3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(7):768-773. doi:10.7150/jca.13894 This issue Cite
Research Paper
HOXC9 Induces Phenotypic Switching between Proliferation and Invasion in Breast Cancer Cells
1. Department of Surgery, National Health Insurance Service Ilsan Hospital, Goyang 410-719, Korea;
2. Department of Anatomy, Embryology Laboratory, and Brain Korea 21 PLUS project for Medical Science, Yonsei University College of Medicine, Seoul 120-752, Korea.
*These authors contributed equally to this work.
Received 2015-9-18; Accepted 2016-3-20; Published 2016-4-10
Abstract
HOX genes encode a family of transcriptional regulators that are involved in pattern formation and organogenesis during embryo development. In addition, these genes play important roles in adult tissues and some of the dysregulated HOX genes are associated with cancer development and metastasis. Like many other HOX genes, HOXC9 is aberrantly expressed in certain breast cancer cell lines and tissues; however, its specific functions in breast cancer progression were not investigated. In the present study, we demonstrated that HOXC9 overexpression in breast cancer cell lines such as MDA-MB-231 and MCF7 increased the invasiveness but reduced the proliferation of cells, resembling a phenotype switch from a proliferative to an invasive state. Furthermore, the reciprocal result was detected in MCF7 and BT474 cells when the expression level of HOXC9 was reduced with siRNA. The clinical impact of HOXC9 in breast cancer was interpreted from the survival analysis data, in which high HOXC9 expression led to considerably poorer disease-free survival and distant metastasis-free survival, especially in lymph node-positive patients. Together, the prognostic relevance of HOXC9 and the HOXC9-derived phenotypic switch between proliferative and invasive states in the breast cancer cell lines suggest that HOXC9 could be a prognostic marker in breast cancer patients with lymph node metastasis and a target for therapeutic intervention in malignant breast cancer.
Keywords: HOXC9, breast cancer, metastasis, proliferation, invasion.
Introduction
HOX genes are homeobox genes that function as transcription factors. In humans, a total of 39 HOX genes have been assigned to 13 paralogous groups in four separate clusters termed HOXA, HOXB, HOXC, and HOXD. Much is known about HOX gene structure and molecular functions of HOX protein in determining body patterning during embryogenesis; in addition to this, many studies have also revealed important roles of HOX genes in adult tissues and cancer [1-3]. In particular, several Hox genes, such as Hoxb6, Hoxc6, Hoxb7, Hoxa9, Hoxb9, Hoxd4, and Hoxd9, were identified as being differentially expressed in a spatial and temporal manner in the mammary glands of subadult and pregnant animals [4, 5].
The murine Hox paralog group 9 genes, Hoxa9, Hoxb9, and Hoxd9 have been reported to function together to control the expansion and/or differentiation of the mammary epithelium ductal system in pregnancy [6]. Previous study has shown that HOXA9 is significantly downregulated in breast cancer and is correlated with disease aggressiveness [7]. In contrast, HOXB9 is overexpressed in breast cancer and promotes disease progression [8]. Aberrant expression of HOXD9 in breast cancer has also been reported [9, 10]; however, its functional consequences had not been investigated yet. HOXC9, another member of the HOX paralog 9 group, is also expressed in adult mammary glands; however, its specific roles in the development of mammary tissue after pregnancy as well as in breast cancer have not yet been determined.
A previous report analyzing DNA methylation patterns demonstrated that the CpG island at the promoter region of HOXC9 was methylated in primary breast cancer samples, implying that HOXC9 might be silenced in this condition [11]. However, we previously found that HOXC9 gene expression was upregulated in breast cancer [12]. This discrepancy may imply that the expression of HOXC9 and its functional role could vary depending on cancer type or aggressiveness. Therefore, in this study, the roles of HOXC9 in breast cancer biology were determined by studying its effect on cell proliferation and invasion.
Materials and Methods
Cell culture, plasmids, siRNA, and transfection
MCF7, BT474, and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; WelGENE Inc., Daegu, Korea) supplemented with 10% fetal bovine serum (FBS; WelGENE Inc.) and 1× antibiotic-antimycotic solution (WelGENE Inc.). For overexpression studies, a pCMV6 expression vector containing full-length cDNA of the HOXC9 gene and Myc-DDK-tag (#RC208833; Origene, Rockville, MD, USA) was transfected in MDA-MB-231 cells using Attractene reagent (Qiagen). As a control, pCMV6 empty vector was transfected. The cells were treated with G418 (Gibco; 300 μg/ml) for 2-3 weeks to generate stable cell lines. The same set of plasmid vector was used for transient transfection of MCF7 cells. For the knockdown experiments, MCF7 and BT474 cells were transfected with ON-TARGET plus SMARTpool siRNA targeting HOXC9 (#L-012341-00-0005; Thermo Scientific) or control siRNA by using G-Fectin transfection reagent (Genolution, Seoul, Korea).
Total RNA isolation and RT-PCR
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was conducted with 1 μg of total RNA using ImProm-llTM Reverse Transcriptase (Promega, Madison, WI, USA). PCR was performed using Taq polymerase (Bioneer, Seongnam, Korea). Primer sequences of HOXC9 were as follows: forward, 5'- GGG AGG GTT CAG TGT TGA GA-3' and reverse 5'- GGG ATG ACC TGG ACC AAA TA-3'. For semiquantitative analysis, the Multi Gauge V3.0 software (Fuji, Tokyo, Japan) was used. β-actin mRNA was used as the invariant control. All experiments were performed in triplicate, and representative examples of the data are shown.
Western blotting
Cells were lysed in Nondet P-40 (NP-40) lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1% NP-40, and protease-inhibitor cocktail). Protein concentrations were estimated by the BCA Protein Assay Kit (Thermo). The primary antibodies used were anti-DDK-tag mouse mAb (Origene) and anti-β-actin (Sigma, St. Louis, MO, USA). After immune blotting, the signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL USA).
MTT assay
Cell proliferation was measured using the MTT assay. Cells were plated in 96-well plates at a density of 5 × 103 cells per well. On the next day, the cells were stained with 20 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 3.5 hours at 37°C, followed by removal of the culture medium and incubation with 100 μl of MTT solvent (4 mM HCl and 0.1% NP40, both in isopropanol). After 15-minute incubation, the absorbance was measured with an ELISA reader (SOFTmax PRO) at 560 nm. All experiments were performed in triplicates.
Matrigel invasion assay
A Matrigel invasion assay was performed using Matrigel™ (BD) as previously described [13]. After 24-72 hours, the invading cells were stained with the fluorochrome 4',6-diamidino-2-phenylindole (DAPI) and observed by fluorescent microscopy. The acquired images were analyzed using the ImageJ software.
In silico analysis
To analyze HOXC9 expression patterns in breast cancer tissues, we used the web-accessible database GENT (Gene Expression across Normal and Tumor tissue), which provides gene expression patterns for diverse human cancer and normal tissues from more than 34,000 samples, profiled by Affymetrix U133A or U133plus2 platforms [14]. In this database, datasets have been pre-processed by the MAS5 algorithm and normalized to a target density of 500. For the survival analysis, the MTCI Breast Cancer Survival Analysis Tool (http://glados.ucd.ie/BreastMark/) was used. The MTCI developed the algorithm BreastMark, which integrates gene expression and survival data from 26 datasets on 12 different microarray platforms corresponding to ~17,000 genes in up to 4,738 samples.
Statistical analysis
Data are expressed as mean values with standard error of the mean. Statistical differences were determined by a Student's t-test. A p value of <0.05 was considered statistically significant.
Results
Switch from a proliferative to an invasive phenotype by HOXC9 expression
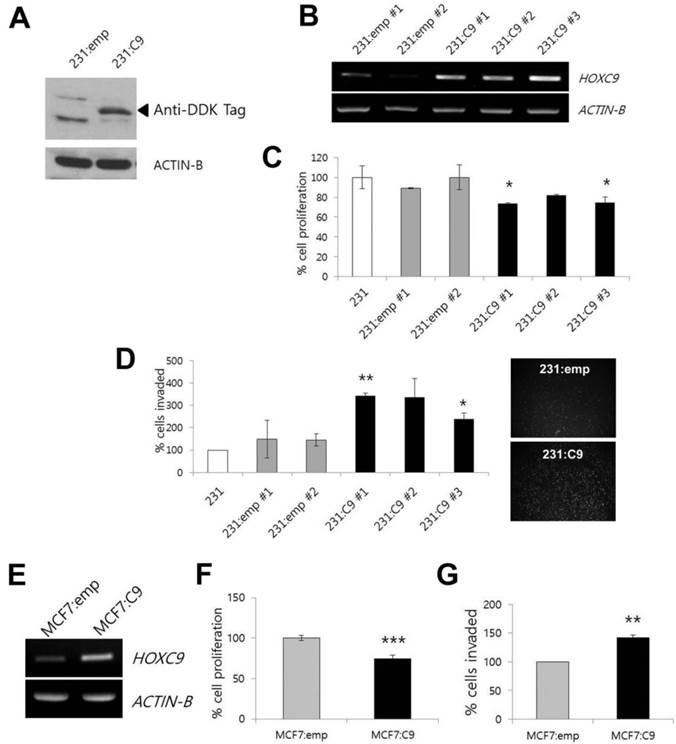
HOXC9 is a candidate gene for dysregulation in breast cancer. In our previous study, most of the breast cancer cell lines studied, except for the MDA-MD-231 cell line, showed higher HOXC9 expression than MCF10A normal breast epithelial cells did [12]. Therefore, for further experiments, we selected the MDA-MB-231 cell line as a model system for gain-of-function studies. The forced expression of HOXC9 in MDA-MB-231 cells upon transfection with the expression plasmid harboring the HOXC9 was initially analyzed by Western blotting after sustained drug selection (Fig. 1A). After that, to assess the effect of HOXC9 overexpression on tumor cell proliferation and invasion, MDA-MB-231 cells stably expressing HOXC9 were generated (see Materials and Methods), and then three stable cell lines (231:C9 #1, 231:C9 #2, and 231:C9 #3) expressing high levels of HOXC9 were selected (Fig. 1B).
Effect of HOXC9 overexpression on cancer cell proliferation and invasion. (A) Western blotting to detect HOXC9 protein expression in MDA-MB-231 cells transfected with empty vector (231:emp) and HOXC9 expression vector (231:C9). Anti-DDK antibody was used. Arrow indicates the expected size of HOXC9-DDK protein. (B) RT-PCR result showing overexpression of HOXC9 in three independent MDA-MB-231 clones (231:C9 #1, 231:C9 #2, and 231:C9 #3). Two clones harboring the empty vector (231:emp #1 and 231:emp #2) were used as controls. (C) The MTT assay was performed to measure cell viability. The proliferation activity was compared between three clonal cells expressing HOXC9 (231:C9 #1, 231:C9 #2, and 231:C9 #3), and three types of control cells (231:emp #1, 231:emp #2, and parent MDA-MB-231 [231] cells). (D) The Matrigel invasion assay was performed to measure invasion ability. Representative images showing the different number of invading cells between 231:emp and 231:C9 cells are presented on the right side. (E) RT-PCR analysis of HOXC9 expression in MCF cells transfected with the empty vector (MCF7:emp) or HOXC9 expression vector (MCF:C9): proliferation (F) and invasion analyses (G) were performed. *; p < 0.05, **; p < 0.01, ***; p < 0.001 vs. parental MDA-MB-231 or MCF7:emp control cells.
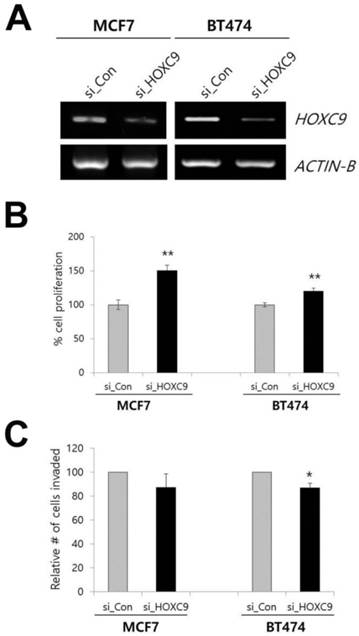
Effect of HOXC9 knockdown on MCF7 and BT474 cells. (A) RT-PCR result showing the suppression of HOXC9 expression in si_HOXC9 treated cells. Nonspecific siRNA (si_Con) was used as the control. The MTT assay (B) and Matrigel invasion assay (C) were performed to measure cell proliferation and invasion ability, respectively. *; p < 0.05, **; p < 0.01 vs. si_Con treated cells.
The MDA-MB-231 cells overexpressing HOXC9 showed statistically significant reduction of cell proliferation as compared to the cells transfected with the empty vector and parental MDA-MB-231 cells (Fig. 1C), whereas HOXC9 overexpression enhanced the invasive capacity compared to control cells (Fig. 1D). When MCF7 cells were tested after HOXC9 overexpression (Fig. 1E), similar results were generated: overexpression of HOXC9 reduced proliferation and increased invasiveness of the cells (Fig. 1F and G).
To determine whether these phenotypes could be reversed when HOXC9 was knocked down, we examined the effect of siRNA targeting HOXC9 on cell proliferation and invasion by using the MCF7 and BT-474 cell lines. The inhibition of HOXC9 expression significantly increased cell proliferation but decreased invasiveness (Fig. 2A-C).
Correlation between HOXC9 expression and prognosis in breast cancer patients with lymph node metastasis
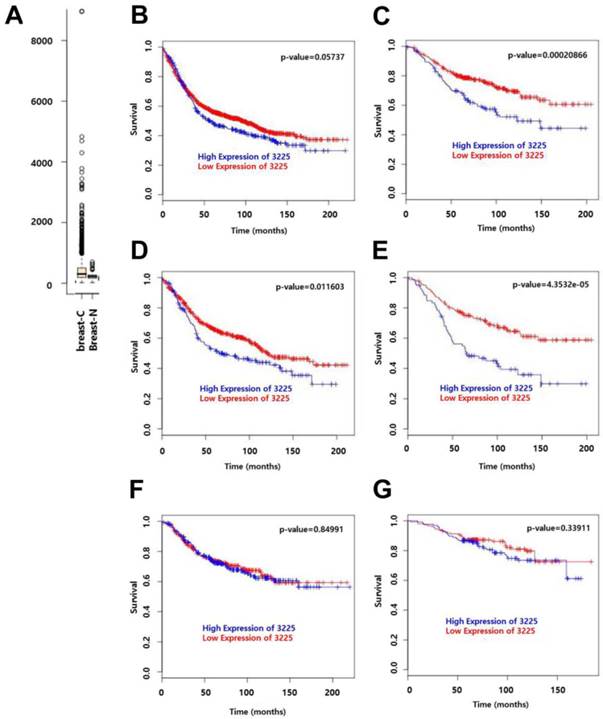
We have previously shown that HOXC9 is a candidate gene that is upregulated in malignant tissues as compared to non-malignant tissues [12]. This is consistent with the information obtained from GENT (http://medical-genome.kribb.re.kr/GENT/) [14], where HOXC9 expression was higher in breast cancer samples as compared to normal samples (Fig. 3A; data from 2, 662 breast-cancer vs. 267 breast-normal, profiled by the Affymetrix U133plus2 platform). To assess the effect of HOXC9 expression on the prognosis of breast cancer patients, disease-free survival (DFS) and distant metastasis-free survival (DMFS) curves were drawn using BreastMark of the MTCI Breast Cancer Survival Analysis Tool (http://glados.ucd.ie/BreastMark/). BreastMark is an algorithm that integrates gene expression and survival data from 26 datasets on 12 different microarray platforms. The number of samples used for analysis depends on how many platforms have probes for an interesting gene and relevant clinical data. A total of 1533 samples, including 767 events, were examined during analysis of DFS for all patients. This analysis revealed that there was no significant difference between the HOXC9-positive and HOXC9-negative groups (p = 0.058) (Fig. 3B). However, high HOXC9 expression had a significant impact on DMFS for all patients (n = 577, number of events = 187, p = 0.000208) (Fig. 3C). Subgroup analysis showed that high HOXC9 expression was correlated with poor prognosis for DFS (number of events = 278) and DMFS (number of events = 109) in lymph node-positive patients (n = 633 for DFS, n = 257 for DMFS) (Fig. 3D and E), but not in lymph node-negative patients (Fig. 3F and G).
Discussion
HOXC9 is one of the HOX genes that are dysregulated in breast cancer cells. In order to understand the functional role of HOXC9 in breast cancer, we performed overexpression and knockdown experiments in breast cancer cell lines and examined the effect of HOXC9 on cancer cell proliferation and invasion. Forced expression of HOXC9 resulted in increased invasiveness, whereas it showed negative effects on cell proliferation. HOXC9 knockdown exhibited opposite effects, promoting cancer cell proliferation. This HOXC9-driven switching phenotype was induced in different breast cancer cell lines, regardless of cell type: the cell lines, MCF7 and MDA-MB-231, that were originally derived from the metastatic site of breast cancer and classified as the same tumor type, that is, metastatic adenocarcinoma, and BT474, originally isolated from the primary tumor, classified as an invasive ductal carcinoma [15].
Previously, high HOXC9 expression has been reported to be associated with neuroblastoma differentiation and as a prognostic marker for better survival in neuroblastoma patients by inducing growth arrest [16]. On the basis of these results, it is interesting to note that the endogenous expression level of HOXC9 in the different breast cancer cell lines, that is, MCF7, BT474, and MDA-MB-231 [12], might explain the properties of each cell line. High expression of HOXC9 in MCF7 and BT474 might account for the noninvasive, differentiated luminal-like features, whereas low expression in MDA-MB-231 could be related to the poorly differentiated basal-like features and highly aggressive properties. Although our knowledge on the regulation of HOXC9 expression in malignant human mammary epithelial cells is still limited, the differences in the expression level of HOXC9 in the MCF7 and MDA-MB-231 cell lines could be partially explained by the different methylation patterns, since the CpG island around the HOXC9 promoter region was reported to be unmethylated in MCF7 but highly methylated in MDA-MB-231 [11].
Prognostic significance of HOXC9 expression in breast cancer patients. (A) Publically available data retrieved from GENT (http://medical-genome.kribb.re.kr/GENT) show that HOXC9 is upregulated in breast cancer tissues. breast-C: breast cancer tissues, breast-N: normal breast tissues. Each circle represents an individual tissue sample. Data are shown as boxplots. The Y axis of the plot indicates normalized expression measures. The box extends from the 25th to 75th percentiles and the line in the middle of the box is plotted at the median. Significance was analyzed using an unpaired t-test (p = 2.913E-11). (B-G) Survival analysis using the MTCI breast cancer survival analysis tool (http://glados.ucd.ie/BreastMark/). Disease free survival (DFS) (n = 1533, number of events = 767, hazard ratio = 1.164 [0.9947 - 1.363], p = 0.058) (B) and distant metastasis-free survival (DMFS) (n = 577, number of events = 187, hazard ratio = 1.758 [1.3-2.379], p = 0.000208) (C) in all patients. DFS (n = 633, number of events = 278, hazard ratio = 1.382 [1.074-1.778] p = 0.0116) (D) and DMFS (n = 257, number of events = 109, hazard ratio = 2.17 [1.483-3.176], p = 4.351e-05) (E) in lymph node-positive patients. DFS (n = 538, number of events = 163, hazard ratio = 1.03 [0.7574-1.401], p = 0.85) (F) and DMFS (n = 276, number of events = 57, hazard ratio = 1.289 [0.7646 - 2.174], p = 0.339) (G) in lymph node-negative patients.
The current finding about the effect of forced expression of HOXC9 in MDA-MB-231 was somewhat unexpected. When HOXC9 was overexpressed, the proliferative activity decreased, as reported in the case of neuroblastoma [16]. HOXC9 has also been reported to induce endothelial cell quiescence, inhibiting proliferation and thus keeping the vasculature in the resting state [17]. When HOXC9 is suppressed by hypoxia, an important physiological stimulus for angiogenesis in the tumor site, the endothelial cells become activated and start to proliferate, thereby promoting the angiogenic activity [17, 18]. However, interestingly, the invasiveness increased here along with the decrease in proliferation, resembling a phenotype switch from a proliferative to an invasive state (Fig. 1). Furthermore, the reciprocal result was detected in MCF7 and BT474 cells when the expression level of HOXC9 was decreased with siRNA (Fig. 2). Recently, this kind of phenotype switching has been reported in melanoma cells, and was considered as a kind of alternative resistance mechanism to the therapeutics [19-21]. Interestingly, a similar reciprocal switching phenotype has also been reported in breast cancer cells [22-24].
Breast cancer-related death is mainly due to metastasis to other organs and the concomitant loss of function of that organ rather than the lesions in the breast itself. Local invasion and migration are the initial steps of the multi-step process of metastasis [25]. The accumulating evidence indicates that epithelial-mesenchymal transition (EMT) plays a critical role in metastasis; the genes involved in EMT promote invasion and migration but suppress proliferation [25, 26]. In the current study, we did not confirm the relatedness to EMT; however, HOXC9 was found to promote the invasion and suppress the proliferation of breast cancer cells. This result is in good agreement with the survival analysis data of breast cancer patients for whom high level expression of HOXC9 correlated well with poor prognosis in DMFS, especially in the case of lymph node-positive patients (Fig. 3). Together, these data suggest that HOXC9 could be a prognostic marker in breast cancer patients with lymph node metastasis and a target for therapeutic interventions in malignant breast cancer.
Acknowledgements
This research was supported by funds from the National Health Insurance Corporation Ilsan Hospital (2014-02) and the Basic Science Research Program through the National Research Foundation (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2008399, NRF-2010-0025149, NRF-2014R1A1A2056986).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Morgan R. Hox genes: a continuation of embryonic patterning? Trends Genet. 2006;22:67-69
2. Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361-371
3. Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med. 2014;92:811-823
4. Lewis MT. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2000;2:158-169
5. Chen H, Sukumar S. Role of homeobox genes in normal mammary gland development and breast tumorigenesis. J Mammary Gland Biol Neoplasia. 2003;8:159-175
6. Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci U S A. 1999;96:541-546
7. Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB. et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535-1550
8. Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N. et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107:1100-1105
9. Cantile M, Pettinato G, Procino A, Feliciello I, Cindolo L. et al. In vivo expression of the whole HOX gene network in human breast cancer. Eur J Cancer. 2003;39:257-264
10. Makiyama K, Hamada JI, Takada M, Murakawa K, Takahashi Y, Tada M. et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol Rep. 2005;13:673-679
11. Kikuyama M, Takeshima H, Kinoshita T, Okochi-Takada E, Wakabayashi M, Akashi-Tanaka S. et al. Development of a novel approach, the epigenome-based outlier approach, to identify tumor-suppressor genes silenced by aberrant DNA methylation. Cancer Lett. 2012;322:204-212
12. Hur H, Lee J-Y, Yun H, Park B, Kim M. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56:64-71
13. Lee J-Y, Hur H, Yun HJ, Kim Y, Yang S, Kim SI. et al. HOXB5 promotes the proliferation and invasion of breast cancer cells. Int J Biol Sci. 2015;11:701-711
14. Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: Gene Expression Database of Normal and Tumor Tissues. Cancer Inform. 2011;10:149-157
15. Kao J, Salari K, Bocanegra M, Choi YL, Girard L, Gandhi J. et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PloS one. 2009;4:e6146
16. Mao L, Ding J, Zha Y, Yang L, McCarthy BA, King W. et al. HOXC9 links cell-cycle exit and neuronal differentiation and is a prognostic marker in neuroblastoma. Cancer Res. 2011;71:4314-4324
17. Stoll SJ, Kroll J. HOXC9: a key regulator of endothelial cell quiescence and vascular morphogenesis. Trends Cardiovasc Med. 2012;22:7-11
18. Stoll SJ, Bartsch S, Augustin HG, Kroll J. The transcription factor HOXC9 regulates endothelial cell quiescence and vascular morphogenesis in zebrafish via inhibition of interleukin 8. Cir Res. 2011;108:1367-1377
19. Hoek KS, Eichhoff OM, Schlegel NC, Döbbeling U, Kobert N, Schaerer L. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650-656
20. Widmer DS, Hoek KS, Cheng PF, Eichhoff OM, Biedermann T, Raaijmakers MI. et al. Hypoxia contributes to melanoma heterogeneity by triggering HIF1α-dependent phenotype switching. J Invest Dermatol. 2013;133:2436-2443
21. Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer Res. 2014;74:5937-5941
22. Grismayer B, Solch S, Seubert B, Kirchner T, Schafer S, Baretton G. et al. Rab31 expression levels modulate tumor-relevant characteristics of breast cancer cells. Mol Cancer. 2012;11:62
23. Qian X, Hulit J, Suyama K, Eugenin EA, Belbin TJ, Loudig O. et al. p21CIP1 mediates reciprocal switching between proliferation and invasion during metastasis. Oncogene. 2013;32:2292-2303
24. Patsialou A, Wang Y, Pignatelli J, Chen X, Entenberg D, Oktay M. et al. Autocrine CSF1R signaling mediates switching between invasion and proliferation downstream of TGFβ in claudin-low breast tumor cells. Oncogene. 2015;34:2721-2731
25. Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192-2206
26. Evdokimova V, Tognon C, Ng T, Sorensen PH. Reduced proliferation and enhanced migration: two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cell Cycle. 2009;8:2901-2906
Author contact
![]() Corresponding author: E-mail: mhkim1ac Tel: +82-2-2228-1647, Fax: +82-2-365-0700
Corresponding author: E-mail: mhkim1ac Tel: +82-2-2228-1647, Fax: +82-2-365-0700

 Global reach, higher impact
Global reach, higher impact