Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(5):576-586. doi:10.7150/jca.13117 This issue Cite
Research Paper
High Expression of the Newly Found Long Noncoding RNA Z38 Promotes Cell Proliferation and Oncogenic Activity in Breast Cancer
1. College of Biology, Hunan University, Changsha, Hunan 410082, China;
2. Department of Biological Engineering, Zunyi Medical College, Zhuhai Campus, Zhuhai, Guangdong 519041, China;
3. College of Life Science, Hunan Normal University, Changsha, Hunan 410000, China;
4. Faculty of Basic Medical Sciences, Medical College, Hunan Normal University, Changsha, Hunan 410013, China.
Received 2015-7-2; Accepted 2016-1-24; Published 2016-3-18
Abstract
The aberrant expression of long noncoding RNAs (lncRNAs) has great impacts on cancer origination and progression. In the current study, a newly found lncRNA Z38, which was identified through combining experiments of suppression subtractive hybridization (SSH) and reverse dot-blotting, was found to have high expression in breast cancer. More importantly, inhibiting Z38 expression by gene silencing greatly suppressed breast cancer cell proliferation and tumorigenesis, and treatment with Z38 siRNAs significantly induced cell apoptosis and inhibited tumor growth. In conclusion, the newly found lncRNA Z38, which plays important roles in breast cancer, may act as a candidate biomarker and therapeutic target in carcinomas.
Keywords: lncRNA Z38, CLDND1, cell proliferation, tumorigenesis, oncogene, siRNA.
Introduction
As one of the most common and deadly diseases in females [1], breast cancer has all the highlighted hallmarks of carcinomas [2, 3], which is greatly influenced by environment [4, 5], food [6], and lactation [7]. Although the mortality rate has declined with the development of medical science, the incidence and mortality of breast cancer are very high at present, especially in many developed countries [8]. Recently, it was reported that the earliest transformed breast cancer cell can diffuse into other organs, thus constituting new challenges for breast cancer therapy [9].
Long noncoding RNAs (lncRNAs), a sequence of non-protein-coding transcripts greater than 200 bases in length [10-13], are enormously involved in physiological and pathological processes including tumor development by acting as decoys [14], scaffolds [15], guides [16], and enhancers [17] in genome regulation. Until now, numerous approaches have been used in the discovery and functional study of lncRNAs, such as functional genomic approaches [18], ChIP-seq analyses [19], and so on. Some research work discovers that lncRNAs can perform regulatory roles by forming ribonucleoprotein (RNP) complexes with numerous chromatin regulators and proteins [10, 20, 21]. For example, the lncRNA NKILA can repress the NF-κB signaling and cancer-related inflammation by forming a stable complex with NF-κB:IκB [22]. However, the significant influences and accurate molecular mechanisms of most lncRNAs on gene regulation have yet to be defined.
The widely studied results about the aberrant expression of lncRNAs in cancers showed that the lncRNA CCAT1-L can act as an oncogene and regulate the long-range chromatin interactions at the MYC locus in human colorectal cancer [23, 24]. In contrast, the lncRNA PCAT29 inhibits the initiation of prostate cancer by acting as an androgen-regulated tumor suppressor [25]. More importantly, some lncRNAs are developed as the potential prognostic biomarkers and therapeutic targets of cancer due to their specific expression pattern in carcinomas [26, 27]. Thus, the study about lncRNAs will certainly bring invaluable benefits to the prevention and treatment of human carcinomas. However, the detailed molecular mechanisms of most lncRNAs in cancer need to be further studied.
Claudins are a family of proteins with at least 26 members and characterized by a common motif (GLWCC; PROSITE ID: PS01346) in a paracellular loop [28-30]. Some major members of this family have been identified as hallmarks of human carcinomas [31-33]. The predicted protein, claudin domain containing 1 (CLDND1), is regarded as claudin-25 of the claudin family [29, 34]. However, our newly found lncRNA Z38 was identified as a protein coding sequence (isoform a) of the CLDND1 mRNAs, which highly expressed in breast cancer. A series of in vitro and in vivo experiments approved the facilitated roles of Z38 on breast cancer cell proliferation and tumorigenesis, which showed the potency for the early diagnosis of carcinomas. Furthermore, the specific siRNAs greatly inhibited Z38 expression in MDA-MB-231 cells and tumor growth in vivo, thus providing therapeutic potential for cancer therapy.
Materials and Methods
Plasmid construction and preparation
The predicted protein coding sequence (isoform a) of CLDND1 was amplified with cDNA that synthesized from total RNA of MCF-7 cells. Recombinant plasmids pEGFP-C1-Z38, pEGFP-N3-Z38, pCMV-Tag2B-Z38, and PBSIIKS-Z38 were constructed through inserting the 762 nucleotides length double-stranded DNA (dsDNA) into the control plasmids pEGFP-C1 (endonucleases: 5'BglII 3'KpnI) (Clontech, CA, USA), pEGFP-N3 (endonucleases: 5'BglII 3'KpnI) (Clontech), pCMV-Tag2B (endonucleases: 5'BamH1 3'XhoI) (Stratagene, CA, USA), and PBSIIKS (endonucleases: 5'XhoI 3'KpnI) (Promega, WI, USA). Plasmids pCMV-Tag2B-Z38b and pCMV-Tag2B-Z38d were constructed in the same way. The terminator codon (TGA) between the sequences of Z38 and EGFP in plasmids pEGFP-C1-Z38(S) and pEGFP-N3-Z38(S) was used to terminate RNA translation. The simulated structures of these plasmids were presented in Figure S1, and the letter 'S' in plasmids represents the insertion of a terminator codon. Plasmid pGEM-Z38-211 was constructed to synthesize the RNA probes of Z38 using the pGEM®-T Vector System I (Promega), and the primers of PCR fragments were as follows: forward primer: AGT GGG ATT GTG GAG ACG GTGT; reverse primer: AGG TAA AAG GAA CTG GCA ACGC. Plasmids shZ38-146 and shZ38-227, that were constructed using pSilencer® 2.1-U6 neo (Life Technologies, CA, USA), were used to inhibit Z38 expression. The inhibitory targets of plasmids were as follows: shZ38-146: GCA TCT GGG ATG AAT TCAT; shZ38-227: GAT TGT GGA GAC GGT GTAT. The negative control plasmid shCtrl was provided in the kit and verified by manufacturer. All recombinant plasmids were transformed and amplified using E.coli DH5α Competent Cells (Takara, Tokyo, Japan) according to the manufacturer's instructions, and sequenced at Sangon Biotech (Shanghai, China). Plasmid isolation was performed using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) according to the handbook. The quality of plasmids was assessed by 1% agarose gel electrophoresis, and the concentration of plasmids was determined using BiophotoMeter Plus Nucleic Acids Detector (Eppendorf, Hamburg, Germany).
Cell culture
Human breast epithelial cell line HBL-100 and cancer cell lines MDA-MB-231, MCF-7, HBL-100, BT20, MDA-MB-453, T47D, A375, Siha, Hela, PL45, SW480, U251, A549, NCI-H358, and Hep G2 were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Human gastric cancer cell lines BGC823 and HGC27 were purchased from ATCC (VA, USA). Cells were cultured in the DMEM or RPMI1640 (Life Technologies) supplemented with 10% (v/v) fetal calf serum (BIOCHROM AG, Berlin, Germany) and Penicillin Streptomycin (Life Technologies). Lipofectamine® 2000 (Life Technologies) was used for cell transfection according to the manufacturer's instructions.
Preparation of RNA and cDNA
Cancer tissues (3 samples each) for RNA extraction were obtained immediately after surgery and maintained in the liquid nitrogen. All samples were identified by two pathologists of Hunan Cancer Hospital, the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University (Changsha, China). TRIzol reagent (Life Technologies) was used to extract total RNA in cells and tissues according to the manufacturer's instructions. The RNA quality was assessed by 1% agarose gel electrophoresis based on the relative abundance of 5S, 18S, and 28S bands. The RNA concentration was determined using NanoDrop 2000 (Thermo Fisher Scientific, MA, USA). Total RNA was pretreated with the RQ1 RNase-Free DNase (Promega) and used to synthesize cDNA using the High-Capacity cDNA Reverse Transcription Kit (Life Technologies) according to manufacturers' instructions. cDNA was stored at 4℃ for a short time or -20℃ for a long time.
Suppression subtractive hybridization (SSH) and reverse dot-blotting
The detailed methods of SSH and reverse dot-blotting were described in one of our published articles [35]. In brief, total RNA and poly (A) tract mRNA was isolated from mammary tumor specimens of MMTV-neu mice and normal mammary tissues of FVB mice. The SSH library was constructed and the subtraction hybridization was carried out using cDNA subtraction kit (Clontech) according to the manufacturer's instructions. The resultant cDNA fragments were purified using the QIAquick PCR purification kit (Qiagen) prior to cloning into the pGEM-T easy vector (Promega). Plasmid DNA of each positive clone was purified. PCR was then carried out to amplify plasmid DNA inserts using nester primers. Next, 5 μL of each PCR product was denatured with equal volumes of 0.6 M NaOH, spotted onto the Hybond N+ membranes (Merck Millipore, MA, USA), and then ultraviolet cross-linked. Subsequently, cDNA probes were prepared using the DIG high prime DNA labeling and detection starter kit II (Roche, Basel, Switzerland) according to the manufacturer's protocol. Immunological detection was carried out using the Anti-Digoxigenin-AP Fab fragments (Roche) and a combination of 5-bromo-4-chloro-3-indoyl phosphate and nitroblue tetrazolium according to the manufacturer's instructions. All the plasmids of the positive clone, that assessed by reverse dot-blotting with an inserted fragment, were sequenced by Sangon Biotech. The DNA sequence was compared with the GenBank database using advanced BLAST.
MTT assay
500 μL of culture medium containing 10,000 cells was added to each well of the 24-well cell culture plate. After incubation for 1, 3, 5, and 7 days, 55.6 µL of 5 mg/mL MTT solution (Sangon Biotech) was added to each well and cultured for 6 hours. Then culture medium was carefully discarded and rinsed in 1×PBS (Life Technologies), and 150 μL of DMSO (Sangon Biotech) was added to each well. After gentle shaking for 20 minutes, the OD value at 490 nm was measured using a micro pore plate detector EnSpire 2300 (Perking Elmer, MA, USA).
In vitro translation assay
Plasmid PBSIIKS-Z38 was constructed for the in vitro translation assays. In vitro translation assays were conducted using the TNT® T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions. The protein products were analyzed by SDS-PAGE, and the transcripts level of RNA was determined through semi-quantitative PCR.
Colony formation assay
Cells were dispersed to 1×106/mL in 2×DMEM (DMEM supplemented with 20% (v/v) FBS, penicillin-streptomycin). 1.2% and 0.7% of Low Melting Point Agarose (Hydragene, NJ, USA) was sterilized and maintained in water at 40℃. 0.7 mL 1:1 ratio mixture of 1.2% agarose solution and 2×DMEM was added to each well of the 12-well cell culture plate. Immediately after condensation, 1.2 mL 1:1 ratio mixture of 0.7% agarose solution and 2×DMEM supplemented with 0.2 mL cell suspension was added to each well. After condensation of the mixture again, cell culture plate was transferred into a cell incubator New Brunswick (Eppendorf) and cultured for 2 weeks. The colony number was counted every 9 random horizons, and every experiment was repeated twice.
siRNA experiments and intratumoral injection
The sequences of two specific siRNAs of Z38 were as follows: siRNA-1 (GGG CUC AGU AAG UUG UUA UUU), siRNA-2 (GGA AAG AGU ACA CCU UAA UUU), and a negative control (NC) was also provided and verified by the manufacturer (GeneCopoeia, Guangzhou, China). After transfection with the siRNAs, colony formation and PCR experiments were performed to verify the interference effect.
The protocol of intratumoral injection was detailed in the reference [36, 37]. In brief, two weeks after subcutaneous injection of 1×107 MDA-MB-231 cells into the BALB/C female nude mice, these mice were arbitrarily assigned into two groups (N = 8 each). 100 μL of double-stranded siRNA (85 nM) in normal saline was administrated into mice by intratumoral injection every 3 days. On the day 20, the mice were killed and their tumors were collected for analysis.
TUNEL assay
After transfecting siRNAs into MDA-MB-231 cells, TUNEL assays were conducted with In Situ Cell Death Detection Kit POD (Roche) according to the manufacturer's instructions. Images were captured using a fluorescence microscope Ti-S (Nikon, Tokyo, Japan).
Tissue H&E staining
All tissue slices for H&E staining were verified by two pathologists of the Second Xiangya Hospital, Central South University (Changsha, China). 50 pairs of breast cancer and paraneoplastic tissues were collected for analysis, and all samples were identified as the invasive breast cancer. Immediately after surgery, fresh tissue samples were collected and stored in the liquid nitrogen, then fixed in 4% paraformaldehyde and embedded in paraffin. Tissues were cut, dewaxed, hydrated, and routinely stained with hematoxylin and eosin. Digital images were captured using an upright microscope BX51WI (Olympus, Tokyo, Japan).
In situ hybridization
All tissue slices for in situ hybridization were identified by two pathologists of the Second Xiangya Hospital, Central South University (Changsha, China). 50 pairs of breast cancer and paraneoplastic tissues were collected for analysis, and all samples were identified as the invasive carcinomas. Tissues were routinely embedded in paraffin, cut into slices, and dewaxed. After incubation in 10 µg/mL proteinase K (Merck Millipore) for a few minutes, slices were pre-hybridized for 4 hours at 65℃. Plasmid pGEM-Z38-211 was used to synthesize the biotin labeled RNA probes of Z38, and the sequence of probes was confirmed with the advanced BLAST on NCBI. The RNA probes were synthesized using Dig-labeling UTP RNA mixes (Roche) and purified using E.Z.N.A.TM MicroElute RNA Clean-up Kit (OMEGA Bio-tek, GA, USA). Slices were hybridized in 1 ng/μL RNA probe solution for 12 hours at 65℃. Then RNA probe solution was removed, and the slices were rinsed in SSCT buffer. After incubation in PBST solution supplemented with 2% goat serum (Life Technologies) and 2 mg/mL BSA (Sigma-Aldrich), slices were incubated in 1:5000 Anti-Digoxigenin-AP Fab fragments (Roche) for 12 hours at 4℃. Then slices were rinsed in TMNT buffer and stained using BCIP staining solution (Sangon Biotech) in dark. Digital images were photographed using an upright microscope BX51WI (Olympus).
Regular PCR and quantitative real-time PCR (qRT-PCR)
Regular PCR was performed using the TaKaRa TaqTM (Takara) and KOD FX (TOYOBO, Osaka, Japan) according to the manufacturers' instruction on the PCR instrument Matercycler (Eppendorf). Expression level of target genes was determined using the SYBR® Premix Ex TaqTM kit (Takara). All gene primers were synthesized at Sangon Biotech, and the primer sequences of genes were listed in Table S1 and Table S2. Results were normalized to β-actin and GAPDH, respectively.
Western blotting
Cells in 6-well cell culture plate were lysed using 150 μL RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitors. Then cell lysates were collected and centrifuged at 10,000 rcf and 4℃ for 15 minutes. The protein concentrations were measured using the Modified BCA Protein Assay Kit (Sangon Biotech). After mixing and boiling with the SDS-PAGE loading buffer, 40 μg of total protein was electrophoresed in 10% SDS-PAGE gels and transferred on to a PVDF membrane (Merck Millipore). Then the PVDF membrane was blocked in PBST buffer supplemented with 5% skim milk and 0.1% Tween 20 for 1 hour at room temperature and incubated with the primary antibodies overnight at 4℃. After incubation with the consistent HRP-conjugated secondary antibodies, the protein bands were detected using SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher Scientific). The following antibodies were used according to the manufacturers' instructions: rabbit anti-CLDND1 (Abcam, MA, USA), rabbit anti-Flag (Sigma-Aldrich), mouse anti-β-actin (CMCTAG, WI, USA), HRP-labeled anti-rabbit IgG (KPL, MD, USA), and HRP-labeled anti-mouse IgG (KPL).
Immunofluorescence
When the coverage of cells on cover slips reached about 90%, cells were fixed for 15 minutes at room temperature in 4% paraformaldehyde. After aspiration of fixative, samples were rinsed three times in 1×PBS for 5 minutes each and blocked in Blocking Buffer (1×PBS supplemented with 0.3% Triton X-100 (Sangon Biotech) and 5% normal goat serum (Life Technologies)) for 60 minutes. After incubation with the primary antibody (1:100) overnight at 4℃, samples were rinsed three times in 1×PBS for 5 minutes each and incubated with the anti-rabbit IgG second antibody (1:500) for 60 minutes at room temperature in dark. The normal rabbit IgG (Life Technologies) was used as negative control. The antibodies for immunofluorescence were as follows: rabbit anti-CLDND1 (Abcam), rabbit anti-IgG (Merck Millipore), and anti-rabbit IgG (Alexa Fluor® 488 Conjugate) (Life Technologies). The nuclei was labeled using Hoechst 33258 (Sangon Biotech), and cells were visualized using a fluorescence microscope Ti-S (Nikon, Tokyo, Japan).
Tumor xenografts
Four-week-old female athymic BALB/C nude mice (SJA, Hunan, China) were fed under standard conditions at the animal care facility. The protocol of animal experiments was approved by the Animal Care and Experiment Committee of Hunan University. These mice were arbitrarily assigned into six groups (N = 8 each). 1×107 MCF-7 or MDA-MB-231 cells, that transfected with different plasmids in 0.2 ml sterile PBS, were administrated into BALB/C nude mice by subcutaneous injection. On the day 26 and 22, the mice were killed and their tumors were collected for analysis. Tumor volume was calculated according to the following formula: tumor volume (mm3) = 0.5×length×width2.
Statistical Analysis
Results were expressed as mean ± SD, and P < 0.05 was considered statistically significant. One-way ANOVA based on the normal distribution and equal variance assumption test were adopted in statistical comparisons. All statistical analysis was determined by using GraphPad Prism 5.0 statistical software (GraphPad Software, CA, USA).
Results
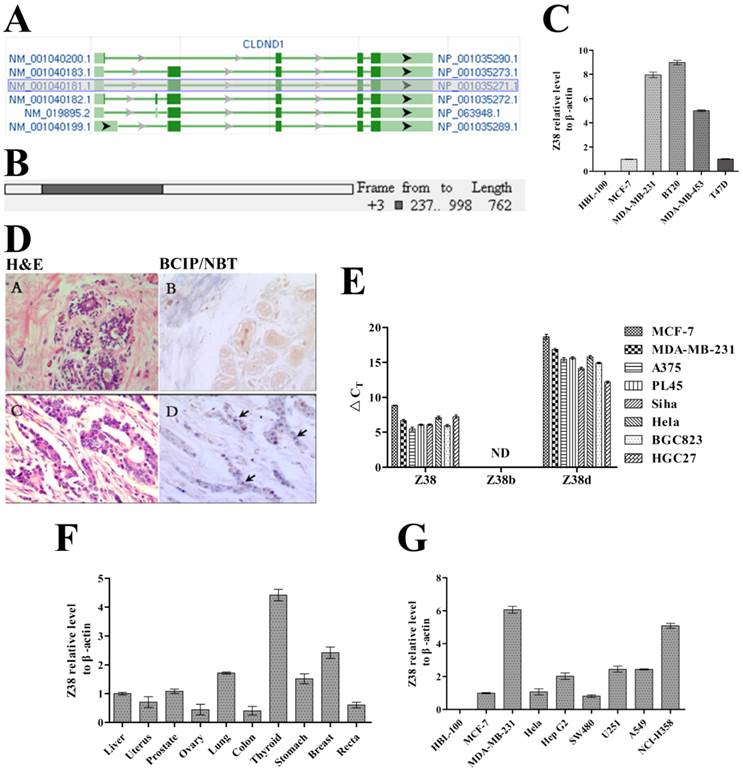
High expression of the newly found RNA gene Z38 in breast cancer
During our premier study, an important RNA gene named Z38 was isolated from breast cancer by combining SSH and reverse dot-blotting [35], and it was one of the identified genes, of which differentially expressed in mammary tumor. However, the discovery of Z38 was not published all the time in consideration of our current research work. Z38 was further identified as one of the most important protein coding sequences of CLDND1 mRNAs on NCBI. The CLDND1 gene, which is located at 3q12.1 on human chromosome, was predicted to transcribe into six mRNAs with three protein coding sequences. Among these predicted mRNAs of CLDND1, NM_001040181.1, NM_001040183.1, NM_001040199.1 and NM_019895.2 share a common 762 nucleotides length protein coding sequence (isoform a). This coding sequence was identified as a newly found RNA gene Z38 (Z38a) (Figure 1A and B). The 834 nucleotides length protein coding sequence (isoform b) of mRNA NM_001040182.1 was identified as RNA gene Z38b, and the 477 nucleotides length protein coding sequence (isoform d) of mRNA NM_001040200.1 was identified as Z38d. Z38 (Z38a), Z38b, and Z38d constitute the RNA gene family Z38, which are highly conserved in human, ape, mouse, dog, and some other mammals. The member Z38 had the highest expression level in cancer cells, and the expression level of Z38d varied a lot according to the type of cancer cell lines (Figure 1E). However, Z38b was not detected by qRT-PCR. Through in situ hybridization experiments, 60% of the breast cancer tissues (total samples: 50 pairs) were found to highly express Z38 compared with the breast paraneoplastic tissues, and not any slices of breast paraneoplastic tissues had Z38 expression in our study (Figure 1D). Meanwhile, Z38 expression profile of breast cancer cell lines MCF-7, MDA-MB-231, BT20, MDA-MB-453, and T47D was much higher than that of breast epithelial cell line HBL-100, which was highly consistent with the degree of cellular malignancy (Figure 1C), thus suggesting the upregulated expression level of Z38 in breast cancer cells. The gene expression profile of different cancer tissues indicated that Z38 was generally expressed in many cancer tissues, and it had relative high expression level in thyroid, breast, lung and stomach cancer tissues, which suggested the significant regulatory roles of Z38 in these cancers (Figure 1F). In addition, the gene expression profile of different cancer cell lines showed that Z38 was generally expressed in many cancer cell lines, especially in breast cancer cell line MDA-MB-231, gastric cancer cell line Hep G2, astrocyte cell line U251, human lung adenocarcinoma cell line A549, and non-small cell lung cancer cell line NCI-H358 (Figure 1G). However, breast epithelial cell line HBL-100, which generated from the milk of a healthy woman [38, 39], didn't express Z38. These results indicate the importance of Z38 on cancer origination and development. Therefore, Z38 may have important regulatory roles in some neoplasms.
Z38 is a long noncoding RNA
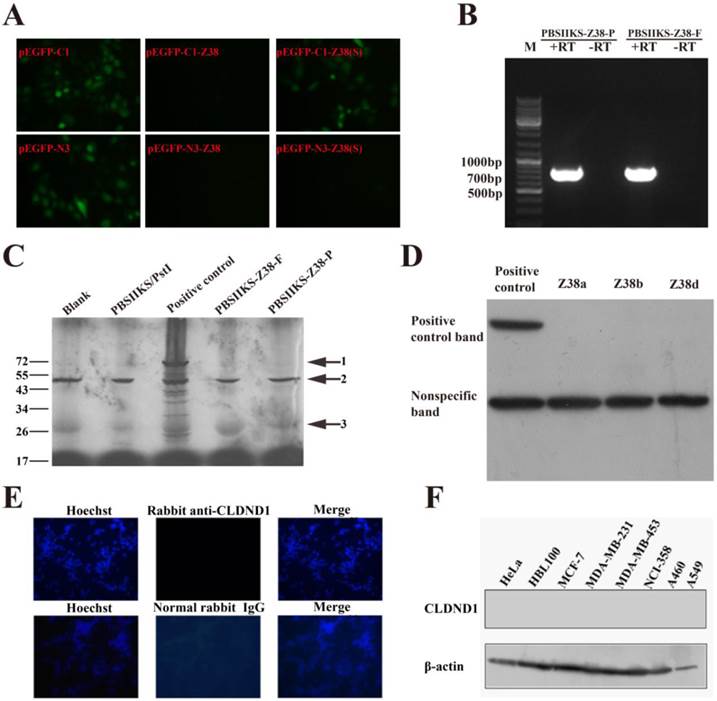
According to the base length, RNA gene Z38 was predicted to translate into protein about 27.83 kDa, i.e. one of the predicted CLDND1 protein variants (isoform a) on NCBI. In order to explore the biological functions of Z38 in cancer, eukaryotic expression vector pEGFP-C1-Z38 was transiently transfected into MCF-7 cells. Out of our expectation, no green fluorescence appeared in these transfected cells (Figure 2A), which led us to suspect the protein coding possibility of Z38. In order to confirm this hypothesis, eukaryotic expression vectors pEGFP-C1-Z38(S), pEGFP-N3-Z38, and pEGFP-N3-Z38(S) were constructed and transiently transfected into MCF-7 cells, respectively. The simulated structures of these plasmids were presented in the Figure S1. Just as we predicted, no green fluorescence was detected in both pEGFP-N3-Z38 and pEGFP-N3-Z38(S) groups, while pEGFP-C1, pEGFP-N3, and pEGFP-C1-Z38(S) groups showed strong green fluorescence (Figure 2A). According to these results, we deduced that the expression of EGFP in plasmids was badly affected by Z38 and RNA gene Z38 was an lncRNA.
In order to validate our deduction, we performed in vitro translation experiments in our study. As a result, RNA gene Z38 did not translate into the predicted CLDND1 or other proteins, though its transcription level was high in experiments (Figure 2B and C). According to these experimental results, we concluded that Z38 is a newly found lncRNA. In order to confirm our conclusion, plasmid pCMV-Tag2B-Z38 was constructed and transfected into MDA-MB-231 cells. However, the expression of FLAG tag in MDA-MB-231 cell lysates was not detected by western blotting, while it was observed in the positive control (Figure 2D). These results strongly supported our conclusion that Z38 is an lncRNA. In addition, no target protein bands were detected in both pCMV-Tag2B-Z38b and pCMV-Tag2B-Z38d groups (Figure 2D), which suggested that Z38b and Z38d may also be lncRNAs.
Our conclusion was further confirmed by western blotting and immunofluorescence using the commercial CLDND1 antibody. In consistent, no green fluorescence in MDA-MB-231 cells was detected in immunofluorescence using the rabbit anti-CLDND1 antibody and the normal rabbit IgG (Figure 2E). Furthermore, western blotting did not detect the expression of CLDND1 proteins in cancer cells (Figure 2F). In summary, these discoveries negated the protein coding ability of RNA gene Z38, which further approved our conclusion that Z38 is an lncRNA.
The structural analysis of RNA gene Z38 and its relative expression level in cancer. (A) The structures of the predicted CLDND1 mRNAs on NCBI. (B) A simulated diagram of Z38 structure. (C) Z38 expression profile of different breast cancer cell lines and breast epithelial cell line HBL-100, assayed by qRT-PCR (Z38 relative expression level of MCF-7 = 1). (D) A (breast paraneoplastic tissues) and C (breast cancer tissues) are images of H&E staining; B (breast paraneoplastic tissues) and D (breast cancer tissues) are images of in situ hybridization. The violet dots and arrows in D represent the expression of Z38. (E) △CT (CT, Target gene-CT, GAPDH) showing the relative expression level of Z38, Z38b, and Z38d in different cancer cell lines. ND: Not Detected. (F and G) Z38 expression profile of different cancer tissues (Z38 relative expression level of liver cancer tissue = 1), and cancer cell lines (Z38 relative expression level of MCF-7 = 1) and breast epithelial cell line HBL-100, assayed by qRT-PCR.
Non-protein-coding features of Z38. (A) The expression of EGFP in MCF-7 cells after transfection with the following plasmids: pEGFP-C1, pEGFP-C1-Z38, pEGFP-C1-Z38(S), pEGFP-N3, pEGFP-N3-Z38, pEGFP-N3-Z38(S). (B) The transcription level of Z38 in the in vitro translation experiments, assayed by semi-quantitative PCR. PBSIIKS-Z38-P represents the circular plasmid, and PBSIIKS-Z38-F represents the linearized plasmid. +RT represents reverse transcription, and -RT represents non-reverse transcription. (C) The results of in vitro translation assays. Arrow 1 indicates the positive control provided in reagent kit, arrow 2 indicates the nonspecific bands detailed by manufacturer, and arrow 3 indicates the target protein. PstI represents a kind of endonuclease. (D) The expression of fusion proteins of FLAG and CLDND1, assayed by western blotting. (E) The expression of CLDND1 proteins in MDA-MB-231 cells, assayed by immunofluorescence. The normal rabbit IgG was used as the negative control. (F) The expression of CLDND1 proteins in cancer cells, assayed by western blotting.
Inhibiting the expression of lncRNA Z38 suppresses breast cancer cell proliferation and in vivo tumorigenesis
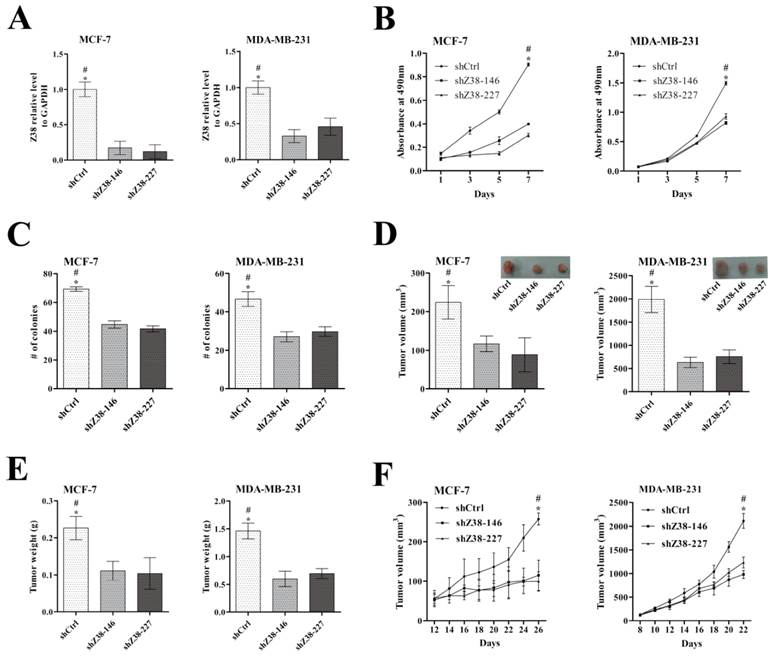
In order to investigate the influences of lncRNA Z38 on cell viability and proliferation, breast cancer cells MCF-7 and MDA-MB-231 were stably transfected with shRNA plasmids to inhibit Z38 expression, respectively. The expression level of Z38 in MCF-7 and MDA-MB-231 cells was significantly reduced at least 50% of both shZ38-146 and shZ38-227 groups compared with that of negative control group shCtrl (Figure 3A). Meanwhile, the cell viability of MCF-7 and MDA-MB-231 cells, that determined by MTT assays, was significantly reduced in experiments (Figure 3B). The number of colony formation in agarose was significantly suppressed in shZ38-146 and shZ38-227 groups (Figure 3C). In conclusion, inhibiting Z38 expression can significantly reduce breast cancer cell viability and proliferation.
In addition, the results of in vivo xenograft assays indicated that tumor weight and tumor volume of both shZ38-146 and shZ38-227 groups were much smaller compared with those of shCtrl group (Figure 3D and E), and the tumor growth rate was significantly inhibited (Figure 3F). These results suggested that inhibiting the expression of lncRNA Z38 in breast cancer cells can efficiently weaken tumorigenesis in vivo, and high expression of Z38 has significance in promoting tumorigenesis. Through combining our above discoveries in cancer cells and tissue samples, we speculated that Z38 may be an important oncogene, and this lncRNA may play significant roles in cancer origination and development. Although the molecular mechanisms are still obscure, Z38 may be an important biomarker in cancer.
The specific siRNAs of lncRNA Z38 efficiently inhibit breast cancer cell proliferation
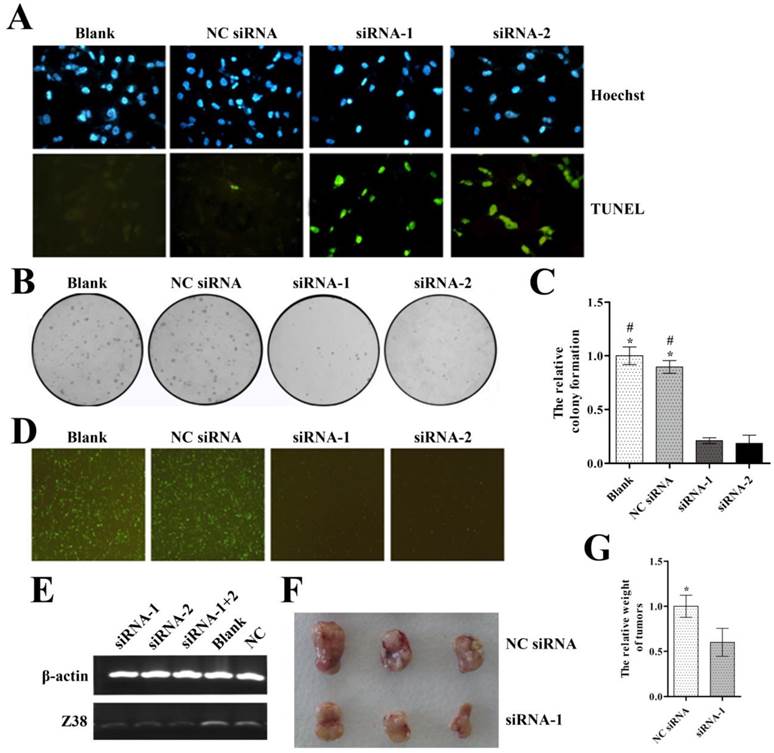
In order to evaluate the therapeutic potential of siRNAs, we next designed two specific siRNAs of Z38 and transfected them into MDA-MB-231 cells, respectively. MDA-MB-231 cells were shown to have high expression level of Z38 in our study (Figure 1C). The inhibitory effects of siRNAs on Z38 expression were examined by semi-quantitative PCR experiments, and the results showed that the Z38 expression of both siRNA-1 and siRNA-2 groups reduced about 60-80% compared with that of control groups (Figure 4E). The results that no synergetic effect appeared in siRNA-1+2 groups suggested the superior inhibitory effects of the specific siRNAs on Z38 expression. We investigated the effects of Z38 siRNAs in cell apoptosis and proliferation. The results of TUNEL assays showed that Z38 siRNAs induced most cell apoptosis (Figure 4A). Meanwhile, the relative colony formation of both siRNA-1 and siRNA-2 groups was lowered to less than 30% compared with that of control groups (Figure 4B and C). These results suggested that Z38 siRNAs can efficiently induce cell apoptosis and inhibit cell proliferation. In addition, the specific siRNAs of Z38 and plasmid pEGFP-C1-Z38(S) were transiently co-transfected into MDA-MB-231 cells. As a result, the green fluorescence of blank and NC siRNA groups was much stronger than that of siRNA-1 and siRNA-2 groups after 24 hours (Figure 4D), thus further approving the superior effects of Z38 siRNAs in inducing breast cancer cell apoptosis.
The inhibitory effects on breast cancer cell proliferation and tumorigenesis after reducing Z38 expression in MCF-7 and MDA-MB-231 cells. (A) Z38 expression level in cells after stable transfection with the following gene silencing plasmids of Z38: shZ38-146, shZ38-227, and the negative control shCtrl. (B) Results of MTT assays. (C) Results of colony formation in agarose. (D and E) Histograms showing the tumor volume and tumor weight of in vivo xenograft experiments, respectively. (F) Histogram showing the tumor growth rate. * (p < 0.05) when compared with shZ38-146; # (P < 0.05) when compared with shZ38-227.
The inhibitory effects of the specific siRNAs of lncRNA Z38 on breast cancer cell MDA-MB-231. (A) Images showing the effects of siRNAs on cell apoptosis, assayed by TUNEL assays. (B) Images of the colony formation in agarose of MDA-MB-231 after transfection with Z38 siRNAs. (C) Histogram showing the relative colony formation. (D) Images showing the effects of siRNAs on cell apoptosis. Cells were transiently co-transfected using siRNAs and plasmid pEGFP-C1-Z38(S). (E) Semi-quantitative PCR measuring Z38 expression level of MDA-MB-231 cells after transfection with siRNAs. (F) Image showing the influence of Z38 siRNAs on tumor growth, assayed by intratumoral injection. (G) Histogram showing the difference of tumor weight, assayed by intratumoral injection. * (p < 0.05) when compared with siRNA-1; # (P < 0.05) when compared with siRNA-2.
The specific siRNA of lncRNA Z38 efficiently inhibits tumor growth in vivo
Next, the therapeutic potential of Z38 siRNAs was further evaluated by intratumoral injection. The relative tumor weight of siRNA-1 group was much smaller than that of control group (Figure 4G). The image showed the visual difference of tumor growth between two groups (Figure 4F). It was clearly demonstrated that the specific siRNAs of Z38 had superior effects in suppressing tumor growth. These results not only demonstrated the therapeutic potentials of Z38 siRNAs in breast cancer, but also further approved the important regulatory roles of lncRNA Z38 in carcinomas.
Discussion
The aberrant expression of some lncRNAs significantly influences cancer origination and progression, and the regulatory roles of lncRNAs in cancer attract wide attentions in the world. For example, the risk-associated lncRNA NBAT-1 regulates cell proliferation and neuronal differentiation and further control the progression of neuroblastoma [40]. The Forkhead box C1 promoter upstream transcript acts as an lncRNA to regulate cell proliferation and migration in basal-like breast cancer [41]. Similarly, the lncRNA ANRIL significantly promotes non-small-cell lung cancer cell proliferation and inhibits cell apoptosis by silencing the expression of KLF2 and P21 [42]. In the present study, the newly found lncRNA Z38 was shown to highly express in the breast cancer. In addition, breast cancer cell proliferation and tumorigenesis were seriously inhibited after inhibiting Z38 expression. All these discoveries indicate the important regulatory mechanisms of Z38 involved in some crucial proteins and transcription of DNA. The relative high expression of Z38 in thyroid, breast, lung, and stomach cancer tissues suggests the significant regulatory roles of Z38 in adenocarcinomas and epithelial neoplasms. However, the detailed mechanisms involved in the regulatory roles of Z38 in carcinomas require further investigation.
Some achievements have been gained in the study of siRNAs, especially in the development of new siRNAs target for tumor therapy [43-46]. Notably, the high efficiency of siRNAs in inducing cell apoptosis and inhibiting tumor growth has been highly praised worldwide [36, 43, 47, 48]. In the current study, the remarkable cancer treatment effects of the specific siRNAs targeting Z38 were approved by in vitro and in vivo experiments, which suggest that Z38 may be a highly efficient target for cancer therapy.
Taken together, the importance of this study mainly embodied in the following aspects: Firstly, the newly found lncRNA Z38 enriches the integrality of genome indirectly. Secondly, the non-protein-coding features of Z38 widen the functional diversity of the claudin family. Thirdly, the high expression of Z38 plays important regulatory roles in improving breast cancer cell proliferation and tumorigenesis. The aberrant expression pattern of Z38 may be an important biomarker for the early diagnosis of tumor. Finally, the capability of Z38 siRNAs on the tumorigenesis inhibition suggests the therapeutic potential of Z38 siRNAs in breast cancer.
More and more studies about the biological regulatory roles of lncRNAs have demonstrated that some lncRNAs can influence the roles of their neighboring protein-coding genes by acting as biological enhancers [49]. For example, the lncRNA HOSTAR was shown to be a modular scaffold of histone modification complexes [50]. The knockout mice study also revealed that lncRNAs can play significant physiological role in the organ development, immunity, organism viability, and some human diseases [51]. In addition, some lncRNAs have been tightly linked between gene regulation and nuclear organization [52]. In the current study, we have focused on the significant influences of lncRNA Z38 on cell proliferation and tumorigenesis. Meanwhile, we have developed research work to identify the interactive proteins and genes of Z38 and found it may interact with heterogeneous nuclear ribonucleoprotein, histone H4, pyruvate kinase, and spermatogenesis associated protein 7 (data not shown). We believe that the regulatory roles and mechanisms of lncRNA Z38 in cancer will be clearly revealed in the future.
Abbreviations
lncRNA: long noncoding RNA; qRT-PCR: quantitative real-time PCR; siRNA: small interfering RNA; CLDND1: claudin domain containing 1; dsDNA: double-stranded DNA; SSH: suppression subtractive hybridization; ND: not detected; RT: reverse transcription; NC: negative control; RNP: ribonucleoprotein.
Supplementary materials
Supplemental Table S1, Table S2, and Figure S1.
Acknowledgements
We gratefully acknowledge the Department of Pathology, the Second Xiangya Hospital, Central South University (Changsha, China) for their help in obtaining breast cancer tissue samples. We thank the Department of Pathology, Hunan Cancer Hospital, the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University (Changsha, China) for their help in providing cancer tissue samples. This research was partly supported by the grant from the “985 project” of Hunan University (No. 531109020011).
Conflict of Interest
The authors declared that they have no conflict of interest in this study.
References
1. Hsiao YH, Chou MC, Fowler C, Mason JT, Man YG. Breast cancer heterogeneity: mechanisms, proofs, and implications. J Cancer. 2010;1:6-13
2. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
3. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674
4. Brody JG, Kripke ML, Kavanaugh-Lynch MH, Rizzo J, Forman MR. Breast cancer and environmental research. Science. 2014;344:577
5. Hill P, Garbaczewski L, Helman P, Walker AR, Garnes H, Wynder EL. Environmental factors and breast and prostatic cancer. Cancer Res. 1981;41:3817-3818
6. Adams C, Glanville NT. The meaning of food to breast cancer survivors. Can J Diet Pract Res. 2005;66:62-66
7. Stendell-Hollis NR, Thompson PA, Thomson CA, O'Sullivan MJ, Ray RM, Chlebowski RT. Investigating the association of lactation history and postmenopausal breast cancer risk in the Women's Health Initiative. Nutr Cancer. 2013;65:969-981
8. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52-62
9. Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E. et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58-68
10. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-914
11. Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348-355
12. Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752-761
13. Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35-43
14. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8
15. Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539-543
16. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-419
17. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120-124
18. Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J. et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344-357
19. Sanchez Y, Segura V, Marin-Bejar O, Athie A, Marchese FP, Gonzalez J. et al. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun. 2014;5:5812
20. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-166
21. Wang L, Fu D, Qiu Y, Xing X, Xu F, Han C. et al. Genome-wide screening and identification of long noncoding RNAs and their interaction with protein coding RNAs in bladder urothelial cell carcinoma. Cancer Lett. 2014;349:77-86
22. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X. et al. A Cytoplasmic NF-kappaB Interacting Long Noncoding RNA Blocks IkappaB Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell. 2015;27:370-381
23. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-531
24. Kam Y, Rubinstein A, Naik S, Djavsarov I, Halle D, Ariel I. et al. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett. 2014;352:90-96
25. Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S. et al. The lncRNA PCAT29 Inhibits Oncogenic Phenotypes in Prostate Cancer. Molecular Cancer Research. 2014;12:1081-1087
26. Zhang Q, Pu R, Du Y, Han Y, Su T, Wang H. et al. Non-coding RNAs in hepatitis B or C-associated hepatocellular carcinoma: Potential diagnostic and prognostic markers and therapeutic targets. Cancer Letters. 2012;321:1-12
27. Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends in Genetics. 2014;30:348-355
28. Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10:235
29. Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K. et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606-612
30. Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403-429
31. Morin PJ. Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res. 2005;65:9603-9606
32. Swisshelm K, Macek R, Kubbies M. Role of claudins in tumorigenesis. Adv Drug Deliv Rev. 2005;57:919-928
33. Ding L, Lu Z, Lu Q, Chen YH. The claudin family of proteins in human malignancy: a clinical perspective. Cancer Manag Res. 2013;5:367-375
34. Hou J, Rajagopal M, Yu ASL. Claudins and the Kidney. Annual Review of Physiology. 2013;75:479-501
35. Wang Y, Peng H, Zhong Y, Li D, Tang M, Ding X. et al. Differential gene expression profiling of human epidermal growth factor receptor 2-overexpressing mammary tumor. Acta Biochimica et Biophysica Sinica. 2008;40:397-405
36. Pille JY, Denoyelle C, Varet J, Bertrand JR, Soria J, Opolon P. et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther. 2005;11:267-274
37. Kim W, Christensen L, Jo S, Yockman J, Jeong J, Kim Y. et al. Cholesteryl Oligoarginine Delivering Vascular Endothelial Growth Factor siRNA Effectively Inhibits Tumor Growth in Colon Adenocarcinoma. Molecular Therapy. 2006;14:343-350
38. Caron de Fromentel C, Nardeux PC, Soussi T, Lavialle C, Estrade S, Carloni G. et al. Epithelial HBL-100 cell line derived from milk of an apparently healthy woman harbours SV40 genetic information. Exp Cell Res. 1985;160:83-94
39. Gaffney EV. A cell line (HBL-100) established from human breast milk. Cell Tissue Res. 1982;227:563-568
40. Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K. et al. The Risk-Associated Long Noncoding RNA NBAT-1 Controls Neuroblastoma Progression by Regulating Cell Proliferation and Neuronal Differentiation. Cancer Cell. 2014;26:722-737
41. Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H. et al. Forkhead box C1 promoter upstream transcript, a novel long non-coding RNA, regulates proliferation and migration in basal-like breast cancer. Mol Med Rep. 2015;11:3155-3159
42. Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R. et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268-277
43. Matheis F, Besch R. Bifunctional siRNAs for tumor therapy. Methods Mol Biol. 2014;1169:181-192
44. Piatek MJ, Werner A. Endogenous siRNAs: regulators of internal affairs. Biochem Soc Trans. 2014;42:1174-1179
45. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811
46. Seton-Rogers S. Therapeutics: siRNAs jump the hurdle. Nat Rev Cancer. 2012;12:376-377
47. Dudek H, Wong DH, Arvan R, Shah A, Wortham K, Ying B. et al. Knockdown of beta-catenin with dicer-substrate siRNAs reduces liver tumor burden in vivo. Mol Ther. 2014;22:92-101
48. Jiang T, Zhou C, Gu J, Liu Y, Zhao L, Li W. et al. Enhanced therapeutic effect of cisplatin on the prostate cancer in tumor-bearing mice by transfecting the attenuated Salmonella carrying a plasmid co-expressing p53 gene and mdm2 siRNA. Cancer Lett. 2013;337:133-142
49. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46-58
50. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693
51. Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594-602
52. Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651-663
Author contact
![]() Corresponding authors: Yinke Yang and Jian Zhang, Tel: +86-0731-8399-8546, Fax: +86-0731-8882-1720, E-mail: ykyang99edu.cn (Yinke Yang), Jhz444com (Jian Zhang).
Corresponding authors: Yinke Yang and Jian Zhang, Tel: +86-0731-8399-8546, Fax: +86-0731-8882-1720, E-mail: ykyang99edu.cn (Yinke Yang), Jhz444com (Jian Zhang).

 Global reach, higher impact
Global reach, higher impact