3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(6):618-625. doi:10.7150/jca.14030 This issue Cite
Research Paper
Increased APOBEC3B Predicts Worse Outcomes in Lung Cancer: A Comprehensive Retrospective Study
1. State Key Laboratory of Oncology in South China and Collaborative Innovation Center for Cancer Medicine, Guangzhou City, Guangdong Province, P.R. China;
2. Department of Pathology, Sun Yat-sen University Cancer Center, Guangzhou City, Guangdong Province, P.R. China;
3. Department of Molecular Oncology, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL 33612, USA;
4. State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou City, Guangdong Province, P.R. China;
5. Department of Biological Sciences, University of Illinois at Chicago, Chicago, Illinois 60607, USA.
* These authors contributed equally to this work and share the first authorship.
Received 2015-10-2; Accepted 2016-1-26; Published 2016-3-19
Abstract
Lung cancer ranks as the most common and lethal malignancy in America and worldwide. APOBEC3B is a newly identified DNA cytosine deaminase, which is supposed to function as a major source of DNA mutation in many different tumors. In this study, we combine the data of online databases and two hundred and twenty-one primary non-small-cell lung carcinoma (NSCLC) specimens from Sun Yat-sen University Cancer Center to investigate, for the first time, the clinical role of APOBEC3B in lung cancer. We found that the APOBEC3 expression was commonly elevated in NSCLC tissues and overexpression of APOBEC3B was correlated with unfavorable prognosis of the patients with NSCLC. Furthermore, APOBEC3B expression was associated with nodal status, TNM staging and adjuvant chemotherapy of the patients with NSCLC. Further research is warranted.
Keywords: APOBEC3B, non-small-cell lung cancer, immunohistochemistry, prognosis.
Introduction
Lung cancer ranks as the most common and lethal malignancy in America and worldwide. In 2013, there were 1.8 million people diagnosed with lung cancer and 1.6 million people died of it[1]. Although gene mutation, air pollution, tobacco use and other oncogenic factors have been proven closely associated with the tumorigenesis of lung cancer, the detailed etiology of lung cancer remains unclear. In lung cancer, the early detection of lung cancer is of great importance since there are hardly any symptoms initially. Advanced medical imaging techniques such as CT and MRI have revolutionized the early diagnosis of lung cancer, but these are too expensive for popularization in many countries [2]. So, the discovery of effective biomarkers might be a realistic approach. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 B (APOBEC3B, AP3B) is a newly defined DNA cytosine deaminase, which belongs to the APOBEC family. It has recently been reported that cytosine deamination resulting cytosine to thymine (C-to-T) transition mutation, catalyzed by APOBEC family, is a major source of DNA mutation in many different tumors [3]. AP3B is located on Chromosome 22 and has been found upregulated in at least 6 different cancers: bladder, cervix, lung, head and neck, and breast cancers [4]. In addition, AP3B's APOBEC3 cytidine deaminases activity has been proved associated with tumor promotion and metastasis [3, 5]. However, the clinical significance of AP3B remains largely unclear and has only been reported in breast cancer [6]. In this manuscript, we investigated, for the first time, whether the expression of AP3B is associated with lung cancer clinical outcomes, combining the results of online databases and two hundred and twenty-one primary non-small-cell lung carcinoma (NSCLC) specimens from Sun Yat-sen University Cancer Center (SYSUCC).
Materials and Methods
Patient selection
This study was approved by the medical ethics committee of SYSUCC. Two hundred and twenty-one primary NSCLC specimens obtained from primary surgery at SYSUCC from October 2000 to April 2007 were recruited in this study. The recruitment criteria are as follows: (a) newly diagnosed NSCLC without previous treatment; (b) histologically confirmed primary NSCLC; (c) no distant metastases, including supraclavicular or celiac lymph nodes metastases, were found; (d) frozen tissue and clinical data were available. To avoid confounding data analyses, patients with neoadjuvant chemotherapy were excluded from the study. The histologic grade and clinical stage of the tumors were defined according to the 7th edition of the TNM classification of the International Union Against Cancer (2009). The survival status of all of the patients were confirmed in December 2013.
Immunohistochemistry (IHC) staining
IHC staining was performed to detect AP3B expression according to the protocol previously described [7, 8]. The tissue sections were deparaffinized with dimethylbenzene and then rehydrated via a graded alcohol series. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 15 minutes. The slides were boiled in tris(hydroxymethyl) aminomethane-EDTA buffer (pH 8.0) in a microwave for 30 min to retrieve antigen. Nonspecific antigens were blocked with 10% normal goat serum for 20 min. Then, the slides were incubated with rabbit APOBEC3B antibody (PAB2474, 1:100 dilution, Abnova) overnight at 4°C in a moist chamber. The controls were treated by replacing the primary antibody with normal goat serum. On the second day, the slides were sequentially incubated with biotinylated rabbit anti-mouse antibody, streptavidin-peroxidase conjugate and 3'-3'diaminobenzidine. Normal alveolar and bronchial epithelial mucosa were utilized as negative controls.
Immunoreactivity score assessment (IRS)
Two independent pathologists (S-M Yan and Y Li) who were blinded to the clinicopathological information performed the IRS assessment for AP3B expression. The scoring criteria are as described previously [7, 9]. The staining result for each case was the average IRS decided by two pathologists. The specimens would be rescored if the difference between the two pathologists was greater than 3.
Statistical analysis
The AP3B IRS cutoff value is determined by median of the staining results of 221 specimens. The correlation between AP3B expression and clinicopathological features was analyzed by Pearson's χ2 test. Overall survival (OS) was defined as the time from surgery to death. Disease-free survival (DFS) was defined as the time from surgery to regional relapse or distant metastasis. The Cox proportional hazards model was used to calculate the hazard ratios (HRs) and their 95 % confidence intervals (95 % CIs) of covariates in the analyses of DFS or OS. Multivariate analysis was performed for all of the parameters that were significant in the univariate analysis. DFS and OS curves of 221 patients in subgroups were constructed by the Kaplan-Meier method. Log-rank test was used to test significant differences between two survival curves. Statistical analysis was performed using SPSS software (standard version 16.0, SPSS, Chicago, IL, USA). A two-sided probability value of less than 0.05 was considered statistically significant.
In addition, we also did online database searching to see results of some microarrays. In Kaplan Meier plotter (www.kmplot.com), we searched the OS curve of breast, lung, gastric and ovarian cancers based on AP3B expression. The patients recruitment criteria are the same as mentioned above except for the certain primary cancer. Log-rank test was used to test significant differences between survival curves.
Results
Patient characteristics
61 females and 160 males, aged from 30 to 79 years (median 59.0 years), were included in the study. The clinicopathological characteristics of the 221 patients are listed in Table 1.
Expression of AP3B in NSCLC
In this study, the median showed that the cutoff value was <8.0. IRS less than this value was considered low and, otherwise, high expression. In this study, AP3B staining of NSCLC tissue and normal alveolar and bronchial epithelial mucosa revealed that immunoreactivity primarily rest in cytoplasm of tumor cells (Fig. 1).
AP3B expression are determined by IHC. (A, B) No or low expression of AP3B protein in the cytoplasm of adjacent normal tissue (magnification: A, ×40; B, ×200). (C, D) Low expression level of AP3B in epithelial NSCLC tissues (magnification: C, ×40; D, ×200). (E, F) High expression levels of AP3B in epithelial NSCLC tissues (magnification: E, ×40; F, ×200). (G, H) Low expression level of AP3B in adeno NSCLC tissues (magnification: G, ×40; H, ×200). (I, J) High expression levels of AP3B in adeno NSCLC tissues (magnification: I, ×40; J, ×200).
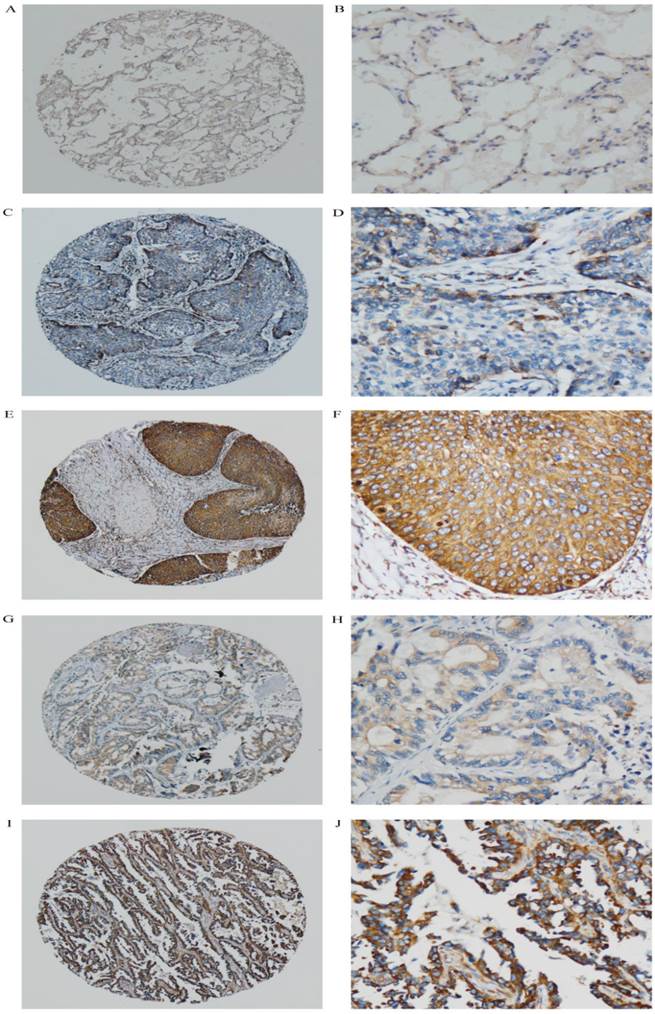
Low expression of AP3B was observed in 49.3% (109/221) of the NSCLC, whereas the high level expression of AP3B is 50.7% (112/221) (Table 1). The associations between clinicopathological features and AP3B expression are summarized in Table 1. The expression of AP3B correlated closely with the nodal status (P=0.017), TNM staging (P=0.046) and adjuvant chemotherapy (P=0.049). No statistical associations were observed between AP3B expression and age, gender, smoking, histology, visceral pleural invasion, tumor grade, tumor status (P=0.841, P=0.239, P=0.762, P=0.679, P=0.473, P= 0.171 and P=0.225 respectively).
Correlation between APOBEC3B expression and clinicopathological variables of NSCLC cases.
| Variables | Cases (n=221) | APOBEC3B protein | ||
|---|---|---|---|---|
| Low expression | High expression | P value | ||
| Age (years) | 0.841 | |||
| Median, Range | 59 30-79 | 60 32-78 | 59 30-79 | |
| <60 | 111 | 54(48.6) | 57(51.4) | |
| ≥60 | 110 | 55(50.0) | 55(50.0) | |
| Gender | 0.239 | |||
| Male | 160 | 75(46.9) | 85(53.1) | |
| Female | 61 | 34(55.7) | 27(44.3) | |
| Smoking | 0.762 | |||
| Yes | 132 | 64(48.5) | 68(51.5) | |
| No | 89 | 45(50.6) | 44(49.4) | |
| Histology | 0.679 | |||
| SCC | 78 | 37(47.4) | 41(52.6) | |
| Non-SCC | 143 | 72(50.3) | 71(49.7) | |
| Visceral Pleural Invasion | 0.473 | |||
| Absent | 68 | 36(52.9) | 32(47.1) | |
| Present | 153 | 73(47.7) | 80(52.3) | |
| Tumor grade | 0.171 | |||
| Grade1 | 31 | 19(61.3) | 12(38.7) | |
| Grade2 | 89 | 38(42.7) | 51(57.3) | |
| Grade3 | 101 | 52(51.5) | 49(48.5) | |
| Tumor status (T) | 0.225 | |||
| T1 | 41 | 25(61.0) | 16(39.0) | |
| T2 | 152 | 72(47.4) | 80(52.6) | |
| T3 | 20 | 10(50.0) | 10(50.0) | |
| T4 | 8 | 2(25.0) | 6(75.0) | |
| Nodal status (N) | 0.017* | |||
| N0 | 124 | 70(56.5) | 54(43.5) | |
| N>0 | 97 | 39(40.2) | 58(59.8) | |
| TNM Staging | 0.046* | |||
| I | 95 | 56(58.9) | 39(41.1) | |
| II | 62 | 26(41.9) | 36(58.1) | |
| III | 64 | 27(42.2) | 37(57.8) | |
| Adjuvant Chemotherapy | 0.049* | |||
| Yes | 104 | 44(42.3) | 60(57.7) | |
| No | 117 | 65(55.6) | 52(44.4) | |
NSCLC: non-small cell lung cance; * p < 0.05, statistically significant.
Non-SCC includes adenocarcinoma, adenosquamous carcinoma, anaplastic large cell carcinoma, adenoid cystic carcinoma, mucoepidermoid carcinoma and???
AP3B expression and survival
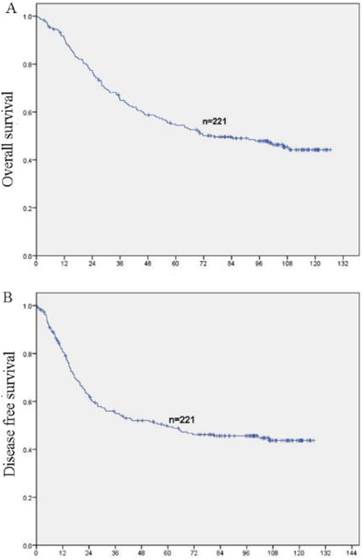
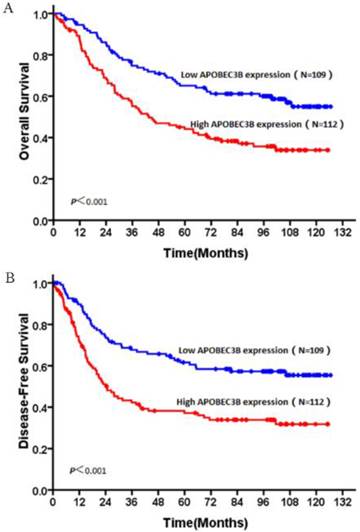
In this study, no patients were lost to follow-up. At the end of the follow-up, 106 (48%) patients were deceased and 115 (52%) patients were still alive. The 5 year survival rates of the whole cohort is 51.6% (114/221). OS and DFS curves for the whole cohort was shown in Figure 3. Patients with high expression of AP3B demonstrated shorter OS and DFS compared with those with low expression of AP3B (OS: mean of 63.6 months versus 88.6 months, P<0.001, DFS: mean of 54.4 months versus 84.4 months, P <0.001, Fig. 4, Table 2). AP3B can also be used to differentiate prognosis of patients in T1-2 categories, N=1/2/3 categories, G1 and G2/3 grades.
Prognostic significance of APOBEC3B expression in NSCLC patients by Kaplan-Meier survival analysis (log-rank test).
| Variable | Case | DFS (months) | OS (months) | ||||
|---|---|---|---|---|---|---|---|
| Mean | Median | P-value | Mean | Median | P-value | ||
| Total | <0.001* | <0.001* | |||||
| Low expression | 109 | 83.4 | NR | 88.6 | NR | ||
| High expression | 112 | 54.4 | 24.6 | 63.6 | 43.5 | ||
| T categories | |||||||
| T1-2 | <0.001* | <0.001* | |||||
| Low expression | 97 | 86.3 | NR | 91.8 | NR | ||
| High expression | 96 | 54.2 | 24.7 | 64.6 | 45.9 | ||
| T3-4 | 0.712 | 0.683 | |||||
| Low expression | 12 | 56.0 | 16.7 | 59.4 | 24.4 | ||
| High expression | 16 | 55.0 | 15.7 | 57.3 | 22.9 | ||
| N categories | |||||||
| N=0 | 0.027* | 0.061 | |||||
| Low expression | 70 | 98.8 | NR | 101.2 | NR | ||
| High expression | 54 | 77.5 | NR | 84.8 | NR | ||
| N=1/2/3 | 0.044* | 0.031* | |||||
| Low expression | 39 | 52.5 | 25.1 | 63.9 | 50.8 | ||
| High expression | 58 | 33.2 | 15.8 | 43.2 | 27.9 | ||
| Histologic grade | |||||||
| G1 | <0.001* | <0.001* | |||||
| Low expression | 19 | 111.2 | NR | 111.8 | NR | ||
| High expression | 12 | 43.5 | 22.0 | 48.2 | 22.9 | ||
| G2-3 | 0.008* | 0.017* | |||||
| Low expression | 90 | 76.4 | 106.2 | 82.8 | 108.7 | ||
| High expression | 100 | 55.6 | 24.7 | 65.1 | 45.1 | ||
NSCLC: non-small cell lung cance; DFS: disease free survival; OS: overall survival; NR: not reached; *:P<0.05.
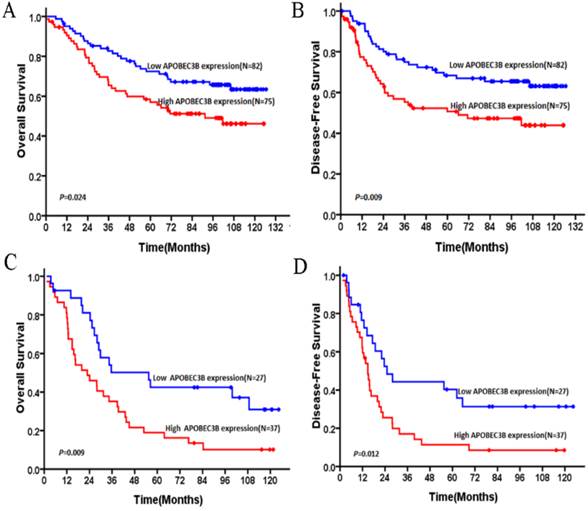
Furthermore, univariate analysis using Cox's proportional hazard model showed that the following parameters correlated significantly with OS: tumor grade, TNM staging and AP3B expression (Table 3). Further analysis was performed with regard to AP3B expression in subsets of NSCLC patients with different stages. The results demonstrated that high expression of AP3B was a prognostic factor in NSCLC patients with stage I + II (Figure 5A and B) and stage III (Figure 5C and D). To exclude the effect of covariate, AP3B expression and clinicopathologic parameters that were significant in univariate analysis were further analyzed in multivariate analysis. The result demonstrated that TNM staging and AP3B expression were independent prognostic factors that affected NSCLC patients' OS (Table 4). High expression of AP3B was a predictor for poor prognosis (hazard ratio, 1.823, 95%CI, 1.250-2.658, P = 0.002; Table 4). Analysis with DFS data were also shown in table 3 and 4.
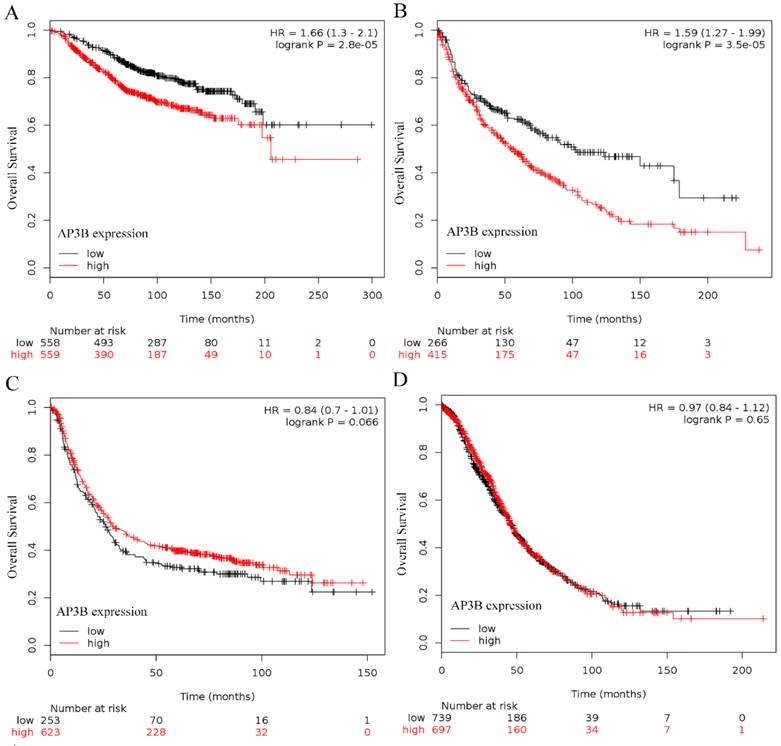
In terms of the results from online databases, 1117 patients were included in breast cancer and high expression of AP3B was a predictor for poor prognosis (hazard ratio, 1.66, 95%CI, 1.3-2.1, P = 2.8e-5; Fig 2A). 681 patients were included in lung cancer and high expression of AP3B was a predictor for poor prognosis (hazard ratio, 1.59, 95%CI, 1.27-1.99, P = 3.5e-5; Fig 2B). No clinical significance of AP3B was found in gastric and ovarian cancer cases (P=0.066 and 0.65, Fig 2C and 2D).
OS curve of breast, lung, gastric and ovarian cancer patients based on different levels of AP3B expression, data from online databases. (A) OS curves: breast cancer patients with high and low levels of AP3B expression. (B) OS curves: lung cancer patients with high and low levels of AP3B expression. (C) OS curves: gastric cancer patients with high and low levels of AP3B expression. (D) OS curves: ovarian cancer patients with high and low levels of AP3B expression.
OS and DFS curves of 221 NSCLC patients (A) OS curves for all patients. (B) DFS curves for all patients.
OS and DFS curves of patients with NSCLC based on their AP3B expression. (A) OS curves: all patients with low and high AP3B expression levels. (B) DFS curves: all patients with low and high AP3B expression levels.
Subgroup analysis of patients with NSCLC based on their AP3B expression. (A) OS curves: patients in TNM stage I + II with high and low levels of AP3B expression. (B) DFS curves: patients in TNM stage I + II with high and low levels of AP3B expression. (C) OS curves: patients in TNM stage III with high and low levels of AP3B expression. (D) DFS curves: patients in TNM stage III with high and low levels of AP3B expression.
Univariate Cox regression Analysis for Disease-free Survival and Overall Survival in Patients with Non-Small Cell Lung Cancer.
| Variables | Disease-free Survival | Overall Survival | ||
|---|---|---|---|---|
| HR(95%CI) | p value | HR(95%CI) | p value | |
| Age (years) | 0.997 | 0.681 | ||
| <60 | Reference | Reference | ||
| ≥60 | 1.001(0.694-1.443) | 1.080(0.749-1.557) | ||
| Gender | 0.559 | 0.759 | ||
| Male | 0.897(0.596-1.348) | 0.938(0.624-1.411) | ||
| Female | Reference | Reference | ||
| Smoking | 0.457 | 0.668 | ||
| Yes | 1.152(0.793-1.674) | 1.085(0.747-1.576) | ||
| No | Reference | Reference | ||
| Histology | 0.298 | 0.317 | ||
| SCC | Reference | Reference | ||
| Non-SCC | 1.231(0.832-1.821) | 1.221(0.826-1.807) | ||
| Visceral Pleural Invasion | 0.241 | 0.188 | ||
| Absent | Reference | Reference | ||
| Present | 1.280(0.847-1.932) | 1.319(0.874-1.992) | ||
| Tumor grade | 0.028 | 0.032 | ||
| Grade1 | Reference | Reference | ||
| Grade2 | 1.405(0.741-2.666) | 1.266(0.668-2.401) | ||
| Grade3 | 2.052(1.104-3.817) | 1.918(1.031-3.566) | ||
| TNM Staging | <0.001 | <0.001 | ||
| I | Reference | Reference | ||
| II | 2.332(1.430-3.803) | 2.307(1.414-3.762) | ||
| III | 4.165(2.655-6.533) | 3.950(2.524-6.183) | ||
| Adjuvant Chemotherapy | 0.024 | 0.124 | ||
| Yes | 1.527(1.058-2.203) | 1.333(0.925-1.922) | ||
| No | Reference | Reference | ||
| APOBEC3B expression | <0.001 | <0.001 | ||
| Low | Reference | Reference | ||
| High | 2.081(1.428-3.031) | 1.973(1.355-2.874) | ||
NSCLC: non-small cell lung cance; * p < 0.05, statistically significant.
Multivariate Cox regression Analysis for Disease-free Survival and Overall Survival in Patients with Non-Small Cell Lung Cancer.
| Variables | Disease-free Survival | Overall Survival | ||
|---|---|---|---|---|
| HR(95%CI) | p value | HR(95%CI) | p value | |
| Tumor grade(G1/2/3) | 1.299(0.964-1.751) | 0.086 | 1.275(0.947-1.718) | 0.110 |
| TNM Staging(I/II/III) | 1.919(1.514-2.433) | <0.001* | 1.874(1.499-2.347) | <0.001* |
| Adjuvant Chemotherapy (Yes/No) | 0.945(0.641-1.393) | 0.775 | ||
| APOBEC3B expression (low/High) | 1.922(1.315-2.809) | 0.001* | 1.823(1.250-2.658) | 0.002* |
NSCLC: non-small cell lung cance; * p < 0.05, statistically significant.
Discussion
Gene mutation is closely involved in oncogene activation, tumor suppressor gene suppression, deregulation of cell signaling, drug resistance and many other critical fields in cancer research. High frequency of cytidine to thymidine conversions have been revealed in the genome of several types of cancer cells, especially in DNA regions with ssDNA and double-strand DNA breaks [10]. Recently studies identified AP3B as a primary source of this type of mutations. Up till now, most of AP3B studies were restricted in genomic sequence level, trying to figure out the mutational power of AP3B [11-13]. Very few studies were implicated in clinical data and prognosis in protein level. We, for the first time, showed that overexpression of AP3B existed in lung cancer and is associated with worse prognosis. Our data also revealed that AP3B expression is correlated with nodal status, TNM staging and adjuvant chemotherapy, implicating the role of AP3B in the progression of NSCLC. Importantly, our data is perfectly consistent with the result from two online databases, which greatly enhanced its reliability.
Initially, AP3 proteins were identified as mutators of viral DNA. Through the synthesis of ssDNA intermediates, almost all AP3 proteins are involved in hypermutation state of viral genomes including retroviruses, hepatitis B virus and human papilloma virus [14, 15]. It is reported that the deletion of AP3B gene leads to the susceptibility to HBV infection and hepatocellular carcinoma [16]. However, the role of AP3B is not restricted in antiviral innate immunity. Recent studies indicated that AP3B is overexpressed in multiple kinds of cancers, including breast, head/neck, lung, bladder and cervical cancers [17]. In addition, genomic sequencing showed that C to T mutations in designated regions, which is regarded to be hallmarks of AP3B activity, are commonly found in many kinds of human cancers [3]. Knockdown of AP3B showed that cytosine deamination mutation is consistent and dependent upon AP3B level in breast and ovarian cancer cell lines, providing further evidence that AP3B is responsible for tumor mutagenesis [18]. Furthermore, scientists began to propose that AP3B functions as the source of intratumor heterozygosity. It is now clear that one solid tumor contains several sub clones of tumor cells, which can be identified with different genotypes. AP3B cytidine deaminase targeted genes' copy number alterations, translocations, and mutations were found closely related with intratumor heterogeneity, particularly prominent in lung adenocarcinomas [19]. All of these emerging evidence suggested that AP3B might act as a mutagenic driver of cancers. With a higher AP3B expression, the tumor cell subclone would accumulate overall mutation level and thus acquire advantages in tumorigenesis, evolution and metastasis. Consistently, our results showed that AP3B expression is associated with TNM staging and lymph node metastasis in NSCLC, which perfectly matches the mutagenic driver hypothesis.
Recently, scientists began to investigate the clinical significance of AP3B. It is reported that increased AP3B expression is associated with worse DFS, MFS, and OS in breast cancer. By detecting AP3B mRNA expression in more than 5000 breast cancer cases, they found that AP3B expression is a marker of pure prognosis and poor outcomes for ER positive breast cancer [6]. Another study showed variations in the expression of AP3B with elevated expression in ovarian cancer cell lines and high-graded ovarian cancers [20]. In chondrosarcoma, knockdown of AP3B expression led to higher percentage of apoptosis than control cells and antitumor activity of RUNX3 was reduced [21]. In our study, by the help of online microarray databases, we evaluated the role of AP3B expression in gastric, ovarian, breast and lung cancer. As shown in figure 2, AP3B expression is commonly elevated and is closely associated with patients' prognosis in breast and lung cancer cases. However, no prognostic significance was found in gastric and ovarian cancers. The data of gastric cancer seemed counterintuitive to the result of a recent study showing that AP3B expression correlated with the unfavorable prognosis of the patients with gastric cancer [22]. To verify this kind of survival significance of AP3B in lung cancer, we assessed the protein level of AP3B in NSCLC specimens and analyzed its prognostic significance by log-rank test. The result is, unsurprisingly, in accordance with online data and previous reports, showing that high AP3B expression predicts worse OS and DFS in NSCLC patients. What's more, we further divided the whole cohort into two subgroups as TNM stage I + II and stage III groups to do survival analysis separately. The results showed that AP3B expression is effective in predicting the prognosis of NSCLC patients in each group, indicating its prognostic significance in both early and late clinical stages. All of these survival analysis of AP3B, on one hand, provided clinical significance of AP3B in NSCLC and lung cancer. On the other hand, these data could also serve as supportive evidence for the mutagenic driver hypothesis of AP3B. Meanwhile, we admit that this study also has some limitations. For example, although we use online databases as verifications, all of the specimens for IHC came from one single institution might cause some geographical bias. Another disadvantage is that it is a retrospective study and no mechanism related experiments were included.
In conclusion, our local and online combined data demonstrate that lung cancer patients with high AP3B expression have worse OS and DFS compared with those with low AP3B expression. AP3B may be involved in the development and metastasis processes of lung cancer cells, and AP3B expression detected by IHC can be an independent predictor of survival for patients with NSCLC. However, further studies are required to elucidate underlying mechanisms.
Conflicts of interest
The authors declare that there is no conflict of interest.
References
1. Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M. et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-27
2. Lung cancer diagnosis. when speed wins the day. Lancet Respir Med. 2015;3:171
3. Nowarski R, Kotler M. APOBEC3 cytidine deaminases in double-strand DNA break repair and cancer promotion. Cancer Res. 2013;73:3494-8
4. Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977-83
5. Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A. 2015;112:2841-6
6. Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, Schlicker A. et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. 2014;5:405-13
7. Feng Y, He F, Wu H, Huang H, Zhang L, Han X. et al. GOLPH3L is a Novel Prognostic Biomarker for Epithelial Ovarian Cancer. J Cancer. 2015;6:893-900
8. Li XH, He F, Yan SM, Li Y, Cao Y, Huang CY. et al. Increased expression of stomatin-like protein 2 (STOML2) predicts decreased survival in gastric adenocarcinoma: a retrospective study. Med Oncol. 2014;31:763
9. Yan SM, Wu HN, He F, Hu XP, Zhang ZY, Huang MY. et al. High expression of zinc-binding protein-89 predicts decreased survival in esophageal squamous cell cancer. Ann Thorac Surg. 2014;97:1966-73
10. Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. 2015;5:704-12
11. Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N. et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487-91
12. Prohaska KM, Bennett RP, Salter JD, Smith HC. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. Wiley Interdiscip Rev RNA. 2014;5:493-508
13. Lada AG, Dhar A, Boissy RJ, Hirano M, Rubel AA, Rogozin IB. et al. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol Direct. 2012;7:47 discussion
14. Janahi EM, McGarvey MJ. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J Viral Hepat. 2013;20:821-8
15. Mori S, Takeuchi T, Ishii Y, Kukimoto I. Identification of APOBEC3B promoter elements responsible for activation by human papillomavirus type 16 E6. Biochem Biophys Res Commun. 2015;460:555-60
16. Zhang T, Cai J, Chang J, Yu D, Wu C, Yan T. et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22:1262-9
17. McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7:283ra54
18. Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B. et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366-70
19. de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251-6
20. Leonard B, Hart SN, Burns MB, Carpenter MA, Temiz NA, Rathore A. et al. APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 2013;73:7222-31
21. Jin Z, Han YX, Han XR. The role of APOBEC3B in chondrosarcoma. Oncol Rep. 2014;32:1867-72
22. Zhang J, Wei W, Jin HC, Ying RC, Zhu AK, Zhang FJ. The roles of APOBEC3B in gastric cancer. Int J Clin Exp Pathol. 2015;8:5089-96
Author contact
![]() Corresponding authors: Qiuliang Wu, MD, Department of Pathology, State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, No. 651 Dongfeng Road East, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China; Tel: +86- 20-87343268, E-mail: wuqlorg.cn Or Yong Li, MD, Department of Pathology, State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, No. 651 Dongfeng Road East, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China; Tel: +86- 20-87343268, E-mail: liyongorg.cn.
Corresponding authors: Qiuliang Wu, MD, Department of Pathology, State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, No. 651 Dongfeng Road East, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China; Tel: +86- 20-87343268, E-mail: wuqlorg.cn Or Yong Li, MD, Department of Pathology, State Key Laboratory of Oncology in South China, Sun Yat-Sen University Cancer Center, No. 651 Dongfeng Road East, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, P.R. China; Tel: +86- 20-87343268, E-mail: liyongorg.cn.

 Global reach, higher impact
Global reach, higher impact