3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(3):324-334. doi:10.7150/jca.13422 This issue Cite
Research Paper
Association of Nuclear PIM1 Expression with Lymph Node Metastasis and Poor Prognosis in Patients with Lung Adenocarcinoma and Squamous Cell Carcinoma
1. Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer;
2. Key Laboratory of Cancer Prevention and Therapy, Tianjin;
3. Department of Thoracic Oncology, Tianjin Lung Cancer Center, Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin 300060, PR China;
4. Department of Radiotherapy, the Second Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu, PR China.
Received 2015-8-2; Accepted 2015-10-17; Published 2016-1-10
Abstract
Increasing evidence indicates that aberrant expression of PIM1, p-STAT3 and c-MYC is involved in the pathogenesis of various solid tumors, but its prognostic value is still unclear in non-small cell lung cancer (NSCLC). Here, we sought to evaluate the expression and prognostic role of these markers in patients with lung adenocarcinoma (AD) and squamous cell carcinoma (SCC). Real time RT-PCR and Western blotting was used to analyze the mRNA and protein expression of PIM1 in NSCLC cell lines, respectively. The expression of PIM1, p-STAT3, and c-MYC was immunohistochemically tested in archival tumor samples from 194 lung AD and SCC patients. High nuclear PIM1 expression was detected in 43.3% of ADs and SCCs, and was significantly correlated with lymph node (LN) metastasis (P = 0.028) and histology (P = 0.003). High nuclear PIM1 expression (P = 0.034), locally advanced stage (P < 0.001), AD (P = 0.007) and poor pathologic differentiation (P = 0.002) were correlated with worse disease-free survival (DFS). High nuclear PIM1 expression (P = 0.009), advanced clinical stage (P < 0.001) and poor pathologic differentiation (P = 0.004) were independent unfavorable prognostic factors for overall survival (OS). High p-STAT3 expression was not associated with OS but significantly correlated with LN metastasis, while c-MYC was not significantly correlated with any clinicopathological parameter or survival. Therefore, in AD and SCC patients, nuclear PIM1 expression level is an independent factor for DFS and OS and it might serve as a predictive biomarker for outcome.
Keywords: PIM1, p-STAT3, c-MYC, prognostic factor, adenocarcinoma, squamous cell carcinoma.
Introduction
Although significant advances in diagnostic and therapeutic approaches have contributed to prolonged survival, NSCLC remains the leading cause of cancer-related death worldwide. Adenocarcinoma (AD) and squamous cell carcinoma (SCC) are the two major histological types of NSCLC. Surgical resection and adjuvant chemotherapy can be effective curative treatments, yet the risk of recurrence of NSCLC still cannot be predicted. Even more, NSCLC is a heterogeneous and complex of disease with alterations of multiple major signaling pathways. Most stimulatory signaling pathways are driven by oncogenes, which shift cells towards malignant transformation, dysregulated proliferation and escape from apoptosis [1]. Therefore, large efforts have explored novel molecular biomarkers in order to predict the outcome of NSCLC and potentially provide a novel targeted therapy approach.
The pim-1 proto-oncogene encodes PIM1 serine/threonine protein kinase which involved in cell survival, proliferation, apoptosis, and tumorigenesis [2]. PIM1 phosphorylated a number of substrates and its overexpression is linked to the development and progression of a wide range of haematological and epithelial tumors. PIM1 has also been shown to contribute to radiation and chemotherapy resistance [3-5]. Elevated levels of PIM1 were discovered in human myeloid and lymphoid leukemia and lymphoma tumors as well as in solid tumors [6]. Anti-PIM1-specific monoclonal antibody can inhibit tumor growth in vitro and in vivo, and synergistically enhance cytotoxic effects in combination with chemotherapy drugs [7]. Previous studies demonstrated PIM1 expression is correlated with poor prognosis in hematopoietic malignancies [8], gastric cancer [9] and squamous cell carcinoma of the head and neck [10]. Conversely, PIM1 overexpression has been correlated with a favorable prognosis in pancreatic ductal carcinoma [11] and prostate adenocarcinoma [12]. In NSCLC, Warnecke-Eberz et al.[13] observed downregulation of PIM1 mRNA and protein expression in lung cancer cells. On the contrary, in Jin et al. and Pang et al.'s studies [14, 15], PIM1 protein expression was found significantly upregulated in NSCLC tissues compared with normal lung tissues. However, the prognostic value of PIM1 in NSCLC remains unknown.
Signal transducer and activator of transcription 3 (STAT3) is a critical signaling mediator which functions as downstream effectors of cytokines and tyrosine kinases [16]. Constitutive activation of STAT3 (phosphorylated STAT3, p-STAT3) has been detected in pancreas, prostate, head and neck, breast and lung cancer [17-20]. Persistent activation of STAT3 may promote tumor angiogenesis, cell proliferation, and resistance to apoptosis. Activated STAT3 upregulates PIM1 gene expression and thus induces resistance to cytotoxic drugs in prostate cancer cells [21]. Furthermore, PIM1 kinase has been long recognized as a highly potent coactivator in MYC-dependent transformation during lymphomagenesis and prostate cancer tumorigenesis [22, 23]. PIM1 kinase is often overexpressed in the context of increased MYC levels in both hematological malignancies and solid tumors [24, 25]. By phosphorylating H3s10, PIM1 contributes to approximately 20% of the MYC-induced gene expression [26]. Overexpression of the myc-proto-oncogene is common in NSCLC, however, the prognostic relevance of c-MYC for patients with NSCLC is controversial.
PIM1, c-MYC and p-STAT3 are individually and cooperatively involved in the pathogenesis of hematological malignancies and solid tumors. However, the expression pattern and clinical significance of PIM1 as well as its interaction with p-STAT3 and c-MYC in NSCLC remain largely unknown. Therefore, we here examined the expression of these markers and further evaluated the association between the expression of these genes with prognosis in patients with lung SCC and AD.
Materials and methods
Study population
Primary formalin-fixed, paraffin-embedded (FFPE) lung tumor samples were obtained from Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin, P. R. China from January 2009 to March 2010. Patients who died within 2 months after surgery or those who died from causes unrelated to the tumor or received previous chemotherapy, radiotherapy or any other anti-tumor therapies prior to surgery were excluded. In total, 101 patients with AD and 93 patients with SCC were included in this study. Additionally, 31 FFPE NSCLC tissues and their matched adjacent normal lung tissues were also included. In the 194 lung cancer cases, there were 122 males and 72 females with ages ranging from 32 to 79 years, and a median age of 59 years. Of the 194 cases, 84 cases were at stage I, 43 were at stage II, while 67 were at stages III. The characteristics of the 194 resected NSCLCs are summarized in Table 1.
As of March 2015, a total of 101 patients had died from lung cancer; the median follow-up was 41 months (range from 2 to 72 months). To use these clinical materials for research purposes, prior consent from the patients and approval from the Research Ethics Committee of Tianjin Cancer Institute & Hospital of Tianjin Medical University were obtained. All specimens were handled and made anonymous according to the ethical and legal standards. Histological classification and clinicopathologic staging of the samples were graded according to the WHO histological classification and the tumor-node-metastasis (TNM) classification system (AJCC, 1997).
Cell culture
A549, PC-9, H1975, H460, SK-MES-1 and human bronchial epithelial (BEAS-2B) cell lines were obtained from American Type Culture Collection (Manassas, VA, U.S.A). A549, PC-9, H1975 and H460 cells were cultured in RPMI 1640 containing 10% fetal calf serum (FBS). SK-MES-1 cells were cultured in Minimum Essential Medium (MEM Eagles with Earle's Balanced Salts) supplemented with 10% FBS. BEAS-2B cells were cultured in LHC-8 Medium. Cell culture reagents were all obtained from Gibco (Carlsbad, CA, U.S.A).
Western blotting
Immunoblotting was performed as previously described [27] after separation by 8-12% polyacrylamide gel electrophoresis. Primary antibodies against PIM1 (Abcam; Cambridge, UK) and α-Tubulin (Cell Signaling Technology; Beverly, MA, U.S.A) were used to confirm protein expression. Data are representative of three independent experiments with nearly identical results.
RNA Preparation and mRNA Detection by Quantitative PCR
Total RNA was extracted from cells by TRIZOL Reagent (Invitrogen Life Technologies; Carlsbad, CA, U.S.A) and first-strand cDNA synthesis was performed with 1.5µg of total RNA using SuperScriptTM III Reverse Transcriptase. For real time quantitative PCR detection of PIM1 gene expression, SYBR-Green master mix was used on an ABI Prism 7900 platform (Applied Biosystems) as described previously.[28] To normalize for equal mRNA/cDNA amounts, PCRs with target-specific and GAPDH-specific primer sets were always run in parallel for each sample. PCR assays were conducted in triplicate for each sample, and each PCR experiment included three nontemplate control wells. The primers sequences used for PIM1 (121 bp) and GAPDH (299 bp) were 5'-TCCGCGTCTCCGACAACTT-3' (forward) and 5'-ACCTTCTTCAGCAGGACCACTT-3'(reverse), and 5'-GGGAAACTGTGGCGTGAT-3' (forward) and 5'-GAGTGGGTGTCGCTGTTGA-3' (reverse), respectively.
Immunofluorescence Staining
A549, PC-9, SK-MES-1 and BEAS-2B cells were fixed with 4% paraformaldehyde for 20 minutes, treated with 0.1% Triton X-100 and blocked with 2% BSA for 60 min, incubated with primary antibody against PIM1 at 4 °C overnight, then incubated with fluoro-conjugated secondary antibody. Nuclei were stained with DAPI and the stained cells were observed under an Olympus 60BX microscope (Hamburg, Germany). Subcellular localization and PIM1 positivity were scored by two independent researchers.
Immunohistochemical Staining and Assessment
The expression of PIM1, c-MYC, and p-STAT3 were evaluated by immunohistochemistry using ChemMate EnVision Detection Kit with DAB substrate (Dako, Carpinteria, CA, USA) following the manufacturer's protocol with modification as described previously.[18] Rabbit monoclonal antibody against PIM1 (Abcam, Cambridge, UK), p-STAT3 [Tyr705] (Cell Signaling Technology, Beverly, MA, U.S.A), or c-MYC (Epitomics, Burlingame, CA, U.S.A) was used for the staining. The slides were blindly examined by two investigators.
Nuclear staining for PIM1, p-STAT3 or c-MYC was regarded as positive signals. Staining intensity was scored as negative = 0, weak = 1, moderate = 2, or strong = 3. The mean percentage of positive tumor cells was determined in at least five areas at 400-fold magnification. The percent of positive cells was scored as ≤ 10% = 0, > 10% to ≤ 25% = 1, >25% to ≤ 50% = 2, > 50% to ≤ 75% = 3 or > 75% = 4. For statistical analysis, the two scores were then multiplied to calculate the final score. An optimal cut-off value for high and low expression was determined on the basis of a measure of heterogeneity with the log-rank test statistical analysis with respect to OS. Expression was considered low if the final score was equal to or less than four; otherwise, the expression was considered high.
Statistical Analysis
Statistical analyses were performed using the SPSS 20.0 (IBM Corporation, Armonch, NY, U.S.A). The Chi-squared test was used to examine possible correlations between gene expression and clinicopathologic factors. A multivariate model was constructed using logistic regression including the possible confounding factors in univariate analysis with P < 0.20. Survival analysis and curves were established using the Kaplan-Meier method and log-rank test was used for comparison. Multivariate analysis was performed according to the Cox proportional hazards model with backward variable elimination to evaluate any possible association between gene expression status and survival after adjusting for other potential confounders. P < 0.05 was considered to be statistically significant.
Results
PIM1, p-STAT3 and c-MYC gene expression in NSCLCs
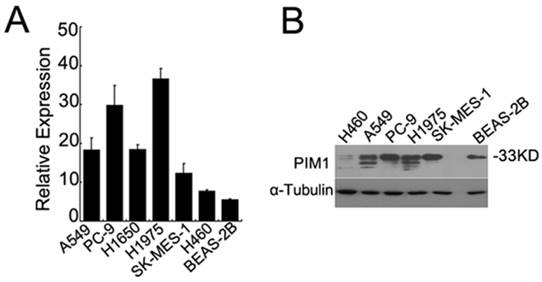
By quantitative real time RT-PCR, we found that PIM1 mRNA expression was much lower in human bronchial epithelial (BEAS-2B) cells than in lung cancer cell lines (Figure 1A). We further detected the protein expression of PIM1 by western blotting analysis. As shown in Figure 1B, high levels of PIM1 (33kDa) were detected in adenocarcinoma cells (A549, PC-9, and H1975) and squamous cell carcinoma cells (SK-MES-1), yet much lower expression was observed in the BEAS-2B cells and large cell carcinoma H460 cells.
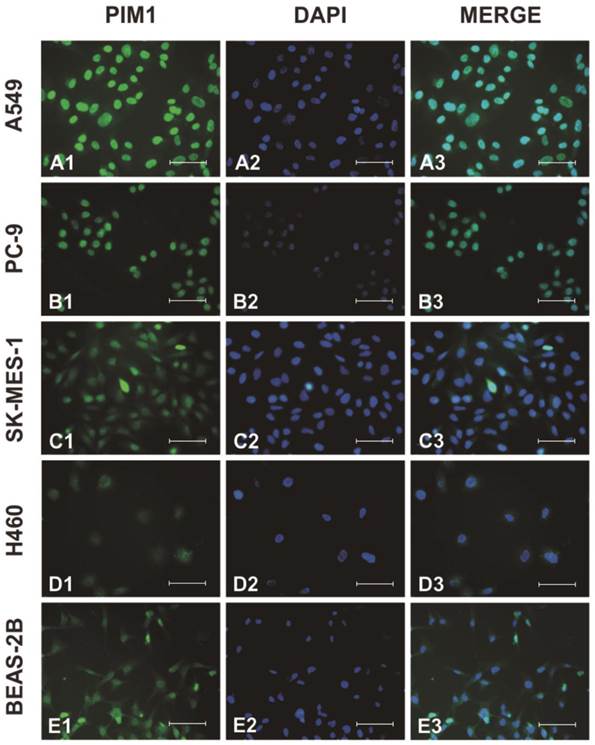
Next, we examined PIM1 localization by immunofluorescence. As shown in Figure 2, although PIM1 signals could be observed in cytosol and nucleus, intense PIM1 staining signals were found predominantly in the nucleus of the cells.
PIM1 expression in human lung cancer and bronchial epithelial cells. (A) qRT-PCR analysis of PIM1 mRNA expression in A549, PC-9, H1975, H460, SK-MES-1 and human bronchial epithelial (BEAS-2B) cells. (B) Western blotting analysis of PIM1 expression in cell lines.
Subcellular localization of PIM1 was examined by confocal microscopy in A549 (A), PC-9 (B), SK-MES-1 (C), H460 (D) and BEAS-2B (E) cells. Endogenous PIM1 (A1, B1, C1, D1, E1), nuclei stained with DAPI (A2, B2, C2, D2, E2) and merged (A3, B3, C3, D3, E3). (Bar = 50µm).
Expression of PIM1 (A), p-STAT3 (B) and c-MYC (C) in lung ADs (A1, A2, B1, B2, C1, C2) and SCCs (A3, A4, B3, B4, C3, C4). Negative staining (A1, A3, B1, B3, C1, C3). Positive expression (A2, A4, B2, B4, C2, C4) (magnification: 200×). (Bar = 100µm).
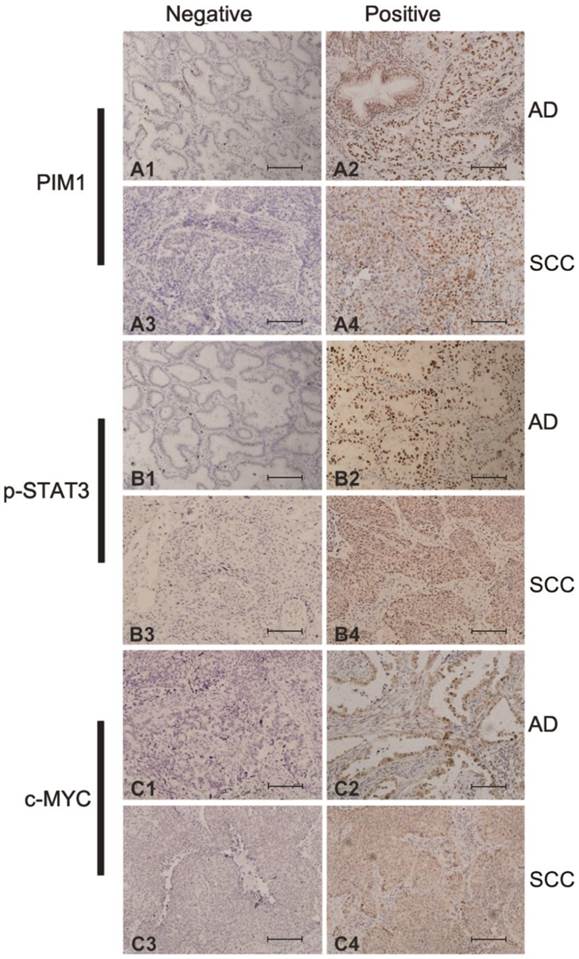
To further examine the expression of PIM1 in archival primary NSCLC, we performed IHC analysis in 194 paraffin-embedded AD and SCC as well as 31 normal lung tissues. Representative IHC results are shown in Figure 3. PIM1 protein expression exhibited a nuclear staining pattern with variable cytoplasmic staining. Based on the known correlation of PIM1 with c-MYC and STAT3, the expression of these two factors was also detected by IHC. As shown in Figure 3, p-STAT3 immunohistochemical staining was predominantly in the nucleus while c-MYC was observed in both cytoplasm and nucleus of malignant cells. In this study, only nuclear staining for PIM1, p-STAT3 and c-MYC were regarded as positive signals. High expression of p-STAT3 was detected in 95 (49.0%) of the 194 lung cancer cases but only in 9 (29.0%) of the 31 normal lung tissues (χ2 = 4.274; P = 0.039), while high c-MYC was prevalent in 101 (52.1%) of cancer cases versus 7 (22.6%) of normal samples (χ2 = 9.801; P = 0.002). Furthermore, high PIM1 expression was detected in 84 (43.3%) of the 194 lung cancers but in only 4 (12.9%) of the 31 normal samples (χ2 = 10.370; P = 0.001). Therefore, the expression ratio of PIM1, p-STAT3 and c-MYC in tumor tissues was significantly higher than that in normal lung tissues.
Correlation between PIM1, p-STAT3 and c-MYC Expression and Clinicopathologic Features of ADs and SCCs
We further analyzed the relationship between the expression of these genes and the clinical characteristics in 194 AD and SCC cases. Univariate analysis revealed a strong correlation between PIM1 expression and the clinicopathologic characteristics including gender (χ2 = 5.508, P = 0.019), tumor size (χ2 = 4.323, P = 0.038), LN metastasis (χ2 = 4.561, P = 0.033), histology (χ2 = 8.870, P = 0.003), and p-STAT3 expression status (χ2 = 3.961, P = 0.047) (Table 1). Multivariate logistic regression analysis for the association between PIM1 expression and various clinicopathologic factors showed that PIM1 expression was significantly with regional LN metastasis (OR = 1.939; 95% CI: 1.073-3.506; P = 0.028) and histologic subtype (OR = 2.480; 95% CI: 1.369-4.491; P = 0.003) (Table 2). Expression of p-STAT3 expression was also found to be significantly correlated with the regional LN metastasis (χ2 = 4.563; P = 0.033) (Supplementary Table S1). No significant correlation was found between c-MYC expression and any clinicopathologic parameter (Supplementary Table S2). Moreover, although there were no strong associations between expressions of these genes, a weak correlation between expression of PIM1 and p-STAT3 expression was detected (r = 0.147; P = 0.047). Additional subset analysis in different histologic subtypes of NSCLC indicated that high expression of PIM1 was significantly associated p-STAT3 expression in AD (r = 0.244; P = 0.015) but not in SCC (r = 0.010; P = 0.920).
Relationship between expression of PIM1, p-STAT3 and c-MYC and patient survival
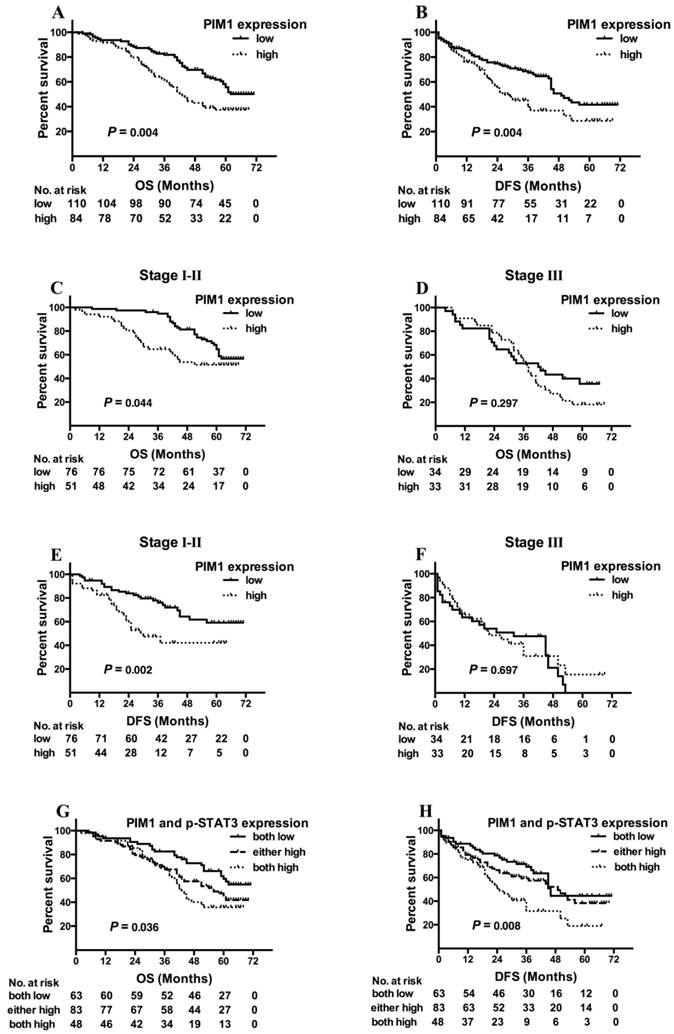
Kaplan-Meier analysis demonstrated that no association was found between p-STAT3 or c-MYC expression and survival (either DFS or OS). However, high PIM1 expression cases showed a shorter OS (Figure 4A, log-rank χ2 =8.370, P = 0.004) and DFS (Figure 4B, log-rank χ2 =8.245, P = 0.004) than those with low PIM1 levels.
Correlation between PIM1 expression and the clinicopathological characteristics.
| Variables | Number of Patients | PIM1 expression (%) | χ2 | *P-value | |
|---|---|---|---|---|---|
| Low/none | High | ||||
| Gender | |||||
| Female | 72 | 33(45.8) | 39(54.2) | 5.508 | 0.019 |
| Male | 122 | 77(63.1) | 45(36.9) | ||
| Age (years) | |||||
| < 60 | 101 | 53(52.5) | 48(47.5) | 1.532 | 0.216 |
| ≥ 60 | 93 | 57(61.3) | 36(38.7) | ||
| Smoking index | |||||
| < 400 | 95 | 55 (54.7) | 43(45.3) | 0.293 | 0.589 |
| ≥ 400 | 99 | 58(58.6) | 41(41.4) | ||
| Clinical Stage | |||||
| I | 84 | 48(57.1) | 36(42.9) | 3.206 | 0.361 |
| II | 43 | 28(65.1) | 15(34.9) | ||
| IIIa | 54 | 29(53.7) | 25(46.3) | ||
| IIIb | 13 | 5(38.5) | 8(61.5) | ||
| Tumor size | |||||
| ≤ 3cm | 92 | 45(48.9) | 47(51.1) | 4.323 | 0.038 |
| > 3cm | 102 | 65(63.7) | 37(36.3) | ||
| Regional LN metastasis | |||||
| No | 107 | 68(63.6) | 39(36.4) | 4.561 | 0.033 |
| Yes | 87 | 42(48.3) | 45(51.7) | ||
| Distant metastasis | |||||
| No | 180 | 105(58.3) | 75(41.7) | 2.707 | 0.100 |
| Yes | 14 | 5(35.7) | 9(64.3) | ||
| Histology | |||||
| AD | 101 | 47(46.5) | 54(53.5) | 8.870 | 0.003 |
| SCC | 93 | 63(67.7) | 30(32.3) | ||
| Pathologic differentiation | |||||
| Well | 64 | 31(53.1) | 33(46.9) | 2.656 | 0.103 |
| Poor | 130 | 79(60.8) | 51(39.2) | ||
| p-STAT3 expression | |||||
| Low/ none | 99 | 63(63.6) | 36(36.4) | 3.961 | 0.047 |
| High | 95 | 47(49.5) | 48(50.5) | ||
| c-MYC expression | |||||
| Low/ none | 93 | 51(54.8) | 42(45.2) | 0.252 | 0.615 |
| High | 101 | 59(58.4) | 42(41.6) | ||
| Total | 194 | 110(56.7) | 84(43.3) | ||
LN: lymph node. SCC: squamous cell carcinoma. AD: adenocarcinoma. *P values < 0.05 in bold.
Multivariate Logistic Regression Analysis for PIM1 expression.
| Variables | OR (95% CI) | P value |
|---|---|---|
| Regional LN metastasis | ||
| No | ||
| Yes | 1.939 (1.073-3.506) | 0.028 |
| Histology | ||
| AD | ||
| SCC | 2.480 (1.369-4.491) | 0.003 |
OR: odd ratio. CI: confidence interval. LN: lymph node. SCC: squamous cell carcinoma. AD: adenocarcinoma.
Kaplan-Meier analysis for outcome of NSCLC cases with different PIM1 expression levels [A, overall survival (OS) of all cases; B, disease-free survival (DFS) of all cases; C, OS of patients at stages I and II; D, OS of patients at stages III; E, DFS of patients at stages I and II; F, DFS of patients at stages III], and outcomes of cases with different PIM1 and p-STAT3 expression levels (G, OS of all cases; H, DFS of all cases).
Predictive and prognostic values of variables by univariate analysis.
| Variables | DFS | OS | ||||
|---|---|---|---|---|---|---|
| 3-year DFS rate (%) | χ2 | *P value | 5-year survival rate (%) | χ2 | *P value | |
| All patients (n=194) | ||||||
| Gender | ||||||
| Male | 58.9 | 0.412 | 0.521 | 45.7 | 1.316 | 0.251 |
| Female | 51.8 | 51.6 | ||||
| Age (years) | ||||||
| ≥ 60 | 54.1 | 2.142 | 0.143 | 41.5 | 1.995 | 0.158 |
| < 60 | 58.3 | 53.6 | ||||
| Smoking index | ||||||
| ≥ 400 | 58.7 | 0.398 | 0.528 | 45.4 | 1.147 | 0.284 |
| < 400 | 53.6 | 50.3 | ||||
| Clinical Stage | ||||||
| IIIa-IIIb | 40.2 | 22.116 | < 0.001 | 26.6 | 22.853 | < 0.001 |
| Ia-IIb | 64.7 | 59 | ||||
| Tumor size | ||||||
| > 3cm | 55 | 1.259 | 0.262 | 42.2 | 2.710 | 0.100 |
| ≤ 3cm | 57.6 | 54 | ||||
| Regional LN metastasis | ||||||
| Yes | 40 | 23.226 | < 0.001 | 31.3 | 20.101 | < 0.001 |
| No | 69.5 | 61.4 | ||||
| Histology | ||||||
| AD | 50.3 | 3.157 | 0.076 | 50.7 | 1.669 | 0.196 |
| SCC | 63.2 | 44.8 | ||||
| Pathologic differentiation | ||||||
| Poor | 50.3 | 2.318 | 0.128 | 41.5 | 5.870 | 0.015 |
| Well | 65.2 | 59.8 | ||||
| PIM1 expression | ||||||
| High | 39.7 | 8.245 | 0.004 | 37.5 | 8.370 | 0.004 |
| Low/ none | 67.4 | 55.7 | ||||
| p-STAT3 expression | ||||||
| High | 49.9 | 2.424 | 0.12 | 43.2 | 1.153 | 0.283 |
| Low/ none | 62.6 | 52.2 | ||||
| c-MYC expression | ||||||
| High | 54 | 0.033 | 0.856 | 52.4 | 0.423 | 0.515 |
| Low/ none | 57.9 | 44.8 | ||||
| PIM1 and pSTAT3 expression | ||||||
| Both high | 31.6 | 9.711 | 0.008 | 35.7 | 6.676 | 0.036 |
| Either high | 59.3 | 41.9 | ||||
| Both low/ none | 69.1 | 59.6 | ||||
| Patients in stage I-II (n=127) | ||||||
| PIM1 expression | ||||||
| High | 42.1 | 9.956 | 0.002 | 51.4 | 4.069 | 0.044 |
| Low/ none | 76 | 64.6 | ||||
| p-STAT3 expression | ||||||
| High | 60 | 1.205 | 0.272 | 53.5 | 1.349 | 0.246 |
| Low/ none | 68.5 | 64.1 | ||||
| c-MYC expression | ||||||
| High | 64 | 0.185 | 0.667 | 59.2 | 0.001 | 0.993 |
| Low/ none | 62.6 | 58.8 | ||||
| Patients in stage III (n=67) | ||||||
| PIM1 expression | ||||||
| High | 30.9 | 0.151 | 0.697 | 18.2 | 1.089 | 0.297 |
| Low/ none | 47.6 | 35.6 | ||||
| p-STAT3 expression | ||||||
| High | 34.7 | 0.04 | 0.841 | 26.2 | 0.114 | 0.736 |
| Low/ none | 48.8 | 26.9 | ||||
| c-MYC expression | ||||||
| High | 36.4 | 0.127 | 0.721 | 16.9 | 1.111 | 0.292 |
| Low/ none | 41.9 | 34.4 | ||||
DFS: disease-free survival. OS: overall survival. SCC: squamous cell carcinoma. AD: adenocarcinoma. *P values < 0.05 in bold.
When stratifying the clinical stages into early stage (stages I and II) and locally advanced stage (stages III), we found that the association between high PIM1 expression and shorter OS were stronger in patients with early stage cancers (Figure 4C, log-rank χ2 = 4.069, P = 0.044) than with stage III tumors (Figure 4D, log-rank χ2 = 1.089, P = 0.297). Similarly to DFS, we also observed that PIM1 expression could predict recurrence in early stage (Figure 4E, log-rank χ2 = 9.956, P = 0.002), but not in stage III (Figure 4F, log-rank χ2 = 0.151, P = 0.697).
Further analysis showed that the overall survival in patients with both low-PIM1 and low-p-STAT3 tumors was the longest among all of these groups (Figure 4G, log-rank χ2 = 6.676; P = 0.036), and these patients tended to have the longest DFS (Figure 4H, log-rank χ2 = 9.711; P = 0.008). However, the predictive value of PIM1 was not significantly improved by combining with p-STAT3 expression status.
By univariate analysis, we found that DFS significantly associated with clinical stage (P < 0.001), LN metastasis (P < 0.001) and PIM1 expression (P = 0.004). OS significantly associated with clinical stage (P < 0.001), LN metastasis (P < 0.001), pathologic differentiation (P = 0.015) as well as PIM1 expression (P = 0.004) (Table 3). In Multivariate Cox Regression analysis, high PIM1 expression (HR = 1.574; P = 0.034), locally advanced clinical stage (HR = 2.341; P < 0.001), AD (HR = 1.890; P = 0.007) and poor pathologic differentiation (HR = 2.192; P = 0.002) correlated with shorter DFS (Table 4). In regards to OS, high PIM1 expression (HR = 1.700; P = 0.009), together with locally advanced clinical stage (HR = 2.386; P < 0.001) and pathologic differentiation (HR = 1.927; P = 0.004), were significantly and independently unfavorable prognostic factors (Table 4).
Predictive and prognostic values of variables by multivariate analysis.
| DFS | OS | ||||
|---|---|---|---|---|---|
| Variables | HR (95% CI) | P value | HR (95% CI) | P value | |
| PIM expression | |||||
| High vs. low/ none | 1.574 (1.034-2.396) | 0.034 | 1.700 (1.139-2.536) | 0.009 | |
| Clinical stage | |||||
| III vs. I/II | 2.341 (1.560-3.514) | < 0.001 | 2.386 (1.600-3.558) | < 0.001 | |
| Histology | |||||
| AD vs. SCC | 1.890 (1.192-2.994) | 0.007 | |||
| Pathologic differentiation | |||||
| Poor vs. Well | 2.192 (1.345-3.574) | 0.002 | 1.927 (1.226-3.028) | 0.004 | |
HR: hazard ratio. CI: confidence interval. SCC: squamous cell carcinoma. AD: adenocarcinoma.
Discussion
The prognostic significance of PIM1, c-MYC and p-STAT3 in NSCLC remains uncertain. The aim of this study was to elucidate the role of these markers as predictive and prognostic markers of survival in AD and SCC patients.
Constitutively activated STAT3 (p-STAT3) is frequently detected in a variety of human malignant tumors including NSCLC. Our current study showed that high expression of p-STAT3 was detected in 49.0% of lung cancer cases compared to in 29.0% of normal lung tissue samples. In addition, here we found slight differences in p-STAT3 expression between our study and others could be attributed to the patient ethnicity, antibodies used, or criteria chosen to categorize positive expression [18, 29-32]. Moreover, we observed that p-STAT3 expression was correlated with LN metastasis in our study, consistent with the finding of Wang et al. that suggested that activation of STAT3 might play an important role in LN metastasis in NSCLC [33]. However, we found no association between p-STAT3 expression and survival in concordance with several previous studies [30, 32].
Previous studies have demonstrated that overexpression of c-MYC is common in NSCLC [34, 35] but its prognostic value in NSCLC is not clear. Our study demonstrated that high expression of c-MYC was detected in 52.1% of tumors, but no association was found between c-MYC expression and clinicopathologic parameters or survival. This suggests that c-MYC might be involved in the pathogenesis of NSCLC but alone might not critically contribute to the progression of NSCLC.
In NSCLC, the prognostic value of PIM1 is largely unknown. Warnecke-Eberz et al. demonstrated that PIM1 mRNA expression in NSCLC was significantly downregulated compared to paired normal tissues, and the down-regulation was closely associated with LN metastasis [13]. Our detection of PIM1 mRNA expression in lung cancer tissues was consistent with their results (data not shown), although we found no association between PIM1 mRNA expression and clinicopathologic parameters. We further used the TCGA database to investigate the association of PIM1 mRNA levels with survival in lung AD and SCC patients. Consistent with our current findings, no association was found between PIM1 mRNA expression and survival (Supplementary Figure S1). We did however observe that PIM1 mRNA levels in Human bronchial epithelial (BEAS-2B) cells were much lower than that in lung cancer cell lines. We cannot exclude the possibility that normal lung tissues or adjacent non-cancerous lung tissues are composed of more mesenchymal cells which might express higher PIM1 mRNA.
It is noteworthy that PIM1 mRNA transcripts have a short half-life because of the presence of multiple copies of the destabilizing Auuu(A) sequence in the 3′untranslated region [2]. Moreover, PIM1 gene encodes two isoforms with molecular weight 33 and 44 kDa by utilizing two alternative translation initiation sites [36]. Both 33 and 44 kDa PIM1 were generated from the single reading frame, therefore, real-time quantitative RT-PCR cannot discriminate the variants. Even more, PIM1 expression in lung cancer might be regulated at the level of transcription, translation, and proteosomal degradation [37]. Downregulation of micro-RNA-1 (miR-1), miR-486-5p and/or the serine/threonine protein phosphatase 2A (PP2A) might contribute to the overexpression of PIM1 protein in lung cancer [15, 38, 39]. All together, since the mRNA level may not faithfully represent the expression of the protein, it is important to define the expression of PIM1 at the protein level in tumors.
Previous studies have shown that different isoforms of PIM1 might distribute to different subcellular localizations and exhibit different functions in tumorigenesis [3, 40, 41], and 33 kDa PIM1 is located uniformly in the nucleus with some staining in the cytoplasm [42, 43]. Kim et al. found that exposure to radiation led to overexpression and translocation of PIM1 into the nucleus in NSCLC cells [4]. Ionov et al. found that PIM1 can be located in the cytoplasm or the nucleus of cells within normal lymph nodes, but is only located in the nucleus of Burkitt's lymphoma cells. And the nuclear location of PIM1 was deemed essential for its biological activities [44]. Therefore, nuclear staining of the 33 kDa PIM1 was chosen as criteria for positive expression in our current study.
In this study we observed that the high nuclear expression of PIM1 was significantly correlated to LN metastasis, histology, worse DFS, OS and 5-year overall survival rate. Moreover, a stronger correlation between high nuclear PIM1 expression and short survivals was found at early stages, suggesting that PIM1 might be involved in the NSCLC recurrence, progression and metastasis, and nuclear PIM1 expression is an independent predictive and prognostic factor for NSCLC patients.
Warnecke-Eberz et al.[13] observed lower PIM1 expression in lung cancer cells compared to normal respiratory bronchioles, artery walls and macrophages. However, in Jin et al. and Pang et al.'s studies [14, 15], higher PIM1 protein expression was detected in the majority of NSCLCs compared to normal lung tissues. Jin et al. found that PIM1 overexpression correlates with poor differentiation, advanced clinical stage (III/IV) and lymph node and distant metastasis. Pang et al. found that PIM1 expression in NSCLC was significantly associated with tumor size, LN metastasis, histological type and clinical staging. Our result also showed that high PIM1 expression was significantly correlated to LN metastasis and histology. However, we found that a strong correlation between high PIM1 expression and short survival existed in early stage tumors rather than advanced cancers. Furthermore, for the first time, we found high expression of PIM1 was associated with higher risk of disease recurrence and shorter OS. It is noteworthy that PIM1 expression was observed primarily in the cytoplasm in Jin et al.'s study [14] while subcellular localization was not mentioned in Warnecke‑Eberz et al.'s research. However, in our study, nuclear PIM1 expression was detected by both immunofluorescence and immunohistology. We confirmed that 33 kDa PIM1 was the predominant variant detected in lung cancer cell lines by western blot. Thus the differences between our results and previous works may be related to the different antibodies used and different proteins detected. Our results are consistent with the study that the gain of chromosome 6p21 in NSCLC is associated with poor survival, as PIM1 is localized on chromosome 6p21.2 [45].
In conclusion, we observed that nuclear overexpression of PIM1 correlated with LN metastasis, histology and poor survival in lung AD and SCC. Our study strongly indicates that nuclear PIM1 expression level is an independent predictive and prognostic indicator of survival in patients with NSCLC. Future investigations are required to develop new treatment strategies for patients with PIM1-overexpression NSCLC.
Supplementary Material
Supplementary Tables S1-S2 and Figure S1.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81372517 and No. 81000899 for Richeng Jiang), the Tianjin Municipal Science and Technology Commission Key Application Research Projects (No. 11JCZDJC18900) for Richeng Jiang, and the Tianjin Municipal Science and Technology Commission Projects (No. 11JCYBJC11300 and No. 12ZCDZSY15600) for Kai Li. We are grateful to Dr. Douglas E. Linn (Brigham & Women's Hospital, Boston, MA, USA) for critical reading of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Brambilla E, Gazdar A. Pathogenesis of lung cancer signalling pathways: roadmap for therapies. The European respiratory journal. 2009;33:1485-97
2. Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nature reviews Cancer. 2011;11:23-34
3. Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H. et al. The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. The Journal of biological chemistry. 2008;283:3349-56
4. Kim W, Youn H, Kwon T, Kang J, Kim E, Son B. et al. PIM1 kinase inhibitors induce radiosensitization in non-small cell lung cancer cells. Pharmacological research: the official journal of the Italian Pharmacological Society. 2013;70:90-101
5. Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R. et al. Pim-1 plays a pivotal role in hypoxia-induced chemoresistance. Oncogene. 2009;28:2581-92
6. Blanco-Aparicio C, Carnero A. Pim kinases in cancer: diagnostic, prognostic and treatment opportunities. Biochemical pharmacology. 2013;85:629-43
7. Hu XF, Li J, Vandervalk S, Wang Z, Magnuson NS, Xing PX. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. The Journal of clinical investigation. 2009;119:362-75
8. Hsi ED, Jung SH, Lai R, Johnson JL, Cook JR, Jones D. et al. Ki67 and PIM1 expression predict outcome in mantle cell lymphoma treated with high dose therapy, stem cell transplantation and rituximab: a Cancer and Leukemia Group B 59909 correlative science study. Leukemia & lymphoma. 2008;49:2081-90
9. Warnecke-Eberz U, Bollschweiler E, Drebber U, Metzger R, Baldus SE, Holscher AH. et al. Prognostic impact of protein overexpression of the proto-oncogene PIM-1 in gastric cancer. Anticancer research. 2009;29:4451-5
10. Peltola K, Hollmen M, Maula SM, Rainio E, Ristamaki R, Luukkaa M. et al. Pim-1 kinase expression predicts radiation response in squamocellular carcinoma of head and neck and is under the control of epidermal growth factor receptor. Neoplasia. 2009;11:629-36
11. Reiser-Erkan C, Erkan M, Pan Z, Bekasi S, Giese NA, Streit S. et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer biology & therapy. 2008;7:1352-9
12. Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K. et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822-6
13. Warnecke-Eberz U, Bollschweiler E, Drebber U, Pohl A, Baldus SE, Hoelscher AH. et al. Frequent down-regulation of pim-1 mRNA expression in non-small cell lung cancer is associated with lymph node metastases. Oncology reports. 2008;20:619-24
14. Jin Y, Tong DY, Chen JN, Feng ZY, Yang JY, Shao CK. et al. Overexpression of osteopontin, alphavbeta3 and Pim-1 associated with prognostically important clinicopathologic variables in non-small cell lung cancer. PloS one. 2012;7:e48575
15. Pang W, Tian X, Bai F, Han R, Wang J, Shen H. et al. Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol Cancer. 2014;13:240
16. Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. 2003;22:4150-65
17. Scholz A, Heinze S, Detjen KM, Peters M, Welzel M, Hauff P. et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125:891-905
18. Jiang R, Jin Z, Liu Z, Sun L, Wang L, Li K. Correlation of activated STAT3 expression with clinicopathologic features in lung adenocarcinoma and squamous cell carcinoma. Molecular diagnosis & therapy. 2011;15:347-52
19. Berclaz G, Altermatt HJ, Rohrbach V, Siragusa A, Dreher E, Smith PD. EGFR dependent expression of STAT3 (but not STAT1) in breast cancer. International journal of oncology. 2001;19:1155-60
20. Song JI, Grandis JR. STAT signaling in head and neck cancer. Oncogene. 2000;19:2489-95
21. Zemskova M, Sahakian E, Bashkirova S, Lilly M. The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. The Journal of biological chemistry. 2008;283:20635-44
22. Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Molecular and cellular biology. 1991;11:1176-9
23. Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R. et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer cell. 2003;4:223-38
24. Narlik-Grassow M, Blanco-Aparicio C, Carnero A. The PIM family of serine/threonine kinases in cancer. Medicinal research reviews. 2014;34:136-59
25. Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML. et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477-87
26. Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nature cell biology. 2007;9:932-44
27. Jiang R, Cabras G, Sheng W, Zeng Y, Ooka T. Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia. 2009;11:964-73
28. Jiang RC, Qin HD, Zeng MS, Huang W, Feng BJ, Zhang F. et al. A functional variant in the transcriptional regulatory region of gene LOC344967 cosegregates with disease phenotype in familial nasopharyngeal carcinoma. Cancer research. 2006;66:693-700
29. van Cruijsen H, Ruiz MG, van der Valk P, de Gruijl TD, Giaccone G. Tissue micro array analysis of ganglioside N-glycolyl GM3 expression and signal transducer and activator of transcription (STAT)-3 activation in relation to dendritic cell infiltration and microvessel density in non-small cell lung cancer. BMC cancer. 2009;9:180
30. Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor-Stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:8288-94
31. Mukohara T, Kudoh S, Yamauchi S, Kimura T, Yoshimura N, Kanazawa H. et al. Expression of epidermal growth factor receptor (EGFR) and downstream-activated peptides in surgically excised non-small-cell lung cancer (NSCLC). Lung cancer. 2003;41:123-30
32. Cortas T, Eisenberg R, Fu P, Kern J, Patrick L, Dowlati A. Activation state EGFR and STAT-3 as prognostic markers in resected non-small cell lung cancer. Lung cancer. 2007;55:349-55
33. Wang M, Chen GY, Song HT, Hong X, Yang ZY, Sui GJ. Significance of CXCR4, phosphorylated STAT3 and VEGF-A expression in resected non-small cell lung cancer. Experimental and therapeutic medicine. 2011;2:517-22
34. Volm M, Koomagi R. Prognostic relevance of c-Myc and caspase-3 for patients with non-small cell lung cancer. Oncology reports. 2000;7:95-8
35. Yoo J, Kim CH, Song SH, Shim BY, Jeong YJ, Ahn MI. et al. Expression of caspase-3 and c-myc in non-small cell lung cancer. Cancer research and treatment: official journal of Korean Cancer Association. 2004;36:303-7
36. Saris CJ, Domen J, Berns A. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. The EMBO journal. 1991;10:655-64
37. Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. The Journal of clinical investigation. 2005;115:2618-24
38. Ma J, Arnold HK, Lilly MB, Sears RC, Kraft AS. Negative regulation of Pim-1 protein kinase levels by the B56beta subunit of PP2A. Oncogene. 2007;26:5145-53
39. Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S. et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. The Journal of biological chemistry. 2008;283:33394-405
40. Xie Y, Xu K, Dai B, Guo Z, Jiang T, Chen H. et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2006;25:70-8
41. Linn DE, Yang X, Xie Y, Alfano A, Deshmukh D, Wang X. et al. Differential regulation of androgen receptor by PIM-1 kinases via phosphorylation-dependent recruitment of distinct ubiquitin E3 ligases. The Journal of biological chemistry. 2012;287:22959-68
42. Ishibashi Y, Maita H, Yano M, Koike N, Tamai K, Ariga H. et al. Pim-1 translocates sorting nexin 6/TRAF4-associated factor 2 from cytoplasm to nucleus. FEBS letters. 2001;506:33-8
43. Wang Z, Bhattacharya N, Mixter PF, Wei W, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochimica et biophysica acta. 2002;1593:45-55
44. Ionov Y, Le X, Tunquist BJ, Sweetenham J, Sachs T, Ryder J. et al. Pim-1 protein kinase is nuclear in Burkitt's lymphoma: nuclear localization is necessary for its biologic effects. Anticancer research. 2003;23:167-78
45. Kim TM, Yim SH, Lee JS, Kwon MS, Ryu JW, Kang HM. et al. Genome-wide screening of genomic alterations and their clinicopathologic implications in non-small cell lung cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:8235-42
Author contact
![]() Corresponding author: Richeng Jiang, Department of Thoracic Oncology, Tianjin Lung Cancer Center, Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin 300060, China. E-mail: jiangrichengcom; and Kai Li, Department of Thoracic Oncology, Tianjin Lung Cancer Center, Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin 300060, China. E-mail: likai5com.cn.
Corresponding author: Richeng Jiang, Department of Thoracic Oncology, Tianjin Lung Cancer Center, Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin 300060, China. E-mail: jiangrichengcom; and Kai Li, Department of Thoracic Oncology, Tianjin Lung Cancer Center, Tianjin Cancer Institute & Hospital, Tianjin Medical University, Tianjin 300060, China. E-mail: likai5com.cn.

 Global reach, higher impact
Global reach, higher impact