.
siRNA-mediated silencing of TMEM207 impaired the processing and secretion of intelectin-1 in colon cancer cells
The observed cytoplasmic colocalization and interaction of TMEM207 and intelectin-1 suggested that TMEM207 might play a role in the processing of the intelectin-1 protein in cytoplasmic organelles, e.g., the ER or Golgi apparatus. Next, we tested whether downregulation of TMEM207 can alter the intelectin-1 status in cultured colon cancer cells. We used siRNA to silence the TMEM207 gene in SW480 and RCM-1 cells and evaluated the status of the intelectin-1 protein. As demonstrated in Figure 4A, silencing of the TMEM207 gene with small interfering RNA (siRNA) significantly downregulated both TMEM207 protein and intact intelelectin-1 expression. By contrast down-regulation of TMEM207 significantly increased the smeared intelectin-1 bands when probed with anti-ubiquitin antibody in the presence of proteasome inhibitor, MG132 (Figure 4A). We concluded that the downregulation of TMEM207 increased polyubiquitination of intelectin-1 and proteasomal degradation of intelectin-1 in SW480 and RCM-1 cells. In addition, downregulation of intelecitn-1 significantly decreased intelectin-1 secretion from RCM-1 cells (Figure 4B). Together with the result of the proximal ligation assay in situ, these data suggested that TMEM207 might be necessary for proper processing of intelectin-1. A loss of TMEM207 may be linked to ER-associated degradation of insufficiently processed intelectin-1 in proteasomes.
Discussion
In the proximal ligation assay in situ, the close localization (the distance of less than 40 nm) of 2 secondary antibodies, which were conjugated with oligonucleotides, can result in formation of a circular template. The latter can be detected by rolling circle amplification. In general, an α-helical protein with molecular weight ~35 kDa is thought to be 45 nm long [24]. The deduced molecular weights of TMEM207 and intelectin-1 are approximately 17 and 36 kDa, respectively. Together with the result of the coimmunoprecipitation assay, the proximal ligation data suggest that TMEM207 and intelectin-1 may be colocalized in the cytoplasm of colorectal carcinoma cells.
Interaction of TMEM207 and intelecitn-1 in colon cancer cells. A: A representative result of immunoprecipitation followed by western immunoblotting. A lysate of RCM-1 colorectal carcinoma cells was subjected to immunoprecipitation with either a control antibody or anti-TMEM207 antibody and then underwent immunoblotting with an anti-intelectin-1 antibody. Note the intelectin-1 band in TMEM207 immunoprecipitates, which is absent in control immunoprecipitates (indicated as Mock). A similar result was obtained with SW480 cells. B: A representative result of the proximal ligation assay in situ. Fine granular positive signals, which represented overlaps between the signals from a rabbit anti-TMEM207 antibody and from murine monoclonal anti-intelectin-1 antibody. These fine granular signals were found in abundance in the cytoplasm of colon cancer cells. C: Prebinding of the anti-TMEM207 antibody with the immunization peptide strongly reduced the signals.
A: Silencing of the TMEM207 gene with small interfering RNA (siRNA) significantly downregulated not only TMEM207 but also the intelectin-1 protein level in RCM-1 cells (as measured in lysates). Immunoprecipitation with an anti-intelectin-1 antibody followed by western immunoblotting with an anti-ubiquitin antibody produced polyubiquitination bands in the presence of MG132 (proteasome inhibitor). Similar results were obtained with a different siRNA (Ambion Silencer Select siRNA, s43652 and s43653). Representative results of the experiments with s43652 siRNA and control GFP siRNA (marked as si-TMEM207 and si-Mock, respectively) are shown. B: Downregulation of TMEM207 significantly decreased secretion of intelectin-1 by RCM-1 cells. Intelectin-1 concentration in the culture supernatant of RCM-1 cells transfected with s43652 (indicated as si-TMEM207) or control GFP siRNA (indicated as si-Mock) is shown as mean and standard deviation (n = 3). Similar results were obtained with a different siRNA (s43653).
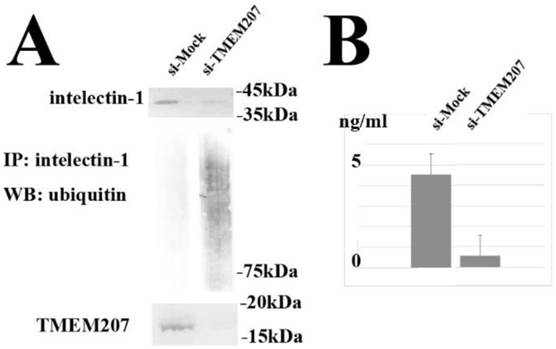
The knockdown of TMEM207 increased polyubiquitination of intelectin-1 in the colorectal carcinoma cells. Moreover, secretion of intelecint-1 seems to depend on TMEM207 in RCM-1 rectal adenocarcinoma cells, which were derived from a patient with a well-differentiated adenocarcinoma harboring mucinous carcinoma components. We believe that TMEM207 plays a role in the cytoplasmic processing of the intelectin-1 protein in colorectal carcinoma cells. TMEM207 contains the PPPY motif, which can bind to WW-domain proteins at the C terminus. Indeed, TMEM207 binds to WW domain-containing oxidoreductase, WWOX. It is possible that TMEM207 serves as a chaperone protein or adaptor for the transfer of intelectin-1 to a cytoplasmic organelle through interaction with the WW-domain containing BAG3 (stress-induced cochaperone Bcl-2-associated athanogene 3) pathways [25]. Downregulation of TMEM207 expression may increase the amount of defective intelectin-1, thus promoting ER-associated proteasomal degradation of intelectin-1 after polyubiquitination. Further research on the interaction of TMEM207 and BAG3 is in progress.
A number of mechanisms have been proposed to explain the association of obesity with increased colorectal cancer risk. Intelectin-1 is an adipokine, and in fact adipokines are downregulated during obesity. The TMEM207-intelectin-1 axis may also participate in the obesity-related colorectal carcinogenesis.
In conclusion, we characterized the hitherto uncharacterized transmembrane protein TMEM207 and showed that expression of this molecule inversely correlates with nodal metastasis and is linked to intelectin-1 processing in colorectal carcinoma. Our findings may facilitate clinical research into intelectin-1 replacement therapies and thus may benefit patients with advanced colorectal carcinoma, especially patients with lymph node metastasis.
Acknowledgements
This study was supported by grants from the Ministry of Education of Japan (KAKEN 24590384).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Clark HF, Gurney AL, Abaya E. et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265-2270
2. Wrzesiński T, Szelag M, Cieślikowski WA, Ida A, Giles R, Zodro E, Szumska J, Poźniak J, Kwias Z, Bluyssen HA, Wesoly J. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer. 2015;15:518
3. Takeuchi T, Adachi Y, Nagayama T. A WWOX-binding molecule, transmembrane protein 207, is related to the invasiveness of gastric signet-ring cell carcinoma. Carcinogenesis. 2012;3:548-554
4. Kito Y, Saigo C, Kurabayashi A, Furihata M, Takeuchi T. Transgenic mouse model of cutaneous adnexal tumors. Dis Model Mech. 2014;12:1379-1383
5. Yang YD1, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210-1215
6. Duvvuri U, Shiwarski DJ, Xiao D, Bertrand C, Huang X, Edinger RS, Rock JR, Harfe BD, Henson BJ, Kunzelmann K, Schreiber R, Seethala RS, Egloff AM, Chen X, Lui VW, Grandis JR, Gollin SM. TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 2012;72:3270-3281
7. Doolan P, Clynes M, Kennedy S, Mehta JP, Germano S, Ehrhardt C, Crown J, O'Driscoll L. TMEM25, REPS2 and Meis 1: favourable prognostic and predictive biomarkers for breast cancer. Tumour Biol. 2009;30:200-209
8. Flamant L1, Roegiers E, Pierre M, Hayez A, Sterpin C, De Backer O, Arnould T, Poumay Y, Michiels C. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 2012;12:391
9. Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H, Park CH, Rakhshan F, Schultz DA, Kolbert CP, Lupu R, Park JY, Harris CC, Yang P. Increased miR-708 expression in NSCLC and its association with poor survival in lung adenocarcinoma from never smokers. Clin Cancer Res. 2012;18:3658-67
10. Wesener DA, Wangkanont K, McBride R, Song X, Kraft MB, Hodges HL, Zarling LC, Splain RA, Smith DF, Cummings RD, Paulson JC, Forest KT, Kiessling LL. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol. 2015;22:603-610
11. Wrackmeyer U, Hansen GH, Seya T, Danielsen EM. Intelectin: a novel lipid raft-associated protein in the enterocyte brush border. Biochemistry. 2006;45:9188-9197
12. Jaikanth C1, Gurumurthy P, Cherian KM, Indhumathi T. Emergence of omentin as a pleiotropic adipocytokine. Exp Clin Endocrinol Diabetes. 2013;121:377-383
13. Zheng L, Weng M, Qi M, Qi T, Tong L, Hou X, Tong Q. Aberrant expression of intelectin-1 in gastric cancer: its relationship with clinicopathological features and prognosis. J Cancer Res Clin Oncol. 2012;138:163-172
14. Kim HJ, Kang UB, Lee H, Jung JH, Lee ST, Yu MH, Kim H, Lee C. Profiling of differentially expressed proteins in stage IV colorectal cancers with good and poor outcomes. J Proteomics. 2012;75:2983-2997
15. Li D, Mei H, Pu J, Xiang X, Zhao X, Qu H, Huang K, Zheng L, Tong Q. Intelectin-1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up-regulation of N-myc downstream regulated gene 2. Mol Cancer. 2015;14:47
16. Li D, Zhao X, Xiao Y, Mei H, Pu J, Xiang X, Jiao W, Song H, Qu H, Huang K, Zheng L, Tong Q. (2015) Intelectin-1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2012;6:16168-16182
17. Kant P, Hull MA. Excess body weight and obesity-the link with gastrointestinal and hepatobiliary cancer. Nat Rev Gastroenterol Hepatol. 2011;8:224-238
18. Washimi K, Yokose T, Yamashita M, Kageyama T, Suzuki K, Yoshihara M, Miyagi Y, Hayashi H, Tsuji S. Specific expression of human intelectin-1 in malignant pleural mesothelioma and gastrointestinal goblet cells. PLoS One. 2012;7:e39889
19. Leibovitz A, Stinson JC, McCombs WBIII, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562-4569
20. Kataoka H, Nabeshima K, Komada N, Koono M. New human colorectal carcinoma cell lines that secrete proteinase inhibitors in vitro. Virchows Arch. 1989;57:157-161
21. Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, Ohtsuki Y. Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489-1497
22. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350-4354
23. Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gústafsdóttir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473-477
24. David E. Metzler Biochemistry, 2nd Edition, The Chemical Reactions of Living Cells. 2003:76.
25. Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, Matchuk ON, Smirnova SG, Orlova NV, Zamulaeva IA, Garcia-Marcos M, Li X, Young ZT, Rauch JN, Gestwicki JE, Takayama S, Sherman MY. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731-4740
Author contact
![]() Corresponding author: Tamotsu Takeuchi, MD, PhD, Department of Pathology and Translational Research, Gifu University Graduate School of Medicine, Yanagido, Gifu 501-1193, Japan, Tel.: +81 058-230-6361, Fax: +81 058-230-6362, Email: takeutit08com
Corresponding author: Tamotsu Takeuchi, MD, PhD, Department of Pathology and Translational Research, Gifu University Graduate School of Medicine, Yanagido, Gifu 501-1193, Japan, Tel.: +81 058-230-6361, Fax: +81 058-230-6362, Email: takeutit08com

 Global reach, higher impact
Global reach, higher impact