Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(9):1105-1113. doi:10.7150/jca.14801 This issue Cite
Research Paper
The Prognostic Value of Circulating Cell-Free DNA in Colorectal Cancer: A Meta-Analysis
1. Department of Gastrointestinal Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China;
2. Department of Research Administration, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China.
Received 2015-12-25; Accepted 2016-3-22; Published 2016-6-4
Abstract
Background: Circulating cell-free DNA (cfDNA) is a promising candidate biomarker for detection, monitoring and survival prediction of colorectal cancer (CRC). However, its prognostic significance for patients with CRC remains controversial. To derive a precise estimation of the prognostic significance of cfDNA, a meta-analysis was performed.
Methods: We made a systematic search in data base of the Science Citation Index Embase and Pubmed for studies reporting prognostic data of cfDNA in CRC patients. The data of cfDNA on recurrences-free survival (RFS) and overall survival (OS) were extracted and measured in hazard rates (HRs) and 95% confident intervals (CIs). Subgroup analyses were carried out as well. Finally, the meta-analysis is accompanied with nine studies including 19 subunits.
Results: The pooled HRs with 95% CIs revealed strong associations between cfDNA and RFS (HR [95%CI]=2.78[2.08-3.72], I2=32.23%, n=7) along with OS (HR [95%CI]=3.03[2.51-3.66], I2=29.24%, n=12) in patients with CRC. Entire subgroup analyses indicated strong prognostic value of cfDNA irrespective tumor stage, study size, tumor markers, detection methods and marker origin.
Conclusions: All the results exhibits that appearance of cfDNA in blood is an indicator for adverse RFS and OS in CRC patients.
Keywords: Plasma DNA, cell-free DNA, colorectal cancer, prognosis, meta-analysis.
Introduction
Colorectal cancer is one of the leading causes responsible for millions of death each year all over the world1,2. Advance techniques has been implemented though no considerable results are observed, and metastatic diseases still suffers from poor prognosis3,4. Early detection of tumor progression and metastases remains the most effective way to improve the outcomes of patients.
Conventionally, carcinoembryonic antigen (CEA) and carbohydrate antigen-19-9 (CA 19-9) etc are clinically used as routine tumor markers to monitor disease progression. Nevertheless, these markers figure-out inadequate information due to their low sensitivity5-11. Thus new concepts are emerging in search of non-invasive biological markers that could be used to evaluate the risk of disease progression. In recent years, plasma cell-free DNA (cfDNA) has led an interest as a screening tool and is regarded as potential biomarkers for tumor burden in patients with many various cancers including CRC12-20. The cfDNA has been implicated as strong diagnostic and prognostic markers because the expression of cfDNA is usually altered in malignancies12-15. It is believed that the sources of cfDNA in blood stream are necrotic and apoptotic cells21-25. However, there are several differences regarding cfDNA of diverse origins. The cfDNA released from necrotic tumor cells differs in size, whereas cfDNA fragments released from apoptotic non-tumor cells are consistent and truncated measuring 185-200 base pairs in length26-29. In affected individuals, the cfDNA is released from necrotic cells while in healthy individual cfDNA is released from apoptotic cells 30-31. Integrity index has been widely accepted to deliberate the presence of cancers, which explains the ratio of longer to shorter fragments of DNAs32,33. Some studies have demonstrated that integrity index is higher in patients with colorectal cancer (CRC) and other cancers 20-23.
Based on the features of cfDNA in biology, it has been considered and investigated quantitatively and qualitatively as a promising non-invasive diagnostic and prognostic marker since long with various results34-39, however the approximated results were not distinctly marked. Two meta-analyses addressing the controversies regarding the diagnostic values of cfDNA in lung cancer24 and hepatocellular Carcinoma25 are recently published. But its prognostic role in cancer is still not well understood and needs to be interpreted statistically. Therefore, we performed a meta-analysis here to clarify the prognostic significance of cfDNA in blood of CRC patients, which is one of the most common malignancies worldwide.
Methods
Search Strategy and study selection
Potentially relevant studies were searched via Pubmed, Embase and Science citation prior to 3-July 2015 without applying start date limit. The term of colon cancer, rectal cancer, cell-free DNA, plasma DNA, serum DNA and prognosis were searched as topics and free words at the same time (see detailed search strategy Table S1-Table S3 in supplementary material). No language restrictions were imposed initially.
To yield relevant publications, we further evaluated the titles, abstract and author information of the collected studies. We contacted the authors for further studies if needed. For the analysis and full text review, only English language articles were considered. Non-research publications such as proceeding papers, editorials, comments, book chapter and other type uninvolved publications were excluded. However, cross references of relevant reviews were assessed for extra potential studies. Researchers would not appraised for detailed assessment unless the studies met the following inclusion criteria: (i) study should investigate the prognostic significance of cfDNA in colorectal cancer patients with at least one outcome (i.e., OS and RFS), (ii) the make-up of cfDNA was serum DNAs and plasma DNAs from peripheral blood (PB) rather than genomic and tumor DNAs, and (iii) studies from the same authors and institutions using the same cases were considered as duplicate publications, and only included the latest study to avoid re-assessment on the same cases.
For the meta-analysis, we endorsed the studies to be included in the final analysis through the following exclusion criteria: (i) duplicate publications, (ii) diagnostic studies, (iii) serum protein markers and genomic studies, (iv) studies with less than 20 patients, and (v) no survival or insufficient data to be statistically extracted.
Data extraction
Two reviewers (Basnet Shiva and Zhen-Yu Zhang) independently assessed the final set of articles and resolved disagreements by consensus. Data retrieved from the eligible study were: surname of the first author, number of patients, country of origin, year of publication, male/female ratio, mean/median age of patients, follow up durations, TNM staging (UICC), methods of detecting cfDNA, tumor locations, markers defining cfDNA, markers origin, sampling time, positive rate, endpoints and survival data including RFS, OS and other endpoints that could be estimated as RSF and OS.
Data processing and subgroup analysis
We used standard methods recommended for meta-analysis to statistically assess the prognostic effects of cfDNA on survival of CRC. HRs and accompanying 95%CIs were extracted individually if available. Apart from that, they were estimated using suggested methods by Parmar 40 and Tierney et al 41 based on survival data or survival curves. Besides, if HRs were mentioned by both univariate and multivariate analyses, multivariate analysis was preferable because of its consideration in possible confounding effects 42
Normally, HR >1 indicated a worse outcome. We pooled the extracted HRs with generic inverse variance method in Comprehensive Meta-analysis program (version2.2, Englewood, NJ, Biostat). Individual measures and overall effects were illustrated by forest plots 43. We used Cochrane's Q statistics and I2 Statistics to test the homogeneity of the studies 44 and applied Random effect model only when the tests were significant (two-tailed P value≤0.1, I2>50%) 45, 46.
For the studies with multiple markers, each of the markers was treated as an independent unit of analysis. If studies contained post-treatment time points, they were also meta-analyzed but also followed by sensitivity test after excluding them from the analysis. To classify the potential confounding factors related to outcomes of patients with CRC, we carried out subgroup analysis. The subgroup was stratified by tumor stages, tumor markers, methods, marker origin and size of the patients. We performed subgroup analysis only when there were two or more studies included.
Quality assessment and sensitivity analysis
Since no standardized tools could be applicable to observational studies, we used the Newcastle-Ottawa scale (NOS) 47 to assess the quality of the studies because it was recommended by the Cochrane library. Sensitivity analysis was performed to test reliability of the results. The combined HRs after random removal of one included study were calculated and explored to observe the influences of a particular study on overall results. Cumulative meta-analysis was performed to test the impact of publication year on stability of final results.
Publication bias
We statistically tested for the potential presence of publication bias using funnel plots along with Begg's and Mazumdar rank correlation method 48. All of the statistical techniques in the meta-analysis mentioned above had followed PRISMA checklist (Table S4).
Results
Global characteristics
From the extensive search made on 3rd July 2015, a total no. of 1282 articles was retrieved. Among the outcomes, 1023 studies were recognized as non-English publication, duplicates and studies out of scope and another 223 articles were reported as non-research articles, all of which were excluded from detailed assessment. Notably, three out of four studies from Spindler et al 49-51 and one out of two studies from Philipp et al 52 were excluded because of significant overlapping (the same author, same institutions, time period and part data were repeated), and only most recent studies (latest submission) were given priority. After the independent full review of the rest 36 studies, nine publications 53-61 dealing with quantitative and qualitative analysis of cfDNA with the prognostic purpose of CRC were considered as eligible studies (Figure 1). The 9 studies included a total of 1022 colorectal patients published between the year 2003 and 2014, eight from Europe 53-59, 61 and one from China 60. The mean patient no. per study was 114. The samples were collected before treatment (n=8) 53, 55-61 and after treatment (n=1) 54 One study reported cancer-related survival 57 which could be regarded as a subset of OS. Molecular techniques like PCR followed by sequencing 53, 54, Spectrophotometry 41, quantitative PCR (q-PCR) 42, 46 mutant-enriched PCR (ME-PCR) 57 and real-time PCR (rt-PCR) 58,59,61 were used to detect cfDNA from PB samples. Quality assessment was performed by NOS scale (Table S5) and all of the studies scored as high quality with 5 points or more. In total, all main characteristics of the eligible studies (19 units of analysis) were summarized (Table 1 and Table S6).
Major characteristics of included studies.
| Study | Country | Stage | Method | Markers | Marker origin | Sampling time | Positive rate | Endpoints | Hazard ratio | Multivariate/univariate analysis | Research quality | Reference No. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID-Name(Year) | n/N | |||||||||||
| Bazan (2006) | Italy | I-III | PCR and sequencing | K-RAS/TP53 | Plasma | Baseline | 8/50 | RFS | Data explorated | NR | High | [53] |
| Ryan (2003) | Netherland | I-III | PCR and sequencing | Mut.KRAS coden-12,13 | Serum | Post-Treatment | 15/85 | RFS | Reported in text | Multivariate | High | [54] |
| Schwarzenbach (2008) | Germany | IV | spectrophotometry | Total cfDNA | Serum | Baseline | 26/55 | RFS | Data explorated | NR | High | [55] |
| Spindler (2014) | Denmark | IV | q-PCR | Mut.KRAS BRAF Total cfDNA | Plasma | Baseline | 29/7/NR/86 | RFS/OS | Reported in text | Multivariate | High | [56] |
| Trevisiol (2012) | Italy | I-IV | ME-PCR | Mut.KRAS coden-12 | Serum | Baseline | 11/86 | OSa | Data explorated | NR | High | [57] |
| Wallner (2006) | Germany | I-IV | Real-Time PCR | mHLTF/ mhMLH1/mHPP1 | Serum | Baseline | 31/24/13/104 | OS | Reported in text | Univariate | High | [58] |
| Herbst (2008) | Germany | I-III | Real-Time PCR | mHLTF/mHPP1 | Seru m | Baseline | 13/6/106 | OS | Reported in text | Multivariate | High | [59] |
| Lin (2014) | China | I-IV | q-PCR | Total cfDNA | Plasma | Baseline | NR/191 | OS | Reported in text | Multivariate | High | [60] |
| Philipp (2014) | Germany | I-IV | Real-Time PCR | mHLTF/mHPP1 | Serum | Baseline | 41/57/259 | OS | Data explorated | NR | High | [61] |
Study selection flow chart.
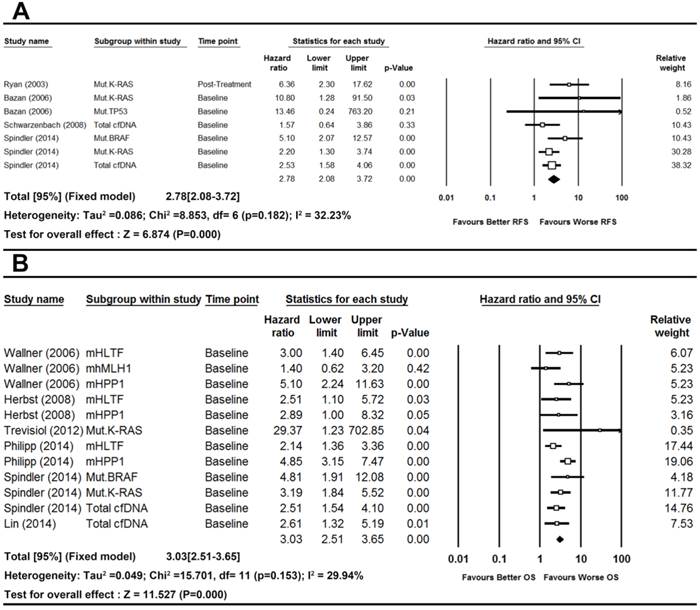
Heterogeneity and overall effects
There was no significant heterogeneity observed in the statistical tests among study group of RFS (I2=32.23%, P-value=0.1820) and OS (I2=29.94%, P-value=0.1530). Thus we used fixed effects model. The pooled results demonstrated that the detection of cfDNA had significant value in predicting RFS (HR [95%CI]=2.78[2.08-3.72], n=7) and OS (HR [95%CI]=3.03[2.51-3.66], n=12) in patients with CRC (Figure 2).
Sensitivity analysis
To test whether the results were stable, we therefore performed sensitivity analysis. First of all, when the fixed effects model was switched to random effects model to re-compute the pooled HRs, the results showed no significant changes in overall effects for RFS (HR [95%CI]=2.99 [2.00-4.47]) and OS (HR [95%CI]=3.02[2.38-3.83]). Secondly, when the single study by Ryan (2003)54 with post-treatment data from RFS group was removed from the analysis, the result remained stable (HR [95%CI]=2.58, [1.90-3.50], n=6, I2=17.86, P-value=0.298). The results showed similar effect after the removal of the study by Schwarzenbach (2008)55 with the method of spectrophotometry from the RFS group because the result was only slightly changed (HR[95%CI]=2.97[2.18-4.04], n=6, I2=29.89%, P-value=0.211). Similarly, sensitivity analysis showed no significant changes when removing total cfDNA from the analysis (HR [95%CI]=3.34, [2.22-5.02], n=5, I2=37.26, P-value=0.173). As per the study in OS group, we excluded the study by Trevisiol (2012) 57 which reported cancer-related survival. The analysis did not lead any obvious changes in the total effects (HR[95%CI]=3.00[2.49-3.62], n=11, I2=27.14%, P-value=0.186). Likewise when total cfDNA was removed from the analysis, no obvious changes was seen in the combined results (HR [95%CI]=3.18, [2.57-3.94], n=10, I2=39.01, P-value=0.098). Finally, the combined effects of the analysis were stable even randomly removing one particular study from RFS and OS groups (Figure S1 and Figure S2). The situations were similar with cumulative meta-analysis of both groups cumulated by the year of publication (Figure S3 and Figure S4).
Forest plots of pooled effects for RFS (a) and OS (b) in CRC patients.
Subgroup Analysis
Although no significant heterogeneity was observed in our including studies either on RFS or OS. It was reported that, tumor markers, tumor stage, marker origin, methods and population size for the presence on cfDNA acted as confounding factors which might exert considerable influences on outcomes of cancer patients62. Therefore, we performed subgroup analysis stratified according to the above mentioned factors. And the findings had confirmed the sensibility. In subgroup analysis (Table 2) followed by patient size, the results remained stable. Particularly, the combined HRs and associated CIs of both group was >1 with narrower CIs in large patient group. The results implied studies containing larger patient number précised a better prediction in survival effects. Similarly, when stratified by stage, we found a similar direction of pooled HRs in predicting survival of patients in both localized (UICC stage I-III) and metastatic (UICC stage IV) cancer patients. However heterogeneity became significant when pooling them together (M0+M1).The subgroup analysis stratified by methods demonstrated that the combined HRs and associated CIs of both RFS and OS seemed stable, nevertheless, qPCR technique showed reliable results with narrower CIs. Finally subgroup analysis based on type of tumor markers showed both quantitative and qualitative detection of cfDNA were predictive in RFS and OS, while contributing to considerable inter-study heterogeneity, the situation was similar with marker origin. In depth analysis showed that methylated, mutant markers and total cfDNA determination were of prognostic significance.
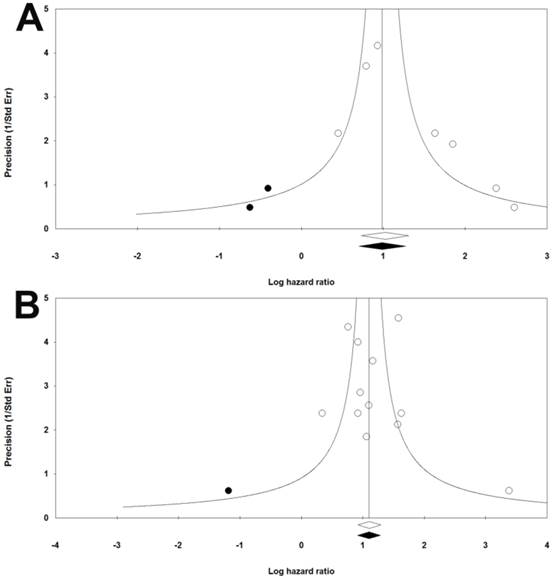
Publication bias
The funnel plots were symmetric. Meanwhile, results of Begg's and Mazumdar rank correlation were not significant for both group (RFS: two-tailed P-value=0.5480, OS: two-tailed P-value=0.4507) (Figure 3). Hence, there were no obvious publication biases in our included studies.
Subgroup analysis of RFS and OS studies.
| Variables | RFS | OS | ||||||
|---|---|---|---|---|---|---|---|---|
| HR[95%CI] | n | I2 (%) | P | HR[95%CI] | n | I2 (%) | P | |
| Overall | 2.78[2.07-3.71] | 7 | 32.23 | 0.182 | 3.03[2.51-3.65] | 12 | 29.94 | 0.153 |
| Marker type | ||||||||
| methylated | / | / | / | / | 2.94[2.04-4.24] | 7 | 49.94 | 0.062 |
| mutant | 3.34[2.22-5.02] | 5 | 37.26 | 0.173 | 3.72[2.33-5.93] | 3 | 10.14 | 0.329 |
| total cfDNA | 2.29[1.51-3.47] | 2 | 0.00 | 0.355 | 2.54[1.71-3.79] | 2 | 0.00 | 0.926 |
| Markers | ||||||||
| mHLTF | / | / | / | / | 2.365[1.67-3.36] | 3 | 0.00 | 0.745 |
| mHPP1 | / | / | / | / | 4.62[3.22-6.61] | 3 | 0.00 | 0.649 |
| mut.K-RAS | 4.04[1.57-10.35] | 3 | 58.06 | 0.092 | 3.40[1.98-5.84] | 2 | 45.16 | 0.177 |
| total cfDNA | 2.29[1.51-3.47] | 2 | 0.00 | 0.355 | 2.54[1.71-3.80] | 2 | 0.00 | 0.926 |
| others | 5.35[2.22-12.89] | 2 | 0.00 | 0.646 | 2.55[0.76-8.51] | 2 | 73.74 | 0.051 |
| Marker origin | ||||||||
| serum | 3.09[0.79-12.19] | 2 | 72.41 | 0.044 | 3.09[2.43-3.93] | 8 | 49.74 | 0.052 |
| plasma | 2.75[1.99-3.80] | 5 | 16.09 | 0.312 | 2.92[2.15-3.96] | 4 | 0.00 | 0.638 |
| Method | ||||||||
| PCR and sequencing | 7.24[2.955-17.77] | 3 | 0.00 | 0.866 | / | / | / | / |
| qPCR | 2.64[1.90-3.66] | 3 | 20.97 | 0.282 | 3.01[2.49-3.62] | 11 | 27.11 | 0.186 |
| others | / | 1 | / | / | / | 1 | / | / |
| Tumor stage | ||||||||
| I-III | 7.25[3.00-17.77] | 3 | 0.00 | 0.866 | 2.65[1.38-5.07] | 2 | 0.00 | 0.838 |
| IV | 2.48[1.82-3.37] | 4 | 17.90 | 0.301 | 3.01[2.14-4.22] | 3 | 0.00 | 0.457 |
| I-IV | / | / | / | / | 3.05[2.03-4.58] | 7 | 56.83 | 0.031 |
| Patient size | ||||||||
| ≤ mediana | 3.39[1.79-6.39] | 4 | 48.73 | 0.119 | 2.98[2.27-3.92] | 7 | 28.64 | 0.210 |
| >median | 2.64[1.90-3.66] | 3 | 20.97 | 0.282 | 3.07[2.37-3.98] | 5 | 44.98 | 0.122 |
a The median patient number in RFS and OS subgroups are 85 and 104, respectively.
Funnel plots of publication bias for RFS (a) and OS (b) in CRC patients.
Discussion
After preliminary pooling 19 subgroups from 9 studies, independently stratified by marker types, markers origin, methods, tumor stage and patient size, it was proven that the presence of cfDNA could be used to predict RFS and OS in CRC.
There are some novels findings in our meta-analysis which should be taken into consideration in future studies investigating prognostic significance of cfDNA. Briefly, there are two schemes to characterized cfDNA, i.e., quantification of total cfDNA by spectrophotometry and detection of tumor specific markers by PCR based techniques along with sequencing. We observed both strategies were effective in detection of cfDNA while the combined HRs seems to be more prominent in subgroups by detection of tumor specific markers rather than those by quantification of cfDNA. It might be the reason that PCR based techniques was more sensitive and specific than non-selective cfDNA quantification. Besides, tumor specific methylations and mutations were also associated with RFS and OS of CRC, although the prognostic influences varied in a marker-dependent manner. For example, we could observe certain differences in overall effects of mHLTF, mHPP1 and mutant K-RAS groups. The subgroup analysis classified by tumor stage exhibited cfDNA could be applied in both localized and metastatic group of CRCs. However, heterogeneity increased in subgroup of stage I-IV indicating that there might be significant difference in prognosis of early and late stages CRCs, which should be treated separately to obtain accurate predictive information when estimating CRC patient survival. Further subgroup analysis based on patient size demonstrated that a larger patient size yielded more precise survival estimations with a narrower 95% CI of HRs. In addition, there was only one study reported post-treatment cfDNA (HR[95%CI]=6.36[2.30-17.62]), although it was significant. The use of cfDNA at post-treatment time points remained to be validated. As to the subgroups stratified by other markers, namely mhMLH1 and mutant BRAF, the combined effects of these two studies was insignificant (HR [95%CI]=2.55[0.76-8.51], I2=73.74, P-value=0.051). The culprit in the studies might be the incapacity of mhMLH1, which could be interpreted as a marker-dependent manner as mention above. Similarly in subgroup stratified by markers origin; namely serum, the result was insignificant (HR [95%CI]=3.09[0.79-12.19], I2=72.41, P-value=0.044).The level of heterogeneity might be due to the sample with different time point. Regarding method analysis, both PCR sequencing and qPCR were significant. However, qPCR showed more precise results with narrower 95%CI (HR [95%CI]=2.64[1.90-3.66], I2=20.97, P-value=0.282). Entirely, types of marker, markers origin, detection methods and tumor stage might contribute to substantial inter-study heterogeneity of included studies.
It should be noted that we encountered some limitations caused by the nature of meta-analysis, which made us to interpret the result with cautions. There were limited studies included in our meta-analysis because cfDNA was a novel marker and reported only recently. This might lead to bias based on limited data. However, sensitivity analysis showed that the combined HRs were stable and reliable for subsequent analysis. Included studies were homogenous, which was probably because of our strict inclusion criteria. We pre-expected there might be a certain degree of heterogeneity, and thus tried to perform subgroup analysis to explore the sources of existing heterogeneity (RFS, I2=32.23, P-value=0.182, OS, I2=29.94, P-value=0.153). We indeed found some confounding factors probably leading to inconsistencies among studies, i.e., methods, type of markers and tumor stage. Lastly, there were no publication biases illustrated by funnel plots and further demonstrated by Begg's and Mazumdar rank correlation tests.
Conclusion
In conclusion, our meta-analysis has revealed the significant prognostic values of cfDNA for RFS and OS in patients with CRC. Further studies should compare the difference between conventional serum tumor markers and cfDNA as alternatives. More studies are expected to investigate sensitive tumor specific markers and compare multiple time points in different tumor stage group in order to prove the clinical utility of cfDNA.
Abbreviations
cfDNA: Cell-free deoxyribonucleic acid; CRC: Colorectal Cancer; RFS: Recurrence free survival; OS: Overall survival; HR: Hazard ratio; CI: Confident Interval; CEA: Carcinoembryonic antigen; CA19-9: Carbohydrate antigen 19-9; PB: peripheral blood; TNM staging: Tumor Nodes Metastasis staging; UICC: Union for International cancer control; NOS: Newcastle-Ottawa Scale; PCR: Polymerase Chain Reaction; q-PCR: quantitative Polymerase Chain Reaction; ME-PCR: Mutant Enriched Polymerase Chain Reaction; rt-PCR: real time Polymerase Chain Reaction.
Supplementary Material
Supplementary tables and figures.
Acknowledgements
The work was supported by grants from the Shanghai Science and Technology Commission (134119b0600), the Shanghai Municipal Health Bureau (20134194), the National Natural Science Fund of China (81272533), Medical Health Plan of Zhejiang province (2016KYB295) and Science and Technology Planning Project of Jiaxing (2015AY23071).
Competing Interests
The authors declare that they have no potential competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5-29
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
3. O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. Journal of the National Cancer Institute. 2004;96:1420-5
4. Moon SH, Kim DY, Park JW, Oh JH, Chang HJ, Kim SY. et al. Can the new American Joint Committee on Cancer staging system predict survival in rectal cancer patients treated with curative surgery following preoperative chemoradiotherapy? Cancer. 2012;118:4961-8
5. Gion M, Mione R, Barioli P, Dittadi R. Dynamic use of tumor markers, rationale-clinical applications and pitfalls. Anticancer research. 1996;16:2279-84
6. Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nature reviews Cancer. 2010;10:371-8
7. Kaneko I, Tanaka S, Oka S, Yoshida S, Hiyama T, Arihiro K. et al. Immunohistochemical molecular markers as predictors of curability of endoscopically resected submucosal colorectal cancer. World J Gastroenterol. 2007;13:3829-35
8. Glasgow SC, Bleier JI, Burgart LJ, Finne CO, Lowry AC. Meta-analysis of histopathological features of primary colorectal cancers that predict lymph node metastases. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2012;16:1019-28
9. Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. The Journal of molecular diagnostics: JMD. 2007;9:305-14
10. Walker AS, Johnson EK, Maykel JA, Stojadinovic A, Nissan A, Brucher B. et al. Future directions for the early detection of colorectal cancer recurrence. Journal of Cancer. 2014;5:272-80
11. Famulski W, Sulkowska M, Wincewicz A, Kedra B, Pawlak K, Zalewski B. et al. P53 correlates positively with VEGF in preoperative sera of colorectal cancer patients. Neoplasma. 2006;53:43-8
12. Fujimoto A, O'Day SJ, Taback B, Elashoff D, Hoon DS. Allelic imbalance on 12q22-23 in serum circulating DNA of melanoma patients predicts disease outcome. Cancer research. 2004;64:4085-8
13. Tanaka R, Koyanagi K, Narita N, Kuo C, Hoon DS. Prognostic molecular biomarkers for cutaneous malignant melanoma. Journal of surgical oncology. 2011;104:438-46
14. Taback B, Hoon DS. Circulating nucleic acids and proteomics of plasma/serum: clinical utility. Annals of the New York Academy of Sciences. 2004;1022:1-8
15. Agostini M, Pucciarelli S, Enzo MV, Del Bianco P, Briarava M, Bedin C. et al. Circulating cell-free DNA: a promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Annals of surgical oncology. 2011;18:2461-8
16. Chan LL, Jiang P. Bioinformatics analysis of circulating cell-free DNA sequencing data. Clinical biochemistry. 2015;48:962-75
17. Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y. et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. British journal of cancer. 2014;111:1482-9
18. Chan KC, Leung SF, Yeung SW, Chan AT, Lo YM. Persistent aberrations in circulating DNA integrity after radiotherapy are associated with poor prognosis in nasopharyngeal carcinoma patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:4141-5
19. Yoruker EE, Ozgur E, Keskin M, Dalay N, Holdenrieder S, Gezer U. Assessment of circulating serum DNA integrity in colorectal cancer patients. Anticancer research. 2015;35:2435-40
20. Vlassov VV, Laktionov PP, Rykova EY. Circulating nucleic acids as a potential source for cancer biomarkers. Current molecular medicine. 2010;10:142-65
21. Ziegler A, Zangemeister-Wittke U, Stahel RA. Circulating DNA: a new diagnostic gold mine? Cancer treatment reviews. 2002;28:255-71
22. Sirera R, Bremnes RM, Cabrera A, Jantus-Lewintre E, Sanmartin E, Blasco A. et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:286-90
23. Linder S, Havelka AM, Ueno T, Shoshan MC. Determining tumor apoptosis and necrosis in patient serum using cytokeratin 18 as a biomarker. Cancer letters. 2004;214:1-9
24. Zhang R, Shao F, Wu X, Ying K. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysis. Lung cancer (Amsterdam, Netherlands). 2010;69:225-31
25. Liao W, Mao Y, Ge P, Yang H, Xu H, Lu X. et al. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine. 2015;94:e722
26. Giacona MB, Ruben GC, Iczkowski KA, Roos TB, Porter DM, Sorenson GD. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89-97
27. Gang F, Guorong L, An Z, Anne GP, Christian G, Jacques T. Prediction of clear cell renal cell carcinoma by integrity of cell-free DNA in serum. Urology. 2010;75:262-5
28. Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ. et al. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clinical chemistry. 2006;52:1062-9
29. Mangano A, Mangano A, Lianos GD, Cassinotti E, Roukos DH, Dionigi G. et al. Circulating free DNA in plasma or serum as biomarkers of carcinogenesis in colon cancer. Future oncology (London, England). 2015;11:1455-8
30. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer research. 2001;61:1659-65
31. Hauser S, Zahalka T, Ellinger J, Fechner G, Heukamp LC, A VONR. et al. Cell-free circulating DNA: Diagnostic value in patients with renal cell cancer. Anticancer research. 2010;30:2785-9
32. Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC. et al. Increased plasma DNA integrity in cancer patients. Cancer research. 2003;63:3966-8
33. Charlot DJ, Bhalla M, Krishnan R, Tu G. Direct quantitative PCR of LINE repeats to quantify CFC-DNA in serum. Cancer research. 2014;74:1522 -
34. Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. British journal of cancer. 2011;105:239-45
35. Chen H, Sun LY, Zheng HQ, Zhang QF, Jin XM. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44:318-24
36. da Silva Filho BF, Gurgel AP, Neto MA, de Azevedo DA, de Freitas AC, Silva Neto Jda C. et al. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. Journal of clinical pathology. 2013;66:775-8
37. Feng J, Gang F, Li X, Jin T, Houbao H, Yu C. et al. Plasma cell-free DNA and its DNA integrity as biomarker to distinguish prostate cancer from benign prostatic hyperplasia in patients with increased serum prostate-specific antigen. International urology and nephrology. 2013;45:1023-8
38. Pinzani P, Salvianti F, Zaccara S, Massi D, De Giorgi V, Pazzagli M. et al. Circulating cell-free DNA in plasma of melanoma patients: qualitative and quantitative considerations. Clinica chimica acta; international journal of clinical chemistry. 2011;412:2141-5
39. Sriram KB, Relan V, Clarke BE, Duhig EE, Windsor MN, Matar KS. et al. Pleural fluid cell-free DNA integrity index to identify cytologically negative malignant pleural effusions including mesotheliomas. BMC cancer. 2012;12:428
40. Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in medicine. 1998;17:2815-34
41. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
42. Smith T, Smith B, Ryan MA. Survival analysis using Cox proportional hazards modeling for single and multiple event time data. Proceedings of the twenty-eighth annual SAS users group international conference, SAS Institute, Inc, Cary. 2003:254-28
43. Song F, Sheldon TA, Sutton AJ, Abrams KR, Jones DR. Methods for exploring heterogeneity in meta-analysis. Evaluation & the health professions. 2001;24:126-51
44. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21:1539-58
45. Petitti DB. Approaches to heterogeneity in meta-analysis. Statistics in medicine. 2001;20:3625-33
46. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327:557-60
47. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European journal of epidemiology. 2010;25:603-5
48. Macaskill P, Walter SD, Irwig L. A comparison of methods to detect publication bias in meta-analysis. Statistics in medicine. 2001;20:641-54
49. Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10:e0108247
50. Spindler KG, Appelt AL, Pallisgaard N, Andersen RF, Jakobsen A. KRAS-mutated plasma DNA as predictor of outcome from irinotecan monotherapy in metastatic colorectal cancer. British journal of cancer. 2013;109:3067-72
51. Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. Changes in mutational status during third-line treatment for metastatic colorectal cancer-results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. International journal of cancer Journal international du cancer. 2014;135:2215-22
52. Philipp AB, Stieber P, Nagel D, Neumann J, Spelsberg F, Jung A. et al. Prognostic role of methylated free circulating DNA in colorectal cancer. International journal of cancer Journal international du cancer. 2012;131:2308-19
53. Bazan V, Bruno L, Augello C, Agnese V, Calo V, Corsale S. et al. Molecular detection of TP53, Ki-Ras and p16INK4A promoter methylation in plasma of patients with colorectal cancer and its association with prognosis. Results of a 3-year GOIM (Gruppo Oncologico dell'Italia Meridionale) prospective study. Annals of Oncology. 2006;17:vii84-vii90
54. Ryan BM, Lefort F, McManus R, Daly J, Keeling PW, Weir DG. et al. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut. 2003;52:101-8
55. Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Annals of the New York Academy of Sciences. 2008;1137:190-6
56. Spindler KLG, Appelt AL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Cell-free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. International Journal of Cancer. 2014;135:2984-91
57. Trevisiol C, Di Fabio F, Nascimbeni R, Peloso L, Salbe C, Ferruzzi E. et al. Prognostic value of circulating KRAS2 gene mutations in colorectal cancer with distant metastases. The International journal of biological markers. 2006;21:223-8
58. Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Goke B. et al. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:7347-52
59. Herbst A, Wallner M, Rahmig K, Stieber P, Crispin A, Lamerz R. et al. Methylation of helicase-like transcription factor in serum of patients with colorectal cancer is an independent predictor of disease recurrence. European journal of gastroenterology & hepatology. 2009;21:565-9
60. Lin JK, Lin PC, Lin CH, Jiang JK, Yang SH, Liang WY. et al. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: based on the mutation spectra detected in primary tumors. Ann Surg Oncol. 2014;21(Suppl 4):S680-6
61. Philipp AB, Nagel D, Stieber P, Lamerz R, Thalhammer I, Herbst A. et al. Circulating cell-free methylated DNA and lactate dehydrogenase release in colorectal cancer. BMC cancer. 2014;14:245
62. Zhang ZY, Dai ZL, Yin XW, Li SH, Li SP, Ge HY. Meta-analysis shows that circulating tumor cells including circulating microRNAs are useful to predict the survival of patients with gastric cancer. BMC cancer. 2014;14:773
Author contact
![]() Corresponding authors: Hai-yan Ge, Email: ghy1333cn Zhen-Yu Zhang Email: zhangsurgerycom. Department of Gastrointestinal Surgery, Shanghai East Hospital, Tongji University School of Medicine, Pudong New District, No. 150, Jimo Road, Shanghai, 200120, China. Tel: +86 21 38804518 Fax: +86 21 58798999.
Corresponding authors: Hai-yan Ge, Email: ghy1333cn Zhen-Yu Zhang Email: zhangsurgerycom. Department of Gastrointestinal Surgery, Shanghai East Hospital, Tongji University School of Medicine, Pudong New District, No. 150, Jimo Road, Shanghai, 200120, China. Tel: +86 21 38804518 Fax: +86 21 58798999.

 Global reach, higher impact
Global reach, higher impact