Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(5):608-617. doi:10.7150/jca.13822 This issue Cite
Research Paper
Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma
1. Department of Radiation Oncology, Nanfang Hospital, Southern Medical University, Guangzhou, GuangDong Province, 510515, China.
2. Hepatology Unit and Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, Guangzhou, GuangDong Province, 510515, China.
Received 2015-9-11; Accepted 2016-2-2; Published 2016-3-19
Abstract
Recent studies indicated that long noncoding RNAs (lncRNAs) played important regulatory roles in carcinogenesis and cancer progression. However, the contribution of small nucleolar RNA host gene 20 (SNHG20) to cancer development remains largely unknown. The aim of the study is to investigate the expression of SNHG20 and its clinical significance in hepatocellular carcinoma (HCC). Our results showed that the expression of SNHG20 was remarkably up-regulated in HCC tissues compared with adjacent non-tumor liver tissues from 49 fresh HCC samples (cohort 1) detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR, P = 0.004). The results were confirmed in 144 formalin-fixed, paraffin-embedded HCC tissues (cohort 2) by in situ hybridization (ISH). Meanwhile, the expression of SNHG20 was associated with tumor size (P = 0.027 for cohort 1 and P = 0.046 for cohort 2) and clinical stage (P = 0.027 for cohort 1 and P = 0.028 for cohort 2). Importantly, patients with high expression of SNHG20 had a shorter overall survival (OS, P < 0.001) and disease-free survival (DFS, P < 0.001) than those with low expression of SNHG20. Univatiate and multivariate analysis showed that SNHG20 was a significant and independent prognostic predictor for OS of HCC patients (hazard ratio = 3.985, 95% CI = 1.981-8.017, P < 0.001). In addition, a total of 331 HCC patients' data from the Caner Genome Atlas project (TCGA) were used to validate our findings. Consistently, the results from TCGA HCC cohort demonstrated that SNHG20 were overexpressed in HCC tissues compared with non-tumor liver tissues (P < 0.001). Patients with higher expression levels of SNHG20 had poorer OS (P = 0.021) and DFS (P < 0.001). Functionally, knockdown of SNHG20 in SK-Hep-1 cells significantly inhibited cellular proliferation, migration, and invasion. In conclusion, SNHG20, up-regulated in patients with HCC, may serve as an independent prognostic predictor for HCC patients.
Keywords: Long Non-Coding RNA, SNHG20, Hepatocellular Carcinoma, prognosis.
Introduction
Hepatocellular Carcinoma (HCC) is a highly aggressive solid malignancy and contributes to the second leading cause of cancer-related mortality worldwide [1, 2]. Unfortunately, HCC is always diagnosed at an advanced stage with severe hepatic dysfunction, thus, results in poor prognosis for HCC patients. Therefore, understanding the molecular mechanisms of HCC and identifying specific prognostic biomarkers would be of great importance to the development of applicable diagnostic or therapeutic strategies earlier during the cause of HCC.
Long noncoding RNAs (lncRNAs) are non-coding RNAs which are more than 200 nucleotides in length without protein coding capacity [3, 4]. These long noncoding transcripts were once regarded as simply transcriptional “noise” or cloning artifacts [5]. However, a growing number of evidence showed that lncRNAs played key roles in cancer cell biology such as cell growth, differentiation, apoptosis and migration [6-8]. Strong evidence have revealed that lncRNAs have oncogenic or tumor suppressor roles in tumorigenesis [9-11]. For example, the lncRNA HOTAIR is highly induced in breast cancer and colorectal cancer [12, 13]; its expression level is positively associated with tumor recurrence and migration. MALAT1 is a conserved non-coding transcript that regulates a group of genes promoting lung cancer cell migration and becomes a hallmark of lung cancer metastasis [14]. However, only a small portion of lncRNAs are well studied in the cause and progression of cancers [15].
In our previously studies, we found that the long intergenic noncoding RNA UFC1 (lincRNA-UFC1) acted as an oncogene which promote HCC malignancy via promoting cell-cycle progression and inhibiting cellular apoptosis [16]. Our and others' researches indicated that lncRNAs could act as potential prognostic indicators for cancers [17, 18]. Thus, previous studies made us to know if there were any other novel long noncoding RNAs with important roles in the onset or progression of HCC that could serve as prognostic biomarkers. Interestingly, from our previous microarray data (the microarray data can be accessible through GEO series accession number GSE58043), we found that small nucleolar RNA host gene 20 (SNHG20; GenBank Accession no. NR_027058.1) was also up-regulated in HCC.
SNHG20 is a long noncoding transcript that is poorly understood in cancers. The potential prognostic value of SNHG20 expression in HCC has not yet been investigated. In this study, we explored the expression of SNHG20 and its localization by qRT-PCR and in situ hybridization (ISH) respectively using patients from 2 HCC cohorts, and validated our findings using data from TCGA dataset. We aimed to determine whether the expression of SNHG20 would serve as a clinically useful prognostic indicator for HCC patients.
Materials and Methods
Patients and Specimens
The study was reviewed and approved by the ethics committee of Nanfang Hospital, Southern Medical University. All patients gave informed consent before using these clinical materials. A total of 193 patients with primary HCC in two cohorts were enrolled in this retrospective study. In cohort 1, fresh HCC tissues and adjacent non-tumor tissues were collected from 49 HCC patients. All the patients in cohort 1initially underwent hepatectomies at Nanfang Hospital between January 2011 and July 2012. Tissue samples were preserved in RNA keeper tissue stabilizer (Vazyme, Nanjing, China) immediately after hepatectomies and stored in -80 o C before RNA extraction. Formalin-fixed, paraffin-embedded samples from 144 HCC patients, who underwent hepatectomies initially between January 2006 and July 2009 in cohort 2, were randomly selected from the same hospital. None of the patients received anti-cancer therapies prior to surgery. Patients from cohort 2 were followed up for 5 years. The clinical information of HCC patients from cohort 1 and cohort 2 is presented in supplementary Table S1 and Table S2.
TCGA dataset
To validate the potential role of SNHG20 in HCC, we analyzed the Caner Genome Atlas project (TCGA dataset, https://tcga-data.nci.nih.gov/tcga/) which provides more than 300 HCC cases including gene expression data and follow-up information. A total of 331 cases of HCC patients with SNHG20 expression data and follow-up information were enrolled in this study (defined as TCGA cohort here). Among these patients, a total of 50 patients have SNHG20 expression data in both HCC tissues and matched adjacent non-tumor tissues. OS and DFS were calculated based on the expression of SNHG20 (Patients were divided into two groups depend on the expression of SNHG20, the expression level greater than median was classified into high expression group, otherwise was classified into low expression group).
Cell Lines
HL-7702, MHCC-97H, HepG2, SK-Hep-1, SMMC-7721, and BEL-7402 cell lines were purchased from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China) and incubated in Dulbecco's modified Eagle medium (Gibco, Geand Island, NY) with 10% fetal bovine serum (Gibco), and maintained at 37 o C in a humidified incubator containing 5% CO2.
In situ hybridization (ISH) Staining and Evaluation of Staining
ISH was performed using the ISH Kit (Boster Bio-Engineering Company, Wuhan, China) according to the manufacturer's instructions. The ISH staining regions for SNHG20 were scored by 2 pathologists blinded to the clinical parameters. The score standard for the staining intensity was as follows: 0 (negative), 1 (weak), 2 (medium), 3 (strong).The score of staining extent was 0 (10%), 1 (11%-25%), 2 (26%-50%), 3 (51%-75%), and 4(76%-100%). The final SNHG20 expression score was calculated with the intensity score + extent score, ranging from 0 to 7. Total scores of 3 or higher were considered as the high-expression group. This scoring system is relative reproducible and gives highly concordant evaluations between independent pathologists, as described in our previous studies [16].
RNA Extraction, Synthesis of cDNA and Quantitative Real-Time PCR (qRT-PCR)
Total RNA from cell lines, HCC tissues and corresponding adjacent non-tumor tissues was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacture's instruction. RNAse-free DNase I was applied to eliminate the genomic DNA contamination. Then, concentration and quality of total RNA were measured by NanoDrop® Spectrophotometer
(ND-100, Thermo Scientific). RNAs were reverse transcribed into cDNA with a reverse transcription (RT) reagent kit (Takara Biotechnology, Dalian, China) in a 20μl reaction volume with 500 ng RNA. Real-time PCR was performed using the SYBR Green PCR Kit purchased from Takara Biotechnology. The following primers were used to amplify SNHG20: sense primer, 5'-ATGGCTATAAATAGATACACGC-3' and anti-sense primer, 5'-GGTACAAACAGGGAGGGA-3'.
β-actin was amplified as an internal control using sense primer, 5'-TCAAGATCATTGCTCCTCCTGA-3' and anti-sense primer, 5'-CTCGTCATACTCCTGCTTGCTG-3'. The gene specific amplification was performed using LightCycler® 480II. Positive and negative control or no template controls were used. The reaction was performed in a 20μl reaction volume by the following condition: preliminary denaturation at 95°C for 30s, followed by 40 cycles of 95 °C for 5s, 60 °C for 20s. All qPCR primers were verified as specific and efficient. Relative quantification (2-ΔΔCt) method was used for calculating fold changes.
RNA interference
Three duplex siRNAs targeting different regions of human SNHG20 and one negative control siRNA were obtained from Genepharma (Shanghai, China). In brief, 50 nM of each siRNA was introduced in SK-Hep-1 cells using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen) according to the manufacturer's instructions. The sequences of siRNAs targeting SNHG20 were as follows: siSNHG20-1, sense, 5'-GCCUAGGAUCAUCCAGGUUTT-3', anti-sense, 5'-AACCUGGAUGAUCCUAGGCTT-3'; si-SNHG20-2, sense, 5'-CCUGUUGUGUAUGGCUAUATT-3', anti-sense, 5'-UAUAGCCAUACACAACAGGTT-3'; si-SNHG20-3, sense, 5'-GCCACUCACAAGAGUGUAUTT-3', anti-sense, 5'-AUACACUCUUGUGAGUGGCTT-3'.
5-ethynyl-20-deoxyuridine (EdU) incorporation assays
SK-Hep-1 cells was seeded in 24-well plates and incubated overnight. Transfection of cells with siRNAs was performed the following day following the protocol described above. 48 hours later, cell proliferation was detected by the EdU incorporation assays using EdU Cell Proliferation Assay Kit (Ribobio, Wuhan, China). In brief, after incubation with 50µM EdU, cells were fixed and stained with EdU according to the manufacturer's recommendations. The EdU incorporation rate was defined as the ratio of EdU positive cells to total DAPI positive cells.
Transwell migration and Boyden chamber invasion assays
The SK-Hep-1 cells were transfected with siRNAs and incubated for 24h. For transwell migration assay, 1×105 cells suspended in 200 µl serum-free DMEM media were seeded into the upper chamber of Transwell (Corning). 600 µl DMEM containing 10% FBS was added into the bottom chamber. After incubation at 37 o C for 4h, the filters were washed twice with PBS. Before staining with crystal violet, cells adhered on the lower surface of filters were fixed with 100% methanol at room temperature. Cell numbers in 5 random fields in each replicate were counted using microscope (NiKon ECLPSE 80i system; magnification: × 200). Each assay was performed at least for three times. For Boyden chamber invasion assay, the procedure was the same with transwell migration assay except that the transwell membrane was precoated with matrigel (BD Biosciences, San Jose, CA) and cells were incubated for 8h.
Statistical Analysis
All statistical analyses were performed using SPSS statistical software package, version 20.0 (Abbott Laboratories, North Chicago, IL).The correction between SNHG20 expression and clinicopathological characteristics was evaluated with Mann-Whitney U-test. Survival curves were calculated using Kaplan-Meier and log-rank tests. Cox proportional-hazard modeling was used for univariate and multivariate analysis to explore the effects of variables on survival. Conventional clinicopathologic variables along with SHNG20 expression were brought into Cox univariate regression model. Each statistical significant variable on univariate was entered into multivariate model. The t- test was performed to analyze the results of qRT-PCR. All statistical tests were two-sided and P < 0.05 was considered to be statistically significant.
Results
SNHG20 is up-regulated in HCC cell lines and HCC Tissue
In our previous study, to identify transcripts that potentially impel liver tumorigenesis, lncRNA and messenger RNA (mRNA) expression profiles were determined by microarray analysis [16]. From that study, we noted that SNHG20 was significantly up-regulated (fold change: 4.88; P = 0.003). To further validated the expression level of SNHG20 in HCC, we examined the expressions of SNHG20 in a normal hepatocyte cell line HL-7702 and HCC cell lines (SK-Hep-1, HepG2, MHCC-97H, BEL-7402, and SMMC-7721) using qRT-PCR. As shown in Fig. 1A, all the five HCC cell lines showed higher expression of SNHG20 compared with HL-7702 cells.
Next, a total of 49 pairs of fresh HCC tissues and adjacent non-tumor tissues were used to detect the relative expression of SNHG20 using qRT-PCR (Cohort 1). SNHG20 expression level was significantly elevated in HCC tissues in comparison with non-tumor tissues (P = 0.004, Fig. 1B, left panel). To avoid the possibility that the significant difference between tumor tissues and non-tumor liver tissues was caused by outlier effect, we re-assessed the expression data of SNHG20 from 44 out of 49 paired HCC tissues and noncancerous after removing 5 paired outliers. The results convinced that SNHG20 was significantly up-regulated in HCC (P = 0.016, Fig. 1B, right panel). Similar result was found in TCGA cohort (P < 0.001, Fig. 1C). To further validate dysregulated expression of SNHG20, we detected its expression in an addition cohort including 144 paraffin-embedded HCC specimens by ISH (cohort 2). Using a cutoff score of ≥ 3, 98 out of 144 (68.1%) cases were classified as high SNHG20 expression (Fig. 2, P < 0.001). Collectively, these findings strongly suggest that SNHG20 is up-regulated in HCC.
Correlation between SNHG20 and Clinicopathological Features of HCC
To investigate the oncogenic role of SNHG20 in HCC progression, we analyzed the association between SNHG20 and clinicopathologic status in HCC patients from cohort 1and cohort 2. As summarized in Table 1 and Table 2, statistical analyses indicated that the SNHG20 expression was positively associated with tumor size (P = 0.027 for cohort 1 and P = 0.046 for cohort 2) and BCLC stage (P = 0.027for cohort 1 and P = 0.028 for cohort 2). However, no statistically significant correction was found between SNHG20 and other clinicopathological features.
SNHG20 overexpression in HCC cell lines and HCC tissues. (A) Analysis of SNHG20 mRNA expression in a normal hepatocyte cell line HL-7702 and five HCC cell lines (SMMC-7721, BEL-7402, SK-Hep-1, MHCC-97H, HepG2) using qRT-PCR. SNHG20 expression level was normalized to that of β-actin. (B) SNHG20 expression level was analyzed by qRT-PCR in 49 paired HCC samples and adjacent non-tumor liver tissues (cohort 1, n=49, P = 0.004, left panel). The expression data of SNHG20 from 44 out of 49 paired of HCC tissues and matched non-tumor liver tissues was recalculated after removing 5 paired outliers (cohort 1, n=44, P = 0.016, right panel). ** P < 0.01, * P < 0.05.(C) Analysis of SNHG20 mRNA expression in TCGA cohort (TCGA cohort, n = 331, P < 0.001, left panel). Re-analysis of SNHG20 mRNA expression in 50 paired HCC samples and adjacent non-tumor tissues from TCGA cohort (TCGA cohort, n = 50, P < 0.001, right panel). ***P < 0.001.
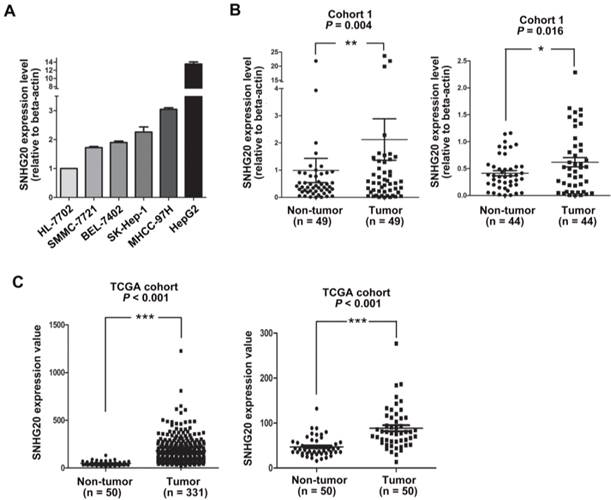
The expression of SNHG20 in HCC tissues and non-cancerous liver tissues were examined in ISH assays. (A-B) Representative images of SNHG20 expression in non-tumor tissues by ISH assays (magnification: A, ×100; B, ×400). (C-D) Representative images of SNHG20 expression in HCC tissues by ISH assays (magnification: A, ×100; B, ×400).
Correlation between SNHG20 expression and HCC clinicopathologic features in 49 patients (cohort 1).
| SNHG20 expression levels | P | ||
|---|---|---|---|
| high expression | low expression | ||
| Gender | 0.152 | ||
| Male | 29 | 16 | |
| Female | 4 | 0 | |
| Age (years) | 0.626 | ||
| ≤55 | 19 | 8 | |
| >55 | 14 | 8 | |
| Edmondson Grade | 0.193 | ||
| I-II | 18 | 4 | |
| III-IV | 15 | 12 | |
| Liver cirrhosis | 0.299 | ||
| With | 22 | 13 | |
| without | 11 | 3 | |
| Portal vein tumor thrombus | 0.057 | ||
| No | 20 | 14 | |
| Yes | 13 | 2 | |
| Tumor size (cm) | 0.027* | ||
| ≤5 | 8 | 9 | |
| >5 | 25 | 7 | |
| BCLC stage | 0.027* | ||
| A | 15 | 12 | |
| B+C | 18 | 4 | |
| Metastasis | 0.152 | ||
| Yes | 4 | 0 | |
| No | 29 | 16 | |
Abbreviations: BCLC: Barcelona Clinic Liver Cancer. *P < 0.05: the values had statistically significant differences.
Correlation between SNHG20 expression and HCC clinicopathologic features in 144 patients (cohort 2).
| SNHG20 expression levels | P | ||
|---|---|---|---|
| high expression | low expression | ||
| Gender | 0.161 | ||
| Male | 78 | 41 | |
| Female | 20 | 5 | |
| Age (years) | 0.436 | ||
| ≤55 | 55 | 29 | |
| >55 | 43 | 17 | |
| Edmondson Grade | 0.597 | ||
| I-II | 78 | 39 | |
| III-IV | 20 | 7 | |
| Liver cirrhosis | 0.629 | ||
| With | 72 | 32 | |
| without | 26 | 14 | |
| Portal vein tumor thrombus | 0.989 | ||
| No | 83 | 39 | |
| Yes | 15 | 7 | |
| No. tumor | 0.064 | ||
| Solitary | 67 | 39 | |
| Multiple | 31 | 7 | |
| Tumor size (cm) | 0.046* | ||
| ≤5 | 36 | 25 | |
| >5 | 62 | 21 | |
| BCLC stage | 0.028* | ||
| A | 56 | 35 | |
| B+C | 42 | 11 | |
| Relapse | 0.196 | ||
| Yes | 34 | 11 | |
| No | 64 | 35 | |
| Metastasis | 0.336 | ||
| Yes | 3 | 3 | |
| No | 95 | 43 | |
Abbreviations: BCLC: Barcelona Clinic Liver Cancer. *P < 0.05: the values had statistically significant differences.
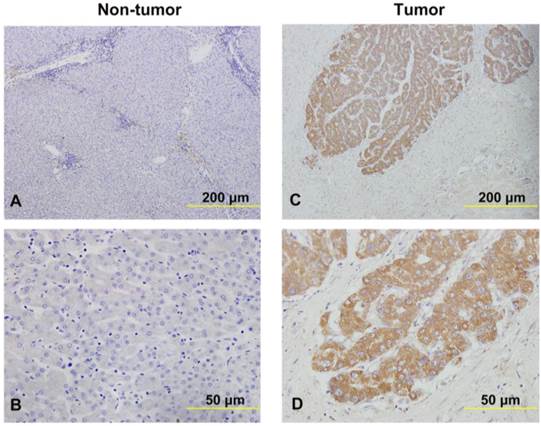
SNHG20 Expression is Associated with Overall Survival and Disease-free survival of HCC
Subsequently, Kaplan-Meier and log-rank test analyses were performed to determine the association between the levels of SNHG20 expression and patients' overall survival (OS) or disease-free survival (DFS). In 144 HCC cases with prognostic information from cohort 2, we observed that the level of SNHG20 was negatively associated with OS (P < 0.001, Fig. 3A) and DFS (P < 0.001, Fig. 3B). Furthermore, a significant inverse correlation was found between SNHG20 expression and OS (P < 0.001, Fig. 3C) or DFS (P <0.001.Fig. 3D) in the subgroup of patients with larger tumor size (tumor size > 5cm). However, SNHG20 expression had no correlation with OS or DFS in the subgroup of patients with smaller tumor size (tumor size < 5cm). To validate our findings, we analyzed the data from TCGA cohort. Consistently, Kaplan-Meier and log-rank test analysis showed that patients with higher expression of SNHG20 had poorer outcome (P = 0.021 for OS and P < 0.001 for DFS, Fig. 4A-B). Taken together, high expression of SNHG20 predicts worse prognosis in patients with HCC.
The correlation between SNHG20 expression and HCC patients' survival in cohort 2. (A-B) Kaplan-Meier survival analysis of OS (P < 0.001, log-rank) and DFS (P < 0.001, log-rank) rate in 144 patients based on the expression level of SNHG20 expression. (C-D) Kaplan-Meier survival analysis of OS (P < 0.001, log-rank) and DFS (P < 0.001, log-rank) rate in the subgroup of patients with tumor size > 5cm.
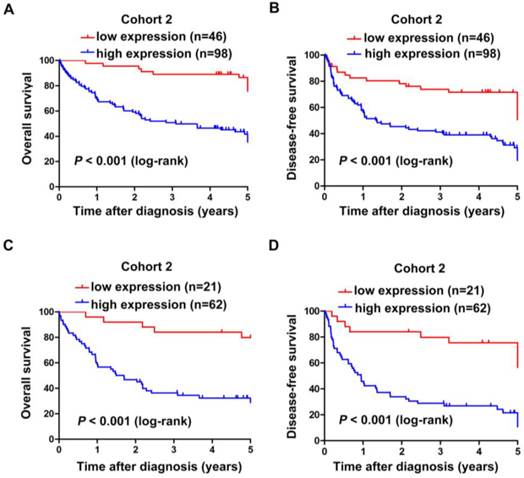
The correlation between SNHG20 expression and HCC patients' survival in TCGA cohort. (A-B) Kaplan-Meier survival analysis of OS (P = 0.021, log-rank) and DFS (P < 0.001, log-rank) rate in TCGA cohort.
SNHG20 is an Independent Prognostic Factor in HCC Patients
Additionally, univariate and multivariate analysis were conducted to assess the prognostic variables in all patients from cohort 2. Univariate analysis showed that SNHG20 expression (P < 0.000), portal vein tumor thrombus (P = 0.004), BCLC stage (P = 0.049), tumor relapse (P < 0.001), and tumor size (P = 0.014) were significantly related to overall survival rate in patients with HCC (Table 3). Multivariate analysis was performed using the Cox Proportional hazards model. The analysis results revealed that SNHG20 expression was a significant independent prognostic factor for HCC patients (95% CI: 1.981-8.017; P < 0.001) in addition to tumor relapse (95% CI: 1.175-3.214; P = 0.01).
Knockdown of SNHG20 inhibits HCC cell proliferation, migration, and invasion
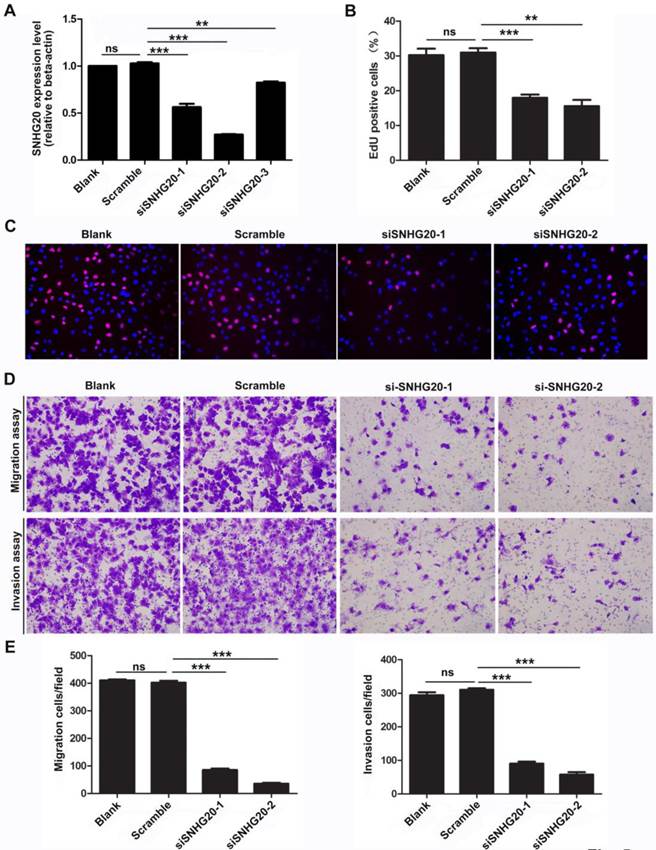
To further investigate the biological function of SNHG20 in HCC development and progression, we knockdown the expression of SNHG20 in SK-Hep-1 cells using siRNAs. Three SNHG20 specific siRNAs targeting different regions of SNHG20 was designed and introduced in SK-Hep-1 cells. As shown in Fig. 5A, siRNA-1 and siRNA-2 effectively silenced the expression of SNHG20 (P < 0.01). Therefore, these two siRNAs were used for further study. To investigate whether endogenous silence of SNHG20 reduced proliferative capacity in SK-Hep-1 cells, EdU assay was performed. The results showed that SNHG20 knockdown significantly decreased cellular proliferation in SK-Hep-1 cells (P < 0.01, Fig. 5B-C). In addition, we observed that SK-Hep-1 cells transfected with siRNA-1 or siRNA-2 exhibited reduced migration ability as determined by transwell migration assays (P < 0.001, Fig. 5D-E). Moreover, using Boyden chamber invasion assays, we found that the cell invasion ability was impaired in SK-Hep-1 cells transfected with siRNA-1 or siRNA-2 (P < 0.001, Fig. 5D-E). Together, the results above reveal that knockdown of SNHG20 significantly inhibits cell proliferation, migration and invasion.
Discussion
To date, this is the first investigation of SNHG20 expression in human cancer. In our study, we investigated the clinical significance of SNHG20 in HCC patients. We found that SNHG20 was up-regulated in HCC tissues relative to corresponding adjacent non-tumor tissues from 2 cohorts using qRT-PCR and ISH. Our results showed that the expression level of SNHG20 was associated with tumor size and BCLC stage. In addition, high expression of SNHG20 was associated with poorer OS or DFS and could be an independent prognostic factor for HCC patients. More importantly, Suppression of SNHG20 distinctly inhibited HCC cell proliferation, migration and invasion in vitro. Overall, SNHG20 may play an important role in development and progression of HCC.
It has been proved that aberrant lncRNA expression levels lead to dysregulation of downstream effectors and that lncRNAs may play crucial roles in a variety of biological functions and provide a cellular growth advantage resulting in HCC. For example, Yuan et al. found that lncRNA-activated by TGF-β (lncRNA-ATB) was up-regulated in HCC. Interestingly, lncRNA-ATB regulated ZEB1and ZEB2 expression by competitively binding the miR-200 family, which led to the EMT-induced cancer cell invasion [19]. lncRNA-hPVT1 was up-regulated in HCC tissues. Its high expression level resulted in upregulation of a series of cell cycle genes [20]. HULC, a long noncoding RNA overexpressed in HCC, contributes to malignant development by activating the acyl-CoA synthetase subunit ACSL1[21].
Univariate and multivariate analyses of OS in 144 HCC patients by Cox regression analysis.
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | CI (95%) | P value | Hazard ratio | CI (95%) | P value | |
| gender | 1.482 | 0.835-2.632 | 0.179 | |||
| Age(year) | 1.295 | 0.804-2.086 | 0.287 | |||
| Edmondson Grade | 1.464 | 0.863-2.483 | 0.158 | |||
| Metastasis(distant) | 1.533 | 0.858-4.216 | 0.408 | |||
| Liver cirrhosis | 0.921 | 0.542-1.566 | 0.761 | |||
| Tumor number | 1.361 | 0.800-2.316 | 0.256 | |||
| Portal vein tumor thrombus | 2.336 | 1.312-4.161 | 0.004* | |||
| BCLC stage | 1.622 | 1.001-2.627 | 0.049* | |||
| Relapse | 2.428 | 1.507-3.912 | 0.000* | 1.944 | 1.175-3.214 | 0.010* |
| Tumor size | 1.883 | 1.138-3.117 | 0.014* | |||
| SNHG20 | 4.440 | 2.254-8.743 | 0.000* | 3.985 | 1.981-8.017 | 0.000* |
Abbreviations: OS: overall survival; BCLC: Barcelona Clinic Liver Cancer; CI: confidence interval; HR: hazard radio; *P < 0.05: the values had statistically significant differences.
Knockdown of SNHG20 obviously inhibits HCC cell proliferation, migration, and invasion. (A) The knockdown efficiency was determined by qRT-PCR in SK-Hep-1 cells. ***P < 0.001, **P < 0.01. (B-C) Suppression of SNHG20 in SK-Hep-1 cells distinctly reduced their proliferative capacities, as determined by EdU assay (magnification: C,×200). ***P < 0.001, **P < 0.01. (D-E) Knockdown of SNHG20 in SK-Hep-1 cells significantly inhibited their migration and invasion abilities, as evaluated by transwell migration assay and Boyden chamber invasion assay (magnification: D,×200). ***P < 0.001, **P < 0.01.
Moreover, accumulating evidence proves that lncRNAs can serve as biomarkers and prognosis factors for many cancer types including HCC [22-25]. Take lncRNA MVIH for example, its overexpression in HCC may serve as a predictor for HCC patients' poor recurrence-free survival after hepatectomy [26]. All these evidence suggest that lncRNAs do play important roles in hepatocarcinogenesis and progression, which leads us to believe that lncRNA SNHG20 may contribute to HCC malignant development and could serve as a predictor for HCC prognosis. However, most of the researches about lncRNAs including our study here are retrospective single center analyses with small sample size. Thus, a prospective, multicenter cohort study with large sample is needed to satisfying better understanding the explicit roles of lncRNAs in HCC.
Herein we have demonstrated that SNHG20 is up-regulated in HCC and may serve as a prognostic predictor for HCC patients, whether SNHG20 may predict outcomes for patients with other tumors remains to be explored. Interestingly, through analyzing the expression data from Oncomine gene expression array datasets, we found that SNHG20 was significantly up-regulated in cancers including colorectal cancer, lung cancer, ovarian cancer, bladder cancer and renal cancer (Fig. S1). The facts of SNHG20 expression in multiple types of cancers suggest that it may be a more general oncogene in cancers. However, lack of clinical follow-up information in these datasets, we could not assess the potential prognostic value of SNHG20 in other human cancers. Therefore, more researches should be performed to investigate the role of SNHG20 in cancers.
The most important finding of the current study was that high expression level of SNHG20 strongly correlated with HCC progression and might serve as a potential prognostic predictor for OS and DFS of HCC patients. However, there are still limitations in our present study. Firstly, all samples were obtained from the same hospital and the sample size was limited. Thus, we used the TCGA dataset to analysis the SNHG20 expression levels of HCC tissues and non-tumor tissues in a larger sample size (including 331 HCC tissues and 50 non-tumor tissues), and then analysis the correlation between SNHG20 expression level and the prognosis of HCC patients. Consistent with our results, the expression level of SNHG20 was higher in HCC tissue compared with non-tumor tissue, and higher expression of SNHG20 predicted poorer prognosis for HCC patients. Secondly, as mentioned above, our study is a retrospective single center analysis, which leads to another limitation. This limitation claims for a prospective, randomized multicenter study in the future. Finally, the underlying mechanisms of SNHG20 in promoting HCC progression remain largely unknown. Numerous evidence have suggested that the regulatory mechanisms of lncRNAs strongly correlate with their localizations [27]. Furthermore, lncRNAs locating in cytoplasm play important roles in post-transcriptional regulation of genes [28]. They may function as partners with miRNA, mRNA s or proteins to post-transcriptional regulate target genes involving in HCC progression [29-31]. Our resulted showed that SNHG20 located in cytoplasm. We speculated that cytoplasmic SNHG20 may act as a sponge for miRNAs or integrate with proteins or mRNAs to regulate downstream molecules. More detailed mechanisms should be experimentally investigated in the future.
Although limitations exit in the present study, the results of the study are meaningful. We believe that SNHG20 will become a novel prognostic biomarker for HCC patients, facilitating earlier diagnosis or improving therapeutic strategies. In summary, a more detailed investigation with larger sample size will be necessary for us to better understanding the role of SNHG20 in hepatocarcinogenesis and progression.
Supplementary materials
Supplementary methods, Tables and Figure.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (Grant Nos. 81372283, 81401180 and 91540111); Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015), and Natural science funding of Guangdong Province (No. 2014A030311013).
Abbreviations
lncRNA: long noncoding RNA; mRNA: messenger RNA; HCC: hepatocellular carcinoma; SNHG20: small nucleolar RNA host gene20; qRT-PCR: quantitative reverse-transcription polymerase chain reaction; ISH: in situ hybridization; TCGA: the Caner Genome Atlas project; OS: overall survival; DFS: disease-free survival; HR: hazard ratio; BCLC: the Barcelona Clinic Liver Cancer.
Competing interests
The authors have declared that no competing interest exists.
References
1. Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B. et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119-24
2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29
3. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-52
4. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
5. Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291-8
6. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cell. 2011;10:38
7. Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C. et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380-8
8. Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C. et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060-76
9. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083-96
10. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097-109
11. Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y. et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110-25
12. Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M. et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142:529-36
13. Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H. et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303-10
14. Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour Biol. 2015;36:2947-55
15. Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH. et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809
16. Cao C, Sun J, Zhang D, Guo X, Xie L, Li X. et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415-26.e18
17. Eades G, Zhang YS, Li QL, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5:134-41
18. Jin K, Luo G, Xiao Z, Liu Z, Liu C, Ji S. et al. Noncoding RNAs as potential biomarkers to predict the outcome in pancreatic cancer. Drug Des Devel Ther. 2015;9:1247-55
19. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666-81
20. Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C. et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278-90
21. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113-21
22. Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A. et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911-23
23. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243-50
24. Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505-15
25. Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303-9
26. Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G. et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231-41
27. Wang XQ, Crutchley JL, Dostie J. Shaping the Genome with Non-Coding RNAs. Curr Genomics. 2011;12:307-21
28. Shi X, Sun M, Wu Y, Yao Y, Liu H, Wu G. et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol. 2015;36:503-13
29. Wang Y, He L, Du Y, Zhu P, Huang G, Luo J. et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413-25
30. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z. et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181-91
31. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol cell. 2014;53:88-100
Author contact
![]() Corresponding author: Dehua Wu, E-mail:wudhcom, fax: +86-020-61642131, and Dr. Li Liu, E-mail: liuli.fimmucom, fax: +86-020-616.
Corresponding author: Dehua Wu, E-mail:wudhcom, fax: +86-020-61642131, and Dr. Li Liu, E-mail: liuli.fimmucom, fax: +86-020-616.

 Global reach, higher impact
Global reach, higher impact