3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(6):650-655. doi:10.7150/jca.13460 This issue Cite
Review
miR-196, an Emerging Cancer Biomarker for Digestive Tract Cancers
1. Department of Medical Biotechnology and Laboratory science, College of Medicine, Chang Gung University, Taoyuan 333, Taiwan;
2. Department of Radiation Oncology, Chang Gung Memorial Hospital, Taoyuan 333, Taiwan;
3. Department of Thoracic Surgery, Chang Gung Memorial Hospital, Taoyuan 333, Taiwan;
4. Department of General Surgery, Chang Gung Memorial Hospital, Taoyuan 333, Taiwan;
5. Department of Colorectal Surgery, Chang Gung Memorial Hospital, Taoyuan 333, Taiwan.
*First and second authors contributed equally.
Received 2015-8-5; Accepted 2016-2-11; Published 2016-3-20
Abstract
Over the past decade, the emergence of microRNA (miRNA) research has firmly established this molecular family as a key component in cells. MiRNAs, which function as negative gene regulators, participate in multiple biological processes and maintain homeostasis in cells. The dysregulation of miRNA may contribute to numerous human disorders, including cancer. Recently, miR-196 was found to be aberrantly expressed in a wide range of malignant diseases, which suggests that it plays important roles in carcinogenesis. Here, we summarize the current knowledge concerning miR-196 family in cancers. This review includes miR-196 gene structure and aberrant expression in various cancers, and current understanding of numerous functions and regulatory targets of miR-196 in specific cancers. Since miR-196 are consistently found over-expressed in digestive tract cancer tissues, we also reviewed the clinical significance and potential applications of miR-196 in these cancers. We highlight that miR-196 may serve as an emerging cancer biomarker for digestive tract cancers.
Keywords: miR-196, cancer, clinical application, regulatory mechanism.
Introduction
Over the past decade, the emergence of miRNA research has firmly established this molecular family as a key component in cells. Under normal physiological conditions, miRNAs implement feedback mechanisms by gene transcripts to regulate cell proliferation, differentiation and apoptosis [1-3]. The dysregulation of miRNAs may facilitate malignant transformation, including tumor initiation, growth, invasion, and therapeutic resistance [1-3]. Recently, several investigations have reported that miR-196 is frequently altered in various cancers, which suggests that it plays an important role in carcinogenesis. Here, we review recent reports on miR-196 family molecules in cancer, whose topics include gene structure, biological functions, and regulatory targets and clinical significance. We highlight that miR-196 may serve as an emerging cancer biomarker for digestive tract cancers.
miRNA discovery, biogenesis and mechanism
miRNAs are endogenous, non-coding small RNA that are approximately 22 nt in length. After the first miRNA was discovered in C. elegans, in 1993, the knowledge of miRNA grew rapidly [1]. Currently, more than 2,500 potential human miRNAs are recorded in the miRBase, and more than 30% of all genes are estimated to be regulated by miRNAs. This family of molecules has important and broad functions in cells, including proliferation, differentiation and apoptosis [2-6].
MiRNAs are encoded in the genome. Their long primary transcripts are produced by RNA polymerase II and are further processed by Drosha and DGCR8, which results in 80- to 100-nt precursor miRNAs (pre-miRNAs) [7, 8]. The pre-miRNAs are transported to the cytoplasm by Expotin5 and are cleaved by Dicer to form double-stranded miRNA duplexes. After unwinding into single-stranded mature miRNAs, the miRNAs bind to Argonaute (Ago2) protein and form the RNA-induced silencing complex (RISC). This complex then binds to the target-gene mRNA, and carries out mRNA degradation or translational repression [2, 9]. More specifically, the mature miRNA recognizes its complementary sequence in the target mRNA 3'-untranslational region (3'-UTR) via the “seed” region of the 5'-miRNA at nucleotide positions 2-7. Recent reports have revealed that the target mRNAs may also be repressed by miRNAs through binding to the coding regions or 5'-UTR [10, 11].
MiRNAs play as negative gene regulators in cells and function based on their target transcripts. Through algorithm analysis of miRNA target prediction, plenty of miRNA-gene relationships have been presumed [12, 13]. Increasing evidences have shown that a single miRNA is able to regulate several transcripts, and a transcript may have multiple miRNA binding sites which allow several miRNAs to bind at a same time [14]. This phenomenon implies a complexity of miRNA-target regulatory network in cellular homeostasis. The dysregulation of miRNA may contribute to numerous disorders, including cancer, immune-related and neurodegenerative diseases [15].
Aberrant expression of miR-196 in various cancers
MiRNAs are frequently found located at the genomic fragile sites or the oncogenic associated regions, which suggests that miRNA molecules participate in malignant transformation [16, 17]. Accumulating studies have demonstrated that miRNAs can serve as either tumor suppressors or oncogenes [18-20]. Recently, miR-196 has been found aberrantly expressed in many cancers. There are three miR-196 genes in human cells: miR-196a-1, miR-196a-2, and miR-196b. They are all located in the HOX gene clusters and produce two mature miRNA family members: miR-196a and miR-196b [21]. Table 1 shows the miR-196 family genes and their mature sequences. The miR-196a-1 and miR-196a-2 genes are located on chromosomes 17 and 12, respectively [22]. Both of these genes transcribe the same mature miR-196a molecule, which has the sequence 5'-UAGGUA- GUUUCA- UGUUGU- UGGG-3'. The miR-196b gene is located on chromosome 7 and transcribes a mature miR-196b molecule with the sequence 5'-UAGGUA- GUUUCC- UGUUGU- UGGG-3' [23]. There is only one nucleotide difference between mature miR-196a and miR-196b, though both possess the same seed region.
Although high similarity exists between miR-196a and miR-196b, these two molecules have a wide range of expression and diverse roles in various types of cancers. For example, miR-196a has been reported to be over-expressed in head-neck [24], oral [25, 26], gastric [27, 28], pancreatic [29, 30], cervical [31, 32], and lung [33] cancers. Similarly, miR-196b has been found to be up-regulated in the cancers of oral [25], gastric [34], colorectal [35, 36], as well as in glioblastoma [37] and in leukemia [23, 38]. The functions of the oncogenic effects of miR-196 family molecules include promoting cell proliferation, migration, invasion and radioresistance [21-34]. However, few reports have indicated that the miR-196 family may act as tumor suppressors. For example, miR-196a suppresses metastasis in melanoma [39] and breast cancers [40], and miR-196b is down-regulated in different types of leukemia cells [41, 42]. Collectively, both miR-196a and miR-196b are frequent aberrations in malignant diseases, and which indicates their close association with disease status.
The information of miR-196 family genes and the mature miR-196 molecules.
| miRNA gene | Accession No. | Gene location | Stem-loop sequence | Mature miRNA | ID number (Mature miRNA) | Accession No. (Mature miRNA) | Mature miRNA Sequence (Seed#) |
|---|---|---|---|---|---|---|---|
| hsa-mir-196a-1 | MI0000238 | Chr17: 48632490-48632559 | GUGAAUUAGGUAGUUUCAUGUUGUUGGGCCUGGGUUUCUGAACACAACAACAUUAAACCACCCGAUUCAC | hsa-mir-196a-5p | hsa-miR-196a | MIMAT0000226 | UAGGUAGUUUCAUGUUGUUGGG |
| hsa-mir-196a-2 | MI0000279 | Chr12: 53991738-53991738 | UGCUCGCUCAGCUGAUCUGUGGCUUAGGUAGUUUCAUGUUGUUGGGAUUGAGUUUUGAACUCGGCAACAAGAAACUGCCUGAGUUACAUCAGUCGGUUUUCGUCGAGGGC | hsa-mir-196a-5p | hsa-miR-196a | MIMAT0000226 | UAGGUAGUUUCAUGUUGUUGGG |
| hsa-mir-196a-3p | hsa-miR-196a* | MIMAT0004562 | CGGCAACAAGAAACUGCCUGAG | ||||
| hsa-mir-196b | MI0001150 | Chr7: 27169480-27169563 | ACUGGUCGGUGAUUUAGGUAGUUUCCUGUUGUUGGGAUCCACCUUUCUCUCGACAGCACGACACUGCCUUCAUUACUUCAGUUG | hsa-miR-196b-5p | hsa-miR-196b | MIMAT0001080 | UAGGUAGUUUCCUGUUGUUGGG |
| hsa-miR-196b-3p | hsa-miR-196b* | MIMAT0009201 | UCGACAGCACGACACUGCCUUC |
# Seed regions are underlined in the mature miRNA sequences.
Summary of the target genes and functions of miR-196 family molecules in diverse cancers.
| miR-196 | Cancers | Family member | Target genes | Function | References |
|---|---|---|---|---|---|
| over- expressed | Head-neck cancer | miR-196a | Annexin A1 | Proliferation, invasion, radioresistance | [24] |
| Oral cancer | miR-196a, miR-196b | NME4 | Migration, invasion | [25, 26] | |
| Esophageal cancer | miR-196a | Annexin A1 | Cell growth | [43] | |
| Gastric cancer | miR-196a,miR-196b | p27kip1,HOXA10,Radaxin | Proliferation, metastasis | [27, 34, 44] | |
| Colorectal cancer | miR-196a, miR-196b | HOX(A7/B8/C8/D8), FAS | Mobility, apoptosis | [36, 45] | |
| Pancreatic cancer | miR-196a | NFKBIA | Proliferation, migration | [29, 30] | |
| Non-small cell lung cancer | miR-196a | HOXA5 | Proliferation, invasion | [33] | |
| Cervical cancer | miR-196a | netrin 4/FOXO1/ p27kip1 | Proliferation, migration | [31, 32] | |
| Acute myeloid leukemia | miR-196a, miR-196b | ERG | Hematopoiesis | [23, 38] | |
| Glioblastoma | miR-196b | -- | Proliferation | [37] | |
| under- expressed | Acute lymphoblastic leukemia | miR-196b | c-Myc | --- | [38] |
| Melanoma | miR-196a | HOXB7/BMP4/HOXC8 | Suppression of migration and invasion | [39, 48] | |
| Chronic myeloid leukemia | miR-196b | BCR-ABL1/HOXA9 | --- | [41] |
Numerous functions of miR-196 in different cancers may be due to various gene targets
Because miRNAs function as gene regulators, identifying miRNA targets allows delineation of the specific function of a miRNA. Several computational software packages have been developed to predict potential miRNA targets by aligning the conserved seed region of a miRNA to the complementary sequences on the 3'-UTR region of a mRNA [43]. Many experiments have further confirmed the regulatory axis. Table 2 summarizes the expression, target genes, and functions of miR-196 family molecules in diverse cancers. The oncogenic functions of these molecules have been found to target diverse tumor suppressors in specific types of cancer tissues. For example, in oral cancer, both miR-196a and miR-196b down-regulate the expression of metastasis suppressor NME4 to accelerate cell migration and invasion [25]. In esophageal and head-neck cancers, miR-196a targets Annexin A1 to promote cell growth, mobility and radioresistance [24, 44]. In gastric cancer, miR-196a down-regulates p27kip1, a cell cycle G1 check-point regulator, to promote cell proliferation [27]. Also in gastric cancer, both miR-196a and miR-196b promote cell metastasis via suppression of radixin [45]. In pancreatic cancer, miR-196a targets NFκB-Iα to promote cancer progression [29]. In colorectal cancer, miR-196a restrict the expression of HoxA7, HoxB8, HoxC8 and HoxD8 genes, leading to the activation of the AKT pathway and increase cell mobility [46]. A high level of miR-196b in colorectal cancer also repressed FAS expression to modulate cell apoptosis [36]. In cervical cancer, miR-196a inhibits p27kip1, FOXO1 or netrin 4 to facilitate cell proliferation or migration [31, 32]. Nevertheless, few studies have reported on the function of the miR-196 family as a tumor suppressor by targeting oncogenes. For example, in melanoma and breast cancer, miR-196a suppresses HOXC8, a transcription factor, to inhibit cell growth, invasion and metastasis [47, 48]. In chronic myeloid leukemia, miR-196b is down-regulated in cells and enhances the functions of oncogenes BCR-ABL1 and HOXA9 [41].
Aside from the miRNA-downstream target gene axis, the upstream regulatory mechanism of miR-196 has also been reported, mainly through epigenetic associated mechanism [49]. The CpG islands in the miR-196b promoter showed more methylation in chronic myeloid leukemia patients than healthy individuals [41]. In gastric cancer, miR-196b has been found regulated by Est2 through an epigenetic pathway [50, 51]. Furthermore, miR-196a2 gene possesses single nucleotide polymorphism in many cancers, including in breast, head-neck, lung and oral cancers, which may result to target binding deficit and increasing cancer susceptibility [26, 52-58]. Taking together, miR-196 family molecules play important roles in homeostasis, and dysregulation of this family closely associated with various malignancy.
Over-expression of miR-196 is commonly found in digestive tract cancer tissues and associated with aggressive clinicopathological status
Although vary in level, miR-196 family molecules are consistently found over-expressed in digestive tract cancer tissues, which comprise of mouth, esophagus, stomach, small intestine, and large intestine. Table 3 summarizes the clinicopathological association of miR-196 family molecules in digestive tract cancers. On the differentiation of normal and cancer tissues, both miR-196a and miR-196b are significantly elevated in oral cancer tissues compared to the adjacent normal counterparts [25], indicating that miR-196 may be a putative diagnostic marker. Consistently, miR-196a is up-regulated in laryngeal cancer and able to discriminate normal and cancer tissues [59]. In esophagus, miR-196a shows correlative expression during cancer progression, as gradually higher from normal tissue, metaplasia, dysplasia, to invasive adenocarcinoma [60]. In addition to the diagnostic potential, miR-196 family molecules have also been reported associated with clinicopathological status of the digestive tract cancers, which implies the potential in using of these molecules in prognostic application. For examples, both miR-196a and miR-196b are up-regulated in oral cancer tissues and associated with lymph node metastasis [25]. The high level of miR-196a in head-neck cancer tissue also contributes to radioresistance [24]. In gastric cancer, miR-196a or miR-196b is linked to advanced pathologic stage, cancer recurrence and shorter survival [27, 34, 45, 61]. Concordantly, both miR-196a and miR-196b in colorectal cancer shows cooperatively correlated with aggressive disease and unfavorable to treatment outcome [35]. Taken together, miR196 family molecules in digestive tract cancers may serve as biomarkers for clinical applications in diagnosis, prognosis, or prediction of treatment efficacy.
Circulating miR-196 may serve as a novel cancer biomarker for digestive tract cancers
Detecting biomolecules in body fluids, including peripheral blood, urine or saliva, provides a less invasive process to facilitate clinical applications. Recently, many reports have revealed that miRNAs can be exosomic released from tumor cells, stably expressed in circulation, which may be suitable to be utilized as promising biomarkers [62-65]. Regarding miR-196 family, several studies have shown that miR-196 molecules are detectable in the peripheral blood from digestive tract cancer patients and associated with clinical significance. For example, miR-196a and miR-196b are significantly elevated in the sera of patients with chronic pancreatitis or pancreatic cancer compared to those from normal individuals [66, 67]. Concordantly, the combined determination of miR-196a and miR-196b produces high sensitivity and specificity for the early detection of pre-malignant or malignant oral diseases [68]. Circulating miR-196a also showed high potential to be a prognostic marker because it correlated with patient survival in recurrence status of gastric cancer [28] or in pancreatic cancer [67].
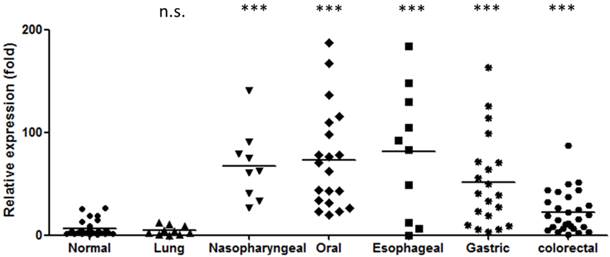
As stated above, miR-196 is elevated in the blood of patients with several types of digestive tract cancers. However, these results were obtained from various investigators who used different assay protocols which may lead to possible bias. To assess the diagnostic potential, we compared the miR-196b level in the plasma from healthy individuals and from patients with several types of cancers, including nasopharyngeal, oral, esophageal, gastric, colorectal and lung cancers. The results are shown in Figure 1. MiR-196b was substantial increased in digestive tract cancer patients, with an average elevation of 3.7- to 25.6-fold (P < 0.001 for all). The detection power of miR-196b for each cancer is summarized in Table 4 (Youden Index [69]). Such high ability to discriminate between healthy individuals and cancer patients indicates that miR-196b may serve as a powerful circulating tumor marker for the detection of digestive tract cancers.
Summary of the clinicopathological significance of miR-196 family molecules in digestive tract cancers.
| Cancers. | Family member | Diagnostic potential | Clinicopathological or prognostic association | References |
|---|---|---|---|---|
| Oral cancer #* | miR-196a, miR-196b | Differentiate normal, pre-cancer and cancer | Associated with lymph node metastasis | [25][68] |
| Head-neck cancer # | miR-196a | Differentiate normal and cancer | Associated with radioresistance | [59, 24] |
| Esophageal cancer # | miR-196a | Differentiate normal, pre-cancer and cancer | -- | [60] |
| Gastric cancer #* | miR-196a,miR-196b | -- | Associated with recurrence and shorter survival | [27,28,34,45,61] |
| Colorectal cancer # | miR-196a, miR-196b | -- | Associated with shorter survival | [35] |
| Pancreatic cancer* | miR-196a | Differentiate normal and cancer | Associated with recurrence and shorter survival | [66, 67] |
#Determination of miR-196 in tissue specimens, * Determination of miR-196 in blood specimens.
The statistical result for each cancer (Mann-Whitney test).
| Plasma from | Sample number | Relative value (Mean) | SEM | P-value (w/normal) | Area under ROC curve (AUC) | (%) Sensitivity/ (%) specificity** |
|---|---|---|---|---|---|---|
| Normal | 30 | 6.1 | 1.345 | -- | -- | -- |
| Lung cancer | 10 | 4.5 | 1.398 | N.S. | 0.552 | 30/97 |
| Oral cancer | 20 | 73.5 | 11.00 | <0.0001 | 0.990 | 100/94 |
| Nasopharyngeal carcinoma | 9 | 67.7 | 11.68 | <0.0001 | 0.996 | 100/97 |
| Esophageal carcinoma | 10 | 156.4 | 55.74 | 0.0008 | 0.858 | 70/100 |
| Gastric cancer | 20 | 78.0 | 24.97 | <0.0001 | 0.924 | 95/77 |
| Colorectal cancer | 28 | 22.6 | 3.806 | <0.0001 | 0.821 | 82/77 |
N.S.: No statistical significance by Mann-Whitney test.
**: The optimal cutoff threshold for diagnosis was obtained by applying the Youden's Index (sensitivity + specificity -1 is maximal) [69].
Elevation of miR-196b levels in the plasma of patients with digestive tract cancer. (n.s.: no statistical significance compared to normal group Mann-Whitney test, ***: P < 0.001 compared to normal group Mann-Whitney test).
Concluding remarks
MiR-196 offers a great potential as a biomarker for cancer detection, diagnosis, prognosis, and therapeutic assessment via both tumor tissues and circulating specimens. It will be imperative to carry out prospective trials in well-defined, large cohort studies to validate its clinical significance. Determining the miR-196 level by using a minimally invasive method may be developed and applied in personalized cancer medicine.
Acknowledgements
This work was supported by grants from National science Counsel (NSC-103-2314-B- 182A-011-MY3), and Chang Gung Memorial Hospital (CMRPG1B0101~3).
Competing interests
The authors have declared that no competing interest exists.
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;755:843-54
2. Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res. 2011;7171-2:1-8
3. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38
4. Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;156:546-54
5. Li H, Yang BB. Friend or foe: the role of microRNA in chemotherapy resistance. Acta Pharmacol Sin. 2013;347:870-9
6. Cellini F, Morganti AG, Genovesi D, Silvestris N, Valentini V. Role of microRNA in response to ionizing radiations: evidences and potential impact on clinical practice for radiotherapy. Molecules. 2014;194:5379-401
7. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;1824:3016-27
8. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;65:376-85
9. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;158:509-24
10. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008:4557216 1124-8
11. Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009;197:1175-83
12. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;1157:787-98
13. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;151:1-19
14. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;1201:15-20
15. Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomic Proteom Bioinform. 2012;105:246-53
16. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;1019:2999-3004
17. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;611:857-66
18. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;3021:1-12
19. Li M, Li J, Ding X, He M, Cheng SY. microRNA and cancer. AAPS J. 2010;123:309-17
20. Ahmad J, Hasnain SE, Siddiqui MA, Ahamed M, Musarrat J, Al-Khedhairy AA. MicroRNA in carcinogenesis & cancer diagnostics: a new paradigm. Indian J Med Res. 2013;1374:680-94
21. Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;151:14-23
22. Tanzer A, Amemiya CT, Kim CB, Stadler PF. Evolution of microRNAs located within Hox gene clusters. J Exp Zoolog B Mol Dev Evol. 2005;3041:75-85
23. Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, Chen J, Rowley JD, Zeleznik-Le NJ. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;11314:3314-22
24. Suh YE, Raulf N, Gaken J, Lawler K, Urbano TG, Bullenkamp J, Gobeil S, Huot J, Odell E, Tavassoli M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int J Cancer. 2014
25. Lu YC, Chang JT, Liao CT, Kang CJ, Huang SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, Wang HM, Yen TC, You GR. et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 2014;131:218
26. Liu CJ, Tsai MM, Tu HF, Lui MT, Cheng HW, Lin SC. miR-196a overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann Surg Oncol. 2013;20(Suppl 3):S406-14
27. Sun M, Liu XH, Li JH, Yang JS, Zhang EB, Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, Wang ZX, Cao XF, De W. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol Cancer Ther. 2012;114:842-52
28. Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, Huang KH, Lin WC. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes Chromosomes Cancer. 2012;514:394-401
29. Huang F, Tang J, Zhuang X, Zhuang Y, Cheng W, Chen W, Yao H, Zhang S. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS One. 2014;92:e87897
30. Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;29:807-13
31. Zhang J, Zheng F, Yu G, Yin Y, Lu Q. miR-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells. Biochem Biophys Res Commun. 2013;4404:582-8
32. Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;1105:1260-8
33. Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, De W, Wang ZX. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348
34. Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, Lee JS, Cho JY. Overexpression of miR-196b and HOXA10 characterize a poor-prognosis gastric cancer subtype. World J Gastroenterol. 2013;1941:7078-88
35. Ge J, Chen Z, Li R, Lu T, Xiao G. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;141:128
36. Mo JS, Alam KJ, Kang IH, Park WC, Seo GS, Choi SC, Kim HS, Moon HB, Yun KJ, Chae SC. MicroRNA 196B regulates FAS-mediated apoptosis in colorectal cancer cells. Oncotarget. 2015;65:2843-55
37. Ma R, Yan W, Zhang G, Lv H, Liu Z, Fang F, Zhang W, Zhang J, Tao T, You Y, Jiang T, Kang X. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One. 2012;76:e38096
38. Coskun E, von der Heide EK, Schlee C, Kuhnl A, Gokbuget N, Hoelzer D, Hofmann WK, Thiel E, Baldus CD. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leukemia Res. 2011;352:208-13
39. Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. 2010;6720:3535-48
40. Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, Thangaraju M, Huang S. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;7020:7894-904
41. Liu Y, Zheng W, Song Y, Ma W, Yin H. Low expression of miR-196b enhances the expression of BCR-ABL1 and HOXA9 oncogenes in chronic myeloid leukemogenesis. PLoS One. 2013;87:e68442
42. Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;3401-2:97-106
43. Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;2672:529-35
44. Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;2752:6667-78
45. Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Lin KH. MicroRNA-196a/-196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett. 2014
46. Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;1517:2089-96
47. Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;125:210
48. Mueller DW, Bosserhoff AK. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer. 2011;1295:1064-74
49. Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C. et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;674:1424-9
50. Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC, Li SC, Lai CH, Ho MR, Fang WL, Huang KH, Lin WC. Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis. 2012;334:760-9
51. Tsai KW, Hu LY, Wu CW, Li SC, Lai CH, Kao HW, Fang WL, Lin WC. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosomes Cancer. 2010;4911:969-80
52. Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;1187:2600-8
53. Tian T, Shu YQ, Chen JP, Hu ZB, Xu L, Jin GF, Liang J, Liu P, Zhou XY, Miao RF, Ma HX, Chen YJ, Shen HB. A Functional Genetic Variant in microRNA-196a2 Is Associated with Increased Susceptiblility of Lung Cancer in Chinese. Cancer Epidem Biomar. 2009;184:1183-7
54. Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, Sun AM, Liu Y. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res. 2011;422:144-8
55. Wang F, Ma YL, Zhang P, Yang JJ, Chen HQ, Liu ZH, Peng JY, Zhou YK, Qin HL. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep. 2012;391:269-75
56. Li T, Niu LJ, Wu L, Gao X, Li M, Liu WX, Yang L, Liu DW. A functional polymorphism in microRNA-196a2 is associated with increased susceptibility to non-Hodgkin lymphoma. Tumor Biology. 2015;365:3279-84
57. Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;6914:5970-7
58. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, McClean MD, Marsit CJ, Kelsey KT. Mature microRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res. 2010;1614:3713-20
59. Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H, Okubo K, Onozaki T, Zama T. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One. 2013;88:e71480
60. Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A. et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;1745:1940-8
61. Mu YP, Tang S, Sun WJ, Gao WM, Wang M, Su XL. Association of miR-193b down-regulation and miR-196a up-regulation with clinicopathological features and prognosis in gastric cancer. Asian Pac J Cancer Prev. 2014;1520:8893-900
62. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;88:467-77
63. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;10530:10513-8
64. Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;103:297-308
65. Hu G, Drescher KM, Chen XM. Exosomal miRNAs: Biological Properties and Therapeutic Potential. Front Genet. 2012;3:56
66. Slater EP, Strauch K, Rospleszcz S, Ramaswamy A, Esposito I, Kloppel G, Matthai E, Heeger K, Fendrich V, Langer P, Bartsch DK. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl Oncol. 2014;74:464-71
67. Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Digest Dis Sci. 2011;562:602-9
68. Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY, Huang BS, Chen YJ, Li HF, Cheng AJ. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 2015;483:115-21
69. Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J. 2008;503:419-30
Author contact
![]() Corresponding author: Ann-Joy Cheng, Professor, Department of Medical Biotechnology, College of Medicine, Chang Gung University, 259 Wen-Hwa 1st Road, Taoyuan 333, Taiwan. Tel: 886-3-2118800 ex 5085 Fax: 886-3-2118247 E-mail: annjoychengcgu.edu.tw.
Corresponding author: Ann-Joy Cheng, Professor, Department of Medical Biotechnology, College of Medicine, Chang Gung University, 259 Wen-Hwa 1st Road, Taoyuan 333, Taiwan. Tel: 886-3-2118800 ex 5085 Fax: 886-3-2118247 E-mail: annjoychengcgu.edu.tw.

 Global reach, higher impact
Global reach, higher impact