3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(8):984-990. doi:10.7150/jca.14237 This issue Cite
Research Paper
The Role of Prion Protein Expression in Predicting Gastric Cancer Prognosis
1. Department of General surgery, Zhongshan Hospital, Fudan University, Shanghai, P.R. China
2. School of Life Sciences, East China Normal University, Shanghai, P.R. China;
3. Department of Physiology, Renji College, Wenzhou Medical University, Wenzhou, P.R. China.
4. Center for Neurodegenerative Science, Van Andel Research Institute, Grand Rapids, Michigan, USA
5. Department of Pathology, Shanghai Pulmonary Hospital, Tongji University, Shanghai, P.R. China
* Both authors contributed equally.
Received 2015-10-26; Accepted 2016-3-15; Published 2016-5-20
Abstract
Previous reports indicated that prion protein (PrP) is involved in gastric cancer (GC) development and progression, but its role in GC prognosis has been poorly characterized. A total of 480 GC patients were recruited in this retrospective study. PrP expression in cancerous and non-cancerous gastric tissues was detected by using the tissue microarray and immunohistochemical staining techniques. Our results showed that the PrP expression in GC was significantly less frequent than that in the non-cancerous gastric tissue (44.4% vs 66.4%, P < 0.001). Cox regression analysis revealed that PrP expression was associated with TNM stage, survival status and survival time. GC patients with higher TNM stages (stages II, III and IV) had significantly lower PrP expression levels in tumors than those with lower TNM stages (stages 0 and I). Kaplan-Meier survival curves revealed that negative PrP expression was associated with poor overall survival (log-rank test: P < 0.001). The mean survival time for patients with negative PrP expression was significant lower than those with positive PrP expression (43.0±28.5m vs. 53.9±31.1m, P<0.001). In multivariate Cox hazard regression, PrP expression was an independent prognostic factor for GC survival, with a HR (hazard ratio) of 0.687 (95%CI:0.520-0.907, P=0.008). Our results revealed that negative PrP expression could independently predict worse outcome in GC and thereby could be used to guide the clinical practice.
Keywords: prion protein, gastric cancer, prognosis, biomarker.
Introduction
Cellular prion protein (PrP) is a highly conserved glycosylphosphatidylinositol-anchored cell surface glycoprotein. It is widely expressed in a variety of tissues with most abundant expression in the central nervous system (CNS). PrP is well known for its involvement in the pathogenesis of transmissible spongiform encephalopathies, also known as ''prion disease'. The conversion of normal host encoded PrP to the pathogenic PrPSc conformer initiates the self-perpetuating replication of PrPSc and causes fatal neurodegenerative disorders [1-3]. Besides prion disease, normal PrP has been found to play a role in CNS development, neurite outgrowth, neuronal survival, early synaptic neuronal transmission and a number of biological processes [4]. However, the exact physiological function of PrP remains unknown.
In addition to the normal and pathological processes in the nervous system, many recent studies have provided strong evidence supporting the involvement of PrP in tumor cell biology [4]. PrP over expression in breast cancer cells was associated with the acquisition of resistance to antitumor drug induced cell death [5, 6]. Knocking down PrP expression significantly reduced the proliferation of DLD-1 and SW480 colon cancer cells [7]. The presence of PrP expression promoted the adhesive, invasive, and metastatic capacities of the gastric cancer (GC) cells [8]. These studies suggested that PrP is a potential target for developing novel anti-cancer therapies. However, this conclusion may depend on the type of cancer cells and the specific type of anti-cancer treatments. We have previously reported that knocking down PrP expression in MDA-MB-435 breast cancer cells increased its susceptibility to serum-deprivation induced apoptosis, but at the same time significantly increased the resistance to doxorubicin, a DNA-interacting chemotherapy drug [9]. Nonetheless, all studies support that PrP was present in many different types of cancers, even though its exact role in cancer biology remains obscure.
The potential diagnostic or prognostic values of PrP expression in cancer have also been explored by several studies. It has been shown that PrP expression was up-regulated in various types of cancers, including GC, osteosarcoma, breast cancer, melanoma, pancreatic cancer and colon cancer [8, 10-14]. PrP overexpression in cancerous tissues was generally associated with poor survival or response to chemotherapy [13, 15-17]. Notably, mRNA level was used to represent PrP expression in several studies, which is not always consistent with the level of protein expression [5, 14, 18]. Moreover, the sample sizes of previous studies were relatively small, preventing them from drawing a powerful conclusion. In this study, we investigated PrP protein expression in cancerous and paired non-cancerous tissues in a large collection of GC patients (480 cases) and analyzed the prognostic value of PrP expression in GC.
Materials and Methods
Patients and Tissue Samples
The study was approved by the ethics committee of Fudan University and informed consent was obtained from all individual participants included in the study. A total of 494 patients who underwent GC surgery at Zhongshan Hospital of Fudan University (Shanghai, China) between February 2000 and September 2005 were included in this retrospective study. None of the patients had received preoperative chemotherapy or radiotherapy. Cancerous gastric tissue specimens and their relative noncancerous tissues (normally > 5cm distant to the malignant tumor) were obtained, formalin-fixed and paraffin-embedded, respectively. Histological diagnosis of each primary cancerous specimen was confirmed with hematoxylin-eosin (H&E) staining by pathologists from Zhongshan Hospital. Patients' clinical and pathological information, such as gender, age, differentiation status, histological type, tumor location and TNM stage was collected. Follow-up information of all participants was updated every 3 month by telephone visit and questionnaire letters. Complete follow-up was updated until death or January 2014, whichever came first. Death of participants was ascertained by reporting from the family and verified by review of public records. The mean follow-up period of enrolled patients was 47.8±30.2months, with the median time of 52.0 months.
Construction of Tissue Microarray and Immunohistochemistry Assay
Tissue microarray (TMA) chips containing the gastric cancers and their paired non-cancerous gastric tissues were prepared by the Department of Pathology, Zhongshan Hospital. Each chip contained 40 tissue dots that were 20 pairs of cancerous and non-cancerous gastric tissues. Each dot was of 2 mm in diameter and 5μm in thickness. Sections were then placed on microscope slides and subject to immunohistochemical (IHC) staining. After sections were deparaffinized, antigen retrieved and blocked, IHC staining was performed using monoclonal mouse antibody against PrP (clone 8H4, 1:200, Sigma, St Louis, MO, USA) at 4℃ overnight. Subsequently, the tissue was exposed to the secondary antibody (biotinylated anti-mouse IgG, 1:400, Sigma) for 1 hour at room temperature. The bound antibodies were visualized using a streptavidin-peroxidase complex, with 3, 3-diaminobenzidine (Sigma) as chromogen. Negative controls were performed by replacing the primary antibody with pre-immune rabbit serum. The chips were then H&E stained for histological verification of the adequacy of the arrayed tumor tissues. Qualified samples were defined as those in which the tumor tissue occupied more than 10% of the core area.
Evaluation of Immunostaining Intensity
Multiple tissue arrays were scored independently by 2 investigators who were blinded to the clinical history, pathologic characteristics of the disease and the clinical outcome of the patient. As shown in supplementary Figure S1, the protein expression was visualized and classified based on the percentage of positive cells and the intensity of staining. The percentage of positive cells was divided into five grades (percentage scores): <1% (0), 1-25% (1), 26-50% (2), 51-75% (3) and >75% (4). The intensity of staining was divided into four grades (intensity scores): negative (0), weak (1), moderate (2) and strong (3). The histological score (H-score) was determined using the following formula: overall scores = percentage score × intensity score. An overall score of 0-12 was calculated and graded as negative (score: 0) or positive (score: 1-12). All stained slides were interpreted by another pathologist unaware of the data. Specimens will be rescored by a third observer if difference of scores from 2 investigators was great than 3.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 for Windows (SPSS, Chicago, IL). To assess the relationships between the PrP expression and the clinicopathological parameters of GC patients, the Cox regression analysis or χ2 test was used. The cumulative survival rate was calculated using a life table method. Univariate survival analysis was performed using the Kaplan-Meier method, and the significance of difference between groups was analyzed with the log-rank test. Variables that were significant in the univariate analysis were included in stepwise multivariate survival analysis using the Cox proportional hazards model. P< 0.05 was considered statistically significant and all P values were 2-sided.
Results
Patient characteristics
A total of 494 GC patients were initially recruited in this study. However, 14 subjects were excluded because their cancerous tissue occupied less than 10% of the core area and could not be successfully examined. Thus, 480 patients were included for our analysis and their general characteristics were summarized in Table 1. The mean age of the group was 61.2±11.7 years (y) and 57.5% of them were more than 60y. About two-thirds (66.5%) of the patients were male. Most of the tumors were localized in distal stomach (58.2%) and low-differentiated (80.8%). The most frequent histological type was poorly differentiated adenocarcinoma (66.9%). According to TNM stage, the patients were ranged from 0 to IV, with 25.2% of stage II and 57.9% of stage III.
Expression of PrP in Cancerous and Noncancerous Gastric tissues
To elucidate the biologic significance of PrP in GC, we examined the immunohistochemical expression of PrP in paired cancerous and non-cancerous tissue samples. First, to select the anti-PrP antibody for this study, we compared 3F4 (Sigma), 8B4(Sigma), and 8H4 antibody for IHC staining in consecutive sections (supplementary Figure S2). The 8H4 antibody [19, 20] was found to be suitable for staining PrP in GC samples because of its low background and clear positive staining. The specificity of 8H4 anti-PrP antibody was further confirmed by IHC and western blot analyses with tissues obtained from PrP knockout and wide-type mice (supplementary Figure S3 and S4) and PrP expression was detected in gastric or brain tissues of wide-type mice, but not knockout mice. We also conducted immunofluorescent staining in frozen tissue of paired cancerous and non-cancerous tissue samples of a GC patient to demonstrate that the 8H4 antibody is able to detect PrP expression in human gastric tissues (supplementary Figure S5). Together, all these results suggested that 8H4 anti-PrP antibody was suitable for IHC detection of PrP expression in GC tissues.
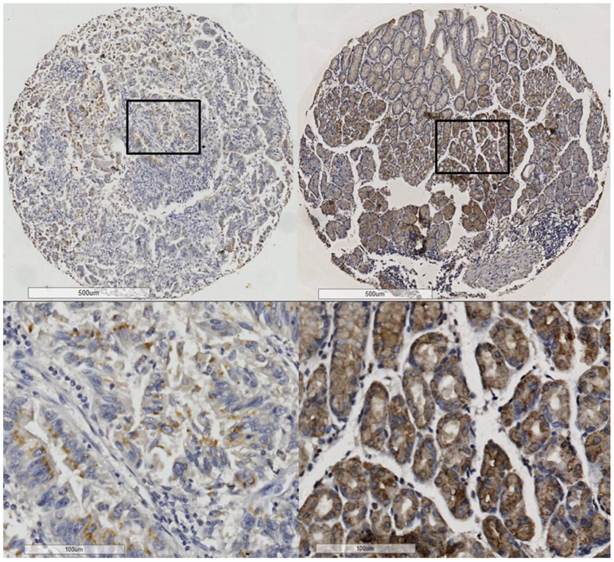
PrP was mainly located in the membranes or cytoplasm of the epithelial cells (Figure 1). Positive PrP protein expression was detected in 213 (44.4%) of the 480 GC tissues and 304 (66.4%) of the 458 non-cancerous gastric tissues (supplementary Table S1). Collectively, the PrP expression in GC was significantly lower than that in the non-cancerous gastric mucosa (P < 0.001).
Characteristics of 480 gastric cancer patients recruited.
| Number | Percentage | |
|---|---|---|
| Age at diagnosis | ||
| <60 | 204 | 42.50% |
| ≥60 | 276 | 57.50% |
| Gender | ||
| Female | 161 | 33.50% |
| Male | 319 | 66.50% |
| Tumor Location | ||
| U: upper | 68 | 14.30% |
| M: middle | 129 | 27.50% |
| D: distal | 280 | 58.20% |
| Multiple | 3 | 0.60% |
| Histological type | ||
| 1: Papillary adenocarcinoma | 9 | 1.90% |
| 2: Tubular adenocarcinoma | 77 | 16.00% |
| 3: Poorly differentiated adenocarcinoma | 321 | 66.90% |
| 4: Mucinous adenocarcinoma | 30 | 6.30% |
| 5: Signet-ring cell carcinoma | 31 | 6.50% |
| 6: Special type | 4 | 0.80% |
| 7: Carcinoma in situ | 8 | 1.70% |
| Differentiation | ||
| L: low | 388 | 80.80% |
| H: high | 92 | 19.20% |
| TNM Stage | ||
| 0 | 6 | 1.30% |
| I | 61 | 12.70% |
| II | 121 | 25.20% |
| III | 278 | 57.90% |
| IV | 14 | 2.90% |
PrP expression in cancerous tissue (left) and noncancerous (right) tissues.
The association between PrP expression and TNM stage.
| PrP Positive | Total (n=480) | |
|---|---|---|
| TNM Stage | ||
| 0 | 4 (66.7%) | 6 |
| I | 37 (60.7%) | 61 |
| II | 54 (44.6%) | 121 |
| III | 111 (39.9%) | 278 |
| IV | 7 (50.0%) | 14 |
| Tumor Depth | ||
| Tis | 4 (50.0%) | 8 |
| T1a | 21 (75.0%) | 28 |
| T1b | 7 (63.6%) | 11 |
| T2 | 24 (45.3%) | 53 |
| T3 | 57 (39.0%) | 146 |
| T4a | 100 (43.5%) | 230 |
| T4b | 0 (0.0%) | 4 |
| Lymph Node Metastasis | ||
| N0 | 64 (50.0%) | 128 |
| N1 | 42 (49.4%) | 85 |
| N2 | 39 (41.5%) | 94 |
| N3a | 42 (38.5%) | 109 |
| N3b | 26 (40.6%) | 64 |
PrP expression is decreased with TNM stage processed (P=0.007, χ2 test). PrP expression positive rate is lower in higher T stage (Tumor characteristics, P=0.008, χ2 test). PrP expression positive rate is lower in higher N stage (Lymph node metastasis, P=0.05, χ2 test)
Association of PrP Expression with Clinicopathological Parameters
To investigate the roles of PrP in GC, we analyzed the relationships between their expressions and the clinicopathological parameters of the GC patients. Firstly, the IHC score distribution was carefully analyzed to define a suitable cut-off value. Negative PrP protein expression, scored as 0, was detected in 55.6% of the 480 GC tissues; while 25.2%, 10.6%, and 8.5% scored as 1-3, 4-6 and 7-12. Considering the sample size in each group should be relatively close to avoid statistical bias, we used score “0” as the cut-off value. In the following analyses, GC patients were stratified into PrP positive group (score: 1-12) and PrP negative group (score: 0). In cox regression analysis, the expression of PrP in GC was associated with TNM stage, survival status, and survival time, but not related to age, gender, tumor location, differentiation and histological type (Supplementary Table S2). GC patients with higher TNM stages (stages II, III and IV) had significantly lower PrP expression than those with lower TNM stages (stages 0 and I) (Table 2). In addition, tumor characteristics and lymph node metastasis was associated with PrP expression. The positive rate of PrP expression was significantly reduced with increased T stage, especially in T2-T4b. Similarly, the PrP expression was decreased with the severity of lymph node metastasis, especially in N2-N3b (Table 2). For distant metastasis, analysis was not performed due to the limited number of cases.
Correlation of PrP Expression with Prognosis
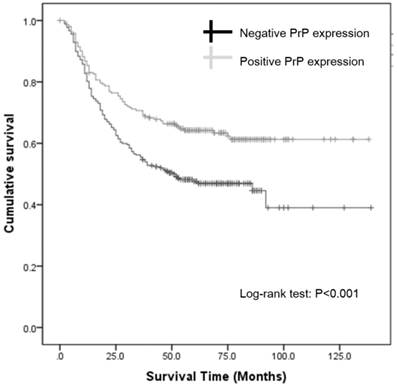
Since the expression of PrP in GC was associated with survival status and survival time in cox regression analysis, we further investigated the association between PrP expression and prognosis using Kaplan-Meier survival curve and log-rank test. Kaplan-Meier survival curves revealed that negative PrP expression was associated with worse overall survival (log-rank test: P < 0.001; Figure 2). The mean survival time for patients with negative PrP expression was significant lower than those with positive expression (43.0±28.5m vs. 53.9± 31.1m, P<0.001). Clearly, GC patients with positive PrP expression had a better prognosis, with the unadjusted hazard ratio (HR) being 0.610 (95%CI: 0.462-0.805, P < 0.001; Table 3). Notably, age and TNM stage were also significant predictors of outcome in univariate Cox hazard regression. When these 3 factors were analyzed again using multivariate Cox hazard regression, our result showed that TNM stage and PrP expression were independent prognostic factors for GC survival, with a HR of 3.095 (95%CI: 2.429-3.944, P < 0.001) for TNM stage and a HR of 0.687 (95%CI:0.520-0.907, P=0.008; Table 3) for PrP expression. Thus, we concluded that negative PrP expression could independently predict worse outcome in GC.
Kaplan-Meier survival curves.
Cox hazard regression.
| Sig. | HR | 95.0% CI | |
|---|---|---|---|
| Univariate analysis | |||
| Gender | 0.292 | 1.166 | 0.877-1.551 |
| Age | 0.026* | 1.013 | 1.002-1.025 |
| Differentiation | 0.12 | 0.753 | 0.526-1.077 |
| Histological type | 0.566 | 0.96 | 0.837-1.103 |
| Tumor location | 0.132 | 0.874 | 0.733-1.042 |
| TNM Stage | <0.001** | 3.144 | 2.474-3.996 |
| PrP expression | <0.001** | 0.61 | 0.462-0.805 |
| Multivariate analysis | |||
| Age | 0.123 | 1.009 | 0.998-1.021 |
| TNM Stage | <0.001** | 3.095 | 2.429-3.944 |
| PrP expression | 0.008** | 0.687 | 0.520-0.907 |
*, P < 0.05; **, P < 0.01. PrP negative expression is an independent risk factor for poor survival.
Discussion
Our results showed that PrP protein level was significantly down-regulated in cancerous tissues of GC and decreased PrP expression correlated with a poor survival of GC patients. Our study is the first study to investigate the prognostic value of PrP expression in such a large collection of patients. Previously, Zhou et al. conducted a clinical study of 238 GC patients and found that high PrP expression correlated to a poorer prognosis [15]. In an earlier study, the same group also investigated PrP expression in 124 GC samples by IHC staining with the monoclonal antibody 3F4. They reported that PrP expression increased in cancerous tissue than in noncancerous tissue and increased PrP expression correlated with the histopathological differentiation and tumor progression [21]. Consistent with their results, we also found that PrP expressed in the crypt epithelial cells and mainly localized in the cytoplasm or plasma membranes. However, in contrast to their results, we found that PrP expression was downregulated instead of upregulated in GC as compared to noncancerous tissues.
The opposite findings might be due to the different specimen resources and antibodies used. Patients in our study were mostly from eastern and southern China; while the majority of participants in their study were from western and northern China. The 3F4 anti-PrP antibody was used in their study, but 8H4 antibody was used in our analysis based on careful evaluation of its specificity and suitability for detecting PrP in gastric tissues (supplementary Figure S2-S5). Actually different antibodies could show different results even in a series of consecutive sections (supplementary Figure S2). Further studies of directly comparing different anti-PrP antibodies on the same cohort of patients are necessary to firmly establish the prognostic value of PrP in GC. In addition, all the above studies were based on the IHC analysis. The method itself may partly influence the results because IHC is always somewhat subjective. Notably, a recent study described a comprehensive molecular evaluation of 295 primary GC cases as part of The Cancer Genome Atlas project (TCGA) [22]. Based on the analysis of its mRNA sequencing data, we found the PrP mRNA level was significantly decreased in the cancerous specimens (P< 0.001, supplementary Figure S6), which is consistent with our findings.
The expression of PrP was previously reported to have prognostic value in other malignancies. Sy et al. found the risk of death was about four times higher (HR = 3.8; 95% CI: 2.2, 6.5) among 108 pancreatic ductal adenocarcinoma (PDAC) cases with PrP-positive tumors compared to the 34 cases with PrP-negative tumors. Meslin et al. reported that PrP was expressed in 113 out of 756 breast cancer patients (15%) and PrP positive frequency was higher in ER-negative breast cancer (BC) than that in ER-positive BC (33% vs 5%, P<0.001). They then analyzed the predicting role of PrP expression in 252 ER-negative BC and found adjuvant chemotherapy decreased the risk for death only in patients with ER-negative/PrP-negative tumors (HR = 0.39, 95% CI 0.2-0.74, P = 0.004) but not in patients with ER-negative/PrP-positive disease. In GC, Zhou et al. reported PrP expression was associated with the depth of invasion and lymph node metastasis of the tumor. Multivariate survival analysis suggested that, combined expression of PrP and MGr1-Ag/37LRP, a protein interacting with PrP, was independent prognostic factors for GC patients [15]. Wang et al. reported that patients with higher PrP expression had a poorer response to chemotherapy (P < 0.05), a lower 2-year survival rate and a higher mortality rate [23]. In contrast to previous literature, our study is the first study to show that negative PrP expression could independently predict worse outcome in cancer. We strongly recommend careful evaluations of the prognostic role of PrP expression globally in large cohorts of patients of various carcinomas.
Since the expression of PrP was altered in malignant tumors, the potential biological function of PrP in tumorigenesis and/or tumor biology is another research area that attracts many interests. A series of studies has been performed using GC cell lines to study the role of PrP in cell proliferation, G1/S transition, invasion, metastasis, drug-resistance, and so on [8, 24-26]. In GC cell line SGC7901, overexpression of PrP could promote cell proliferation and tumorigenesis at least partially through activation of PI3K/Akt pathway and subsequent transcriptional activation of CyclinD1 to regulate the G1/S phase transition [24]. Knocking down of PrP by siRNA in SGC7901and MKN45 cells decreased their adhesive, invasive, and in vivo metastatic capabilities at least partially through activation of MEK/ERK pathway and consequent trans activation of MMP11 [8]. Moreover, in adriamycin-resistant gastric carcinoma cell line SGC7901/ADR, PrP was upregulated compared to its parental cell line SGC7901. Inhibition of PrP expression by antisense or RNAi technology could partially reverse its multidrug resistant phenotype [25]. PrP may have certain effects on drug resistance through upregulation of P-glycoprotein and activation of the PI3K/Akt pathway [26]. Besides GC, inhibition of PrP expression by shRNA in the PDAC cell lines reduced cell proliferation and invasion in vitro as well as tumor growth in vivo [13]. However, we have found that knocking down of PrP in MDA-MB-435 breast cancer cell line reduced its susceptibility to doxorubicin-induced cell death [9]. Thus, depending on the cancer cell type or particular cancer cell line, the role of PrP may differ. Furthermore, previous studies mostly focused on the growth characteristics of the GC cells. However, epithelial cell-cell junctions are also very important to prevent tumorigenesis, changes may contribute to epithelial-mesenchymal transition which plays important role in tumor progression and metastasis. PrP was proved to contribute to the intestinal epithelial cell-cell junctions and barrier function [27]. PrP knock-out mice were more sensitive to dextran sulfate-induced colitis known as one of precancerous lesions (unpublished). Therefore, it is possible that PrP may have similar functions in stomach and reduced expression of PrP may be associated with impaired gastric barrier, increased tumorigenesis and metastasis leading to worse outcome. Future in vivo studies comparing tumor formation, invasion and metastasis in isogenic PrP knock-out and wide-type mice using standard genetic or chemical models would help to clarify the biological roles of PrP in cancers, including GC.
In summary, our study showed PrP expression was down-regulated in gastric cancerous tissue and more importantly, decreased PrP expression could predict worse outcome. Our results support the prognostic role of PrP expression in GC.
Supplementary Material
Supplementary tables and figures.
Acknowledgements
The analysis was supported by National Natural Science Foundation of China (NSFC 30800401, 31172347, 31470257, 31472213, and 31470794), the Ministry of Education of China Project 985 and the Shanghai Municipal Health Bureau #2009016. We thank the following persons for their help in the preparation of manuscript: Dr. Chengjin Huang and Dr. Xiaoli Zhu from Peking Union Medical College Hospital, and Dr. Yi Zhang from East China Normal University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Griffith JS. Self-replication and scrapie. Nature. 1967;215:1043-1044
2. Prusiner SB. Prions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13363-13383
3. Ma J, Wang F. Prion disease and the 'protein-only hypothesis'. Essays in biochemistry. 2014;56:181-191
4. Yang X, Zhang Y, Zhang L. et al. Prion protein and cancers. Acta biochimica et biophysica Sinica. 2014;46:431-440
5. Roucou X, Giannopoulos PN, Zhang Y. et al. Cellular prion protein inhibits proapoptotic bax conformational change in human neurons and in breast carcinoma mcf-7 cells. Cell death and differentiation. 2005;12:783-795
6. Meslin F, Hamai A, Gao P. et al. Silencing of prion protein sensitizes breast adriamycin-resistant carcinoma cells to trail-mediated cell death. Cancer research. 2007;67:10910-10919
7. Li QQ, Sun YP, Ruan CP. et al. Cellular prion protein promotes glucose uptake through the fyn-hif-2alpha-glut1 pathway to support colorectal cancer cell survival. Cancer science. 2011;102:400-406
8. Pan Y, Zhao L, Liang J. et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20:1886-1888
9. Yu G, Jiang L, Xu Y. et al. Silencing prion protein in mda-mb-435 breast cancer cells leads to pleiotropic cellular responses to cytotoxic stimuli. PloS one. 2012;7:e48146
10. Sollazzo V, Galasso M, Volinia S. et al. Prion proteins (prnp and prnd) are over-expressed in osteosarcoma. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2012;30:1004-1012
11. Li C, Yu S, Nakamura F. et al. Pro-prion binds filamin a, facilitating its interaction with integrin beta1, and contributes to melanomagenesis. The Journal of biological chemistry. 2010;285:30328-30339
12. Meslin F, Conforti R, Mazouni C. et al. Efficacy of adjuvant chemotherapy according to prion protein expression in patients with estrogen receptor-negative breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2007;18:1793-1798
13. Li C, Yu S, Nakamura F. et al. Binding of pro-prion to filamin a disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. The Journal of clinical investigation. 2009;119:2725-2736
14. Antonacopoulou AG, Grivas PD, Skarlas L. et al. Polr2f, atp6v0a1 and prnp expression in colorectal cancer: New molecules with prognostic significance? Anticancer research. 2008;28:1221-1227
15. Zhou L, Shang Y, Liu C. et al. Overexpression of prpc, combined with mgr1-ag/37lrp, is predictive of poor prognosis in gastric cancer. International journal of cancer Journal international du cancer. 2014;135:2329-2337
16. Cheng Y, Tao L, Xu J. et al. Cd44/cellular prion protein interact in multidrug resistant breast cancer cells and correlate with responses to neoadjuvant chemotherapy in breast cancer patients. Molecular carcinogenesis. 2014;53:686-697
17. Sy MS, Altekruse SF, Li C. et al. Association of prion protein expression with pancreatic adenocarcinoma survival in the seer residual tissue repository. Cancer biomarkers: section A of Disease markers. 2011;10:251-258
18. Han H, Bearss DJ, Browne LW. et al. Identification of differentially expressed genes in pancreatic cancer cells using cdna microarray. Cancer research. 2002;62:2890-2896
19. Zanusso G, Liu D, Ferrari S. et al. Prion protein expression in different species: Analysis with a panel of new mabs. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8812-8816
20. Pan T, Li R, Kang SC. et al. Epitope scanning reveals gain and loss of strain specific antibody binding epitopes associated with the conversion of normal cellular prion to scrapie prion. Journal of neurochemistry. 2004;90:1205-1217
21. Liang J, Pan YL, Ning XX. et al. Overexpression of prpc and its antiapoptosis function in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2006;27:84-91
22. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209
23. Wang JH, Du JP, Zhang YH. et al. Dynamic changes and surveillance function of prion protein expression in gastric cancer drug resistance. World journal of gastroenterology: WJG. 2011;17:3986-3993
24. Liang J, Pan Y, Zhang D. et al. Cellular prion protein promotes proliferation and g1/s transition of human gastric cancer cells sgc7901 and ags. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:2247-2256
25. Du J, Pan Y, Shi Y. et al. Overexpression and significance of prion protein in gastric cancer and multidrug-resistant gastric carcinoma cell line sgc7901/adr. International journal of cancer Journal international du cancer. 2005;113:213-220
26. Liang J, Ge F, Guo C. et al. Inhibition of pi3k/akt partially leads to the inhibition of prp(c)-induced drug resistance in gastric cancer cells. The FEBS journal. 2009;276:685-694
27. Petit CS, Barreau F, Besnier L. et al. Requirement of cellular prion protein for intestinal barrier function and mislocalization in patients with inflammatory bowel disease. Gastroenterology. 2012;143:122-132 e115
Author contact
![]() Corresponding authors: Fenglin Liu, MD, Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai. P. R. China, 200032. Tel: 86-21-64041990-2910. Fax: 86-21-64041990-2910. Email: Liu.fenglinsh.cn or Yulan Zhao, MD, PhD, School of Life Science, East China Normal University, North Zhongshan Road #3663, Shanghai City, P.R. China, 200062. Tel: 86-21-32530498. Fax: 86-21-32530498. Email: ylzhaoecnu.edu.cn
Corresponding authors: Fenglin Liu, MD, Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai. P. R. China, 200032. Tel: 86-21-64041990-2910. Fax: 86-21-64041990-2910. Email: Liu.fenglinsh.cn or Yulan Zhao, MD, PhD, School of Life Science, East China Normal University, North Zhongshan Road #3663, Shanghai City, P.R. China, 200062. Tel: 86-21-32530498. Fax: 86-21-32530498. Email: ylzhaoecnu.edu.cn

 Global reach, higher impact
Global reach, higher impact