3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(10):1233-1241. doi:10.7150/jca.14616 This issue Cite
Review
The Role of Mir-148a in Cancer
1. Department of Pathology and Pathophysiology, Hunan Normal University Medical School, Changsha 410013, Hunan, China;
2. Department of Statistics and Epidemiology, Public Health School, Central South University, Changsha 410078, Hunan, China.
Received 2015-12-7; Accepted 2016-5-7; Published 2016-6-21
Abstract
MicroRNAs (miRNAs) are highly conserved noncoding RNAs of about 19-25 nucleotides. Through specifically pairing with complementary sites in 3' untranslated regions (UTRs) of target mRNAs, they mediate post-transcriptional silencing. MicroRNAs have been implicated in many physiological processes including proliferation, differentiation, development, apoptosis, and metabolism. In recent years many studies have revealed that the aberrant expression of miRNA is closely related to oncogenesis and is now an intense field of study. Mir-148a is aberrantly expressed in various cancers and has been identified as an oncogenic or tumor suppressor with crucial roles in the molecular mechanisms of oncogenesis. In this review, we have summarized the role of mir-148a in the oncogenic pathways of gastric, liver, breast and urogenital cancers, and in neurogliocytoma oncogenesis. Studying the functional role of mir-148a is crucial in discovering novel tumor molecular markers and identifying potential therapeutic targets.
Keywords: miRNA, mir-148a, cancer, oncogene, tumor suppressor gene, biological function.
Introduction
MicroRNA (miRNA) is a category of small, noncoding, single-stranded RNAs of 19-25 nucleotides that are widely found in both animals and plants. Since Lee1 identified lin-4 as the first miRNA member in C. elegans as far back as 1993, more than 28, 000 miRNAs have been found in animals, plants and viruses. MiRNAs are a novel group of gene regulators wherein their 5' seed sequences bind to complementary sequences in the 3' untranslated region (UTR) of their target mRNAs, whereupon they induce negative regulation. MiRNAs that completely bind to complementary sequences of their target mRNAs will induce mRNA degradation (common in plants). Under situations where miRNAs bind incompletely, translational repression is induced, which downregulates protein expression but not mRNA stability (common in mammals).2
Human miRNAs are mainly located in the introns of genes or the noncoding RNA transcript regions.3 MiRNAs can mediate about 30% protein-coding genes in humans.4 Any particular miRNA species can regulate multiple mRNAs and different miRNAs can coordinately mediate regulation of the same mRNA target. Thus, a complicated regulatory network is formed by many miRNAs and their target genes are involved in various cell functions and processes including early development, proliferation, differentiation, and apoptosis.5,6
Accumulating evidence has shown that miRNAs play important roles in tumorigenesis, development and clinical therapy. About 50% of miRNAs are located in tumor-related fragile sites or genomic regions, which indicate their potential crucial roles.7 MiRNAs that are aberrantly expressed are related to most tumors, and they interact directly or indirectly with oncogenes and tumor suppressor genes to regulate their expression. Moreover, miRNAs, as oncogenes or tumor suppressor genes, are involved in various cell functions including proliferation, apoptosis, angiogenesis, invasion and tumor metastasis.5,8,9 MiRNAs has revealed themselves as potential tumor markers and are predicted to be novel therapeutic targets.
mir-148a gene
Location and expression of mir-148a
The mir-148/mir-152 family is composed of three highly conserved and mature miRNAs with similar sequences, structures and the same seed region (i.e., UCAGUGCA), including mir-148a, mir-148b and mir-152. Precursor mir-148/mir-152 with a stem-loop structure is cleaved by a series of enzymes in the nucleus and cytoplasm to form mir-148a, mir-148b and mir-152 sequences. In human chromosomes, mir-148a with 68 nucleotide sequences locates to chromosome 7p15.2 and its stem-loop structure sequence locates to the negative strand of chromosome 7: 25949919-25949986; mir-148b with 99 nucleotide sequences locates to 12q13.13 and its stem-loop structure sequence locates to the positive strand of chromosome 12: 54337216-54337314; mir-152 with 87 nucleotide sequences locates to 17q21.32 and its stem-loop structure sequence locates to the negative strand of chromosome 17: 48037161-48037247. Studies have shown that mir-148/152 is close to the human homeobox genes (HOX), and mir-148a, mir-148b and mir-152 is close to the HOXA, HOXC and HOXB respectively.7
Under normal physiological conditions, the mir-148a gene is expressed in various human tissues including cerebral, heart, liver, thymus, pancreas, renal, placenta, uterus, testis, and the hematopoietic system.10-12 Fu et al.13 found high expression levels of mir-148a in the liver, but no significant difference in expression was found between mature and embryonic liver. Ribeiro-dos-Santos et al.14 found that mir-148a was expressed in normal gastric tissues and was most highly expressed in gastric cardia biopsies. Additionally, highly expressed mir-148a may be involved in regulation to maintain gene expression in gastric tissue stability, and its abnormal expression might induce gastric neoplasms. Nielsen et al.15 detected mir-148a expression and found that exercise could affect its expression levels in blood. Following 12-week continuous exercise, the expression level of mir-148a declined significantly in the blood circulation.
Biological function of mir-148a gene
Mir-148a has common functions of many miRNA species, and is implicated in a series of biological processes, including cellular differentiation and development. Porstner et al.16 demonstrated that mir-148a expression increased after pre-B cell activation. Next, upregulated mir-148a expression promotes the differentiation of activated B cells to plasma cells, and enhances the survival of plasma cells by inhibiting the transcription factors BACH2 and MITF and the pro-apoptotic factors BIM and PTEN.
Mir-148a is a novel muscle-derived miRNA that can facilitate the differentiation of the C2C12 myoblast and skeletal muscle cells by targeted regulatory Rho-associated coiled-coil containing protein kinase 1 (ROCK1). Mir-148a can also promote C2C12 myoblasts to grow in the G1 phase of the cell cycle and can shorten the S phase that prompts myoblast differentiation into the myotube.17 Van Wijnen et al. 18 observed that mir-148a was involved in osteoclast formation and induced transformation of monocytes to osteoclasts by inhibiting the transcription factor V-maf musculo-aponeurotic fibrosarcoma oncogene homolog B (MAFB). Gailhouste et al.19 proposed that enhanced expression of mir-148a could induce the differentiation and maturation of liver cells by inhibiting DNA (cytosine-5-)-methyltransferase 1 (DNMT1). In addition, mir-148a could also promote primary adipocytes to differentiate to mature adipocytes.20,21 Mir-148a can also affect the development of the nervous system by the targeted regulation of ENAH, STX3 and HMBS during neural development.22 Others have shown that mir-148a regulates several phenotypes including those seen in embryonic stem cells;23 moreover, downregulated expression of mir-148a could control the phenotype of mesenchymal stem cells (MSC) by facilitating the expression of endothelial PAS domain-containing protein 1 (EPAS1) transforming factor.24
Relationship between mir-148a and tumor
Mir-148a expression in tumor and the clinical significance
The downregulated expression of mir-148a can be detected in various cancers including gastric, colorectal, pancreatic, liver, esophageal, breast, non-small cell lung and urogenital system cancers. However, upregulated expression of mir-148a can also be seen in glioma and osteosarcoma. The expression levels of mir-148a have been linked to the clinical classification, efficacy and prognosis of the tumor.
Gastric cancer (GC)
GC is one of the most common malignant diseases of the digestive system. In recent years, studies have indicated that aberrantly expressed miRNAs contributed to the initiation and progression of GC.25,26 Mir-148a was significantly downregulated in GC cell-lines and in GC tissue samples, and thus served as a tumor suppressor.27,28 The downregulated expression of miR-148a was significantly associated with tumor size, TNM stage and lymph node metastasis, progression and prognosis.29,30 By contrast, Song et al.31 found that mir-148a targeted the single nucleotide polymorphism (SNP) rs6976789 (C>T) to affect the risk and prognosis of gastric cancer. The survival rate of patients with the TT mutation was significantly lower than patients expressing the CC/CT mutation in intestinal-type GC.
Colorectal cancer (CC)
CC is one of the most common malignant diseases of the digestive system in which the prevalence has increased worldwide due to rising living standards and an altered dietary structure. CC is a polygenic, multi-factorial and multi-step disease process, in which miRNAs are involved. Takahashi et al.32 discovered that mir-148a was significantly downregulated in CC cells and tissues. The downregulated expression of mir-148a was associated with the poor prognosis associated with stage III patients and the treatment response of stage IV patients after adjuvant chemotherapy. Tsai et al.33 showed that miR-148a had substantially different expression levels in early and non-early relapsed stage II and III CRC tissues and lower levels of mir-148a expression were associated with significantly shorter disease-free survival(DFS) rates and poorer overall survival(OS) rates.
Pancreatic cancer (PC)
The downregulated expression of mir-148a was determined in PC tissues and various PC cell-lines.34 Hanoun et al.35 believed that downregulated mir-148a was linked to hypermethylation of the coding regions in PC and in epithelial tumor tissues. In addition, detection of mir-148a expression and its methylation could be an auxiliary indicator of PC development and chronic pancreatitis.
Hepatocellular carcinoma (HCC)
Some special abnormally expressed miRNA species were linked to certain clinic-pathological characteristics including metastasis, recurrence and prognosis.36 Heo et al.37 found that mir-148a was clearly downregulated in HCC tissues as compared to the adjacent control group. The expression levels of mir-148a with microvascular metastasis was lowerer than those mir-148a levels lacking metastasis. The suppressed expression of mir-148a was associated with the high levels of AFP, poor TNM stage, low OS and the recurrence-free survival rate in patients with hepatocellular carcinoma. Pan et al.38 found that mir-148a was negatively correlated with HCC TNM stage and capsular infiltration. Yan et al.39 confirmed that the expression of mir-148a in poorly differentiated HCC was lower than that found in well-differentiated HCC.
Esophagus cancer (EC)
Wijnhoven et al.40 found that mir-148a was downregulated in esophageal adenocarcinoma tissues when exploring the expression levels of miRNA in normal squamous epithelium, gastric epithelium, Barrett's intestinal metaplastic epithelium and adenomatous carcinoma tissues with gene chips. Hummel et al.41 discovered that mir-148a expression was negatively correlated with esophageal adenocarcinoma differentiation, recurrence of esophageal squamous cell carcinoma and survival time after surgery. Moreover, upregulated expression of mir-148a could improve drug sensitivity of esophageal adenocarcinoma and squamous cell carcinoma to cisplatin and 5-FU.42
Breast cancer (BC)
BC is a very common malignant tumor of female patients that markedly threatens their health. Yu et al.43 argued that mir-148a was downregulated in BC and served as a suppressor gene. Mir-148a was negatively associated with the tumor grade and lymph node metastasis.44 Xu et al.45 verified that the downregulation of mir-148a was related to hypermethylation of its promoter region and presence of the triple negative breast cancer (TNBC) phenotype. Mir-148a could make BC free from immune evasion by controlling the expression of human leukocyte antigen G (HLA-G).46 Thus, it could serve an important role in immunoregulation.
Non-small cell lung cancer (NSCLC)
Mir-148a expression was attenuated in 80% of NSCLC tissues as compared to adjacent normal lung tissues.47 Mir-148a was downregulated in the blood of patients that was linked to tumor size and lymph node metastasis.48 Mir-148a was apparently downregulated if the size of the tumor exceeded 3 cm or during metastasis.49 Chen et al.50 suggested that mir-148a suppression was associated with advanced clinical stages, low disease free and overall survival rates.
Urogenital system tumor
Fujita et al.51 expected that mir-148a was significantly downregulated in PC3 and DU145 hormone-refractory prostate cancer (PC) cell-lines. MiR-148a attenuated paclitaxel-resistance of PC3PR cells by targeting the inhibition of the expression of mitogen- and stress-activated kinase 1 (MSK1) in paclitaxel-resistant PC3 (PC3PR) cells. Lombard et al.52 discovered that mir-148a expression was suppressed in urothelial cell carcinoma (UCC) cell-lines and that overexpression of mir-148a and use of cisplatin and doxorubicin could synergistically facilitate apoptosis of UCC. Zhou et al.53 found that mir-148a expression was down-regulated in ovarian cancer cell lines, and also decreased in ovarian cancer tissues. Gu et al.54 demonstrated that downregulated mir-148a expression identified low OS and short survival time of patients with OC, while upregulated mir-148a predicted both elevated OS and prolonged survival time.
Glioma
This particular cancer is one of the most common primary brain tumors in human subjects. Research has shown that various miRNAs, including that of mir-148a was involved in regulating the development and progression of glioma.55,56 Some studies have illustrated that mir-148a served as a negative risk factor in glioblastoma. Upregulated mir-148a could accelerate the malignant process and was negatively correlated with the survival rate.57,58 In addition, Li et al.59 found that mir-148a was a tumor suppressor gene that was downregulated in the isocitrate dehydrogenase 1 (IDH1) mutant form of glioma due to methylation of the mir-148a promoter, which could suppress its expression and facilitate the malignant process.
Osteosarcoma
Ma et al.60 found that mir-148a was especially upregulated in samples of distant metastases by performing blood analysis of 89 osteosarcoma patients. Mir-148a expression was negatively correlated with the tumor size and presence of distant metastases. The high expression levels of mir-148a predicted a poor OS.
The expression regulation of mir-148a in tumors
Recent research has shown that regulation of mir-148a expression occurs predominantly at the levels of the chromosome, transcription and methylation.
Regulation at chromosome level
The amplification and loss of heterozygosity (LOH) in miRNA sites could cause an abnormal miRNA gene copy number.61 Arslantas et al. found that in glioblastoma, the chromosome 7p region gene was amplified.62,63 Brezinova et al.64 discovered that gene deletions frequently occurred in the chromosomal region of 7p 13.2-15.2 of marrow malignant tumors. Therefore, this might explain why the expression of mir-148a is increased or decreased. In addition, Kulkarni et al.65 found that the mir-148a gene expresses the SNP rs735316 and rs111299611, and that a mutation in this region could affect mir-148a expression. The rs736316G/G phenotype or rs111299611 deletion mutation could increase mir-148a expression levels; by contrast, the rs735316A/A phenotype or the rs111299611 insertion mutation could decrease mir-148a expression levels.
Transcriptional regulation
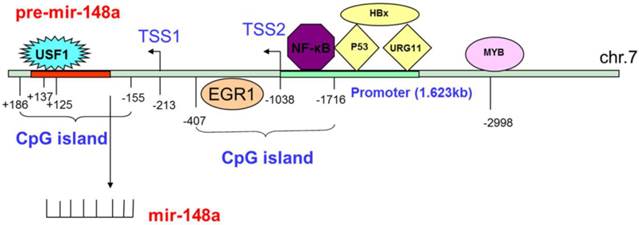
Tao et al. believed that mir-148a expression was suppressed by the action of estrogen (E2) on the G protein-coupled estrogen receptor (GPER).46 The transcription factor MYB was amplified in various tumors, and played an important role in tumor proliferation.66,67 For CC, MYB that bound to the -2998 site upstream of mir-148a suppressed mir-148a expression, and MYB silencing led to recovery of mir-148a expression.68 HBV coding X antigen (HBx), which is an activator of transcription, served as a transcriptional coregulator and led to hepatocellular carcinoma by stimulating cytoplasmic signal transduction. Yuan et al.69 held that HBx could promote UGR11 binding to the mir-148a promoter, and enhanced mir-148a expression in HBV-related HCC. In addition, Xu et al.70 expected that HBx could suppress mir-148a expression by preventing the tumor suppressor gene p53 from gathering to the mir-148a promoter region. The overexpression of p53 could also increase mir-148a expression. Li et al.71 considered that the upstream stimulatory factor 1 (USF1) could bind to the mir-148a promoter (+125 - +137) to activate expression in normal nasopharyngeal cells, while promoter methylation inhibited the bond to suppress expression in nasopharyngeal cancer (NC). For BC, binding of the transcription factor EGR1 to the mir-148a promoter could directly regulate mir-148a expression. Upregulated EGR1 could induce expression of mir-148a, while downregulated EGR1 could inhibit it.45 For glioma, direct binding of NF-κB to the promoter region could increase mir-148a expression (Fig. 1).58
Methylation regulation
Silencing of miRNAs was triggered by the aberrant methylation of CpG islands in or adjacent to promoter regions. The methylation of mir-148a was identified in pancreatic, colorectal, and nasopharyngeal cancers, and in IDH1 mutant glioma.32,35,59,71,72 72.6% methylation of CpG islands in the pre-mir-148a upstream sequence (i.e., -155-+186) was observed. In addition, Li et al.71 also studied the downregulated expression of mir-148a, and found that it resulted from the methylation region of the CpG island in NC. By contrast, the mir-148a CpG island was found in the pre-mir-148a upstream sequence (i.e., -1716—-407) including 12 CpG islands and two transcriptional start sites (TSS) (i.e., TSS1:-213 and TSS2:-1038) in IDH1 mutant glioma. Furthermore, mir-148a was downregulated because of the hypermethylation of 97% of the IDH1 mutant glioma (Fig 1).59
DNA methylation is catalyzed by DNA methyl transferase (DNMT), which can maintain methyl metastases and methylation patterns in mitotic replication. The methyl groups in CpG that mediate nucleotide sequences can be transferred from the S-adenosylmethionine (SAM) to the C-5 position in cytosine to affect both tumorigenesis and cancer cell development.73,74 Many studies have found that mir-148a can directly target the inhibition of DNMT1 expression due to the negative feedback regulatory pathway between them. By contrast, the downregulated expression of mir-148a is due to the negative feedback regulation of suppressed DNMT1.75-78
The role and mechanism of action of mir-148a in tumorigenesis and cancer cell development
MiRNA mainly interacts with target mRNA via the seed region of seven nucleotide sequences. Thus, mir-148a can act on different target genes to affect their function in various tumors (Table 1).
Mir-148a targeted genes and its function.
| Targets | Effect of mir-148a to target genes | Relevant pathway | Functions driven by mir-148a | Cancer types | Ref. | ||
|---|---|---|---|---|---|---|---|
| 3'UTR-reporter assay | mRNA | Protein | |||||
| CCKBR | ↓ | ↓ | ↓ | inhibition of cell proliferation, induction of cell apoptosis | Gastric Cancer, Pancreatic Cancer | [86] | |
| ROCK1 | ↓ | ↓ | ↓ | inhibition of cell proliferation, EMT, migration and invasion | Gastric Cancer, NSCLC,NPC | [47][71] [84] | |
| DNMT1 | ↓ | ↓ | ↓ | inhibition of cell proliferation and migration, induction of cell apoptosis | Gastric Cancer, HCC,Pancreatic Cancer, IDH1-Mutant Gliomas, Bladder Cancer, | [52][75]- [78][87] | |
| SMAD2 | ↓ | ↓ | ↓ | TGF-β/SMAD2 pathway | inhibition of cell migration and invasion, inhibition of CSCs-like properties | Gastric Cancer, HCC | [92] |
| MMP7 | ↓ | ↓ | ↓ | inhibition of cell migration and invasion | Gastric Cancer | [98] | |
| BCL-2 | ↓ | ↓ | ↓ | mitochondrial apoptosis pathway | induction of cell apoptosis | Gastric Cancer, Colorectal Cancer | [68][86] |
| HPIP | ↓ | ↓ | ↓ | AKT/ERK/FOXO4/ATFS pathway, mTOR pathway | inhibition of cell proliferation, migration and invasion | HCC | [70] |
| MET | ↓ | ↓ | ↓ | HGF/Met/Snail pathway | inhibition of EMT and metastasis | HCC | [95] |
| USP4 | ↓ | ↓ | ND | inhibition of cell growth, migration | HCC | [37] | |
| CDC2B | ↓ | ↓ | ↓ | inhibition of cell growth and survival | Pancreatic Cancer | [85] | |
| ERBB3 | ↓ | ↓ | ↓ | AKT/ERK pathway | inhibition of angiogenesis | Breast Cancer | [43] |
| IGF-IR | ↓ | ↓ | ↓ | IGF-IR/PKM2 | inhibition of cell growth | Breast Cancer | [44][45] |
| IRS1 | ↓ | ↓ | ↓ | inhibition of cell growth | Breast Cancer | [44] | |
| HOTAIR | ↓ | ↓ | ND | inhibition of cell migration and invasion | Breast Cancer | [99] | |
| HLA-G | ↓ | ↓ | ND | inhibition of immune evasion | Breast Cancer | [46] | |
| MSK1 | ↓ | ↓ | ↓ | attenuation of drug-resistant | Prostate Cancer | [51] | |
| CAND1 | ↓ | ↓ | ↓ | promotion of cell proliferation | Prostate Cancer | [82] | |
| WNT10B | ↓ | ↓ | ↓ | WNT/β-catenin pathway | inhibition of mirgration | Endometrial Cancer | [93][94] |
| WNT1 | ↓ | ↓ | ND | WNT/β-catenin pathway | inhibition of cell metastasis,CSCs-like properties, block EMT | HCC | [39] |
| ITGA11 | ↓ | ↓ | ND | integrin-activated signaling pathway | inhibition of mirgration | NPC | [71] |
| ITGB8 | ↓ | ND | ND | integrin-activated signaling pathway | inhibition of mirgration | NPC | [71] |
| VAV2 | ↓ | ↓ | ↓ | integrin-activated signaling pathway | NPC | [71] | |
| WASL | ↓ | ↓ | ↓ | integrin-activated signaling pathway | NPC | [71] | |
| BIM | ↓ | ↓ | ↓ | inhibition of cell growth and survival | Glioblastoma | [57] | |
| MIG6 | ↓ | ↓ | ↓ | inhibition of cell growth and survival | Glioblastoma | [57] | |
| QKI | ↓ | ↓ | ↓ | TGF-β/SMAD pathway | inhibition of cell migration and angiogenesis | Glioblastoma | [58] |
| SKP1 | ↓ | ↓ | ↓ | TGF-β/SMAD pathway | inhibition of cell migration and angiogenesis | Glioblastoma | [58] |
↓:downregulation, ND: not downregulation, NSCLC: non-small cell lung cancer, NPC:nasppharyngeal carcinoma.
HCC: hepatocellular carcinoma, EMT:epithelial-mesenchymal transition.
The regulation of mir-148a.
Regulation of tumor cell proliferation
Hematopoietic PBX-interacting protein (HPIP) can interact with the estrogen receptor (ER) and recruit Src kinases and the p85 subunit of PI3K to the ER complex. This process leads to the activation of the AKT/ERK pathway, and the mTOR signal transduction pathway and eventually affects tumor cell proliferation.79,80 Mir-148a overexpression can target the inhibition of HPIP expression to suppress the PI3K/AKT/mTOR signaling pathway, p70S6 kinase expression and HCC proliferation.70
Insulin receptor substrate 1 (IRS-1) and the insulin-like growth factor-I receptor (IGF- IR) can lead to multiple signal cascade pathways including PI3K/AKT and MAPK/ERK that control tumorigenesis and development.81 Xu et al.44 showed that mir-148a could suppress proliferation and colony cell formation of BC cells by targeted regulation of IRS1 and IGF-IR. Upregulated mir-148a could target the inhibition of IGF-IR/PKM2 expression to suppress the proliferation and colony cell formation of BC.45 Mir-148a could inhibit BC cell proliferation by depressing erb-b2 receptor tyrosine-protein kinase erbB-3.43
Cullin related protein (CAND1) is a scaffold protein in E3 ubiquitin ligase. The ubiquitin ligase-cullin protein complex plays a key role in regulating the cell cycle and differentiation. Mir-148a can depress the PC cell proliferation by dampening functional expression of CAND1.82 The mitogen-inducible gene-6 (MIG6) was implicated in the regulation of the epidermal growth factor receptor (EGFR) and led to EGFR-induced proliferation.83 Mir-148a inhibited glioma cell proliferation by suppressing MIG6 expression.57 Highly expressed mir-148a reduced the expression of the target gene ROCK1 to inhibit the proliferation of the GC cell-lines BGC823 and SGC7901.84 Mir-148a could also target to inhibit the expression of CDC25B,85 cholecystokinin B receptor (CCKBR),86 and DNMT187 to suppress both proliferation and colony formation of PC cells. Mir-148a also inhibited tumor cell proliferation by dampening DNMT1 expression in UCC (Fig 2).52
The role of mir-148a in cancer.
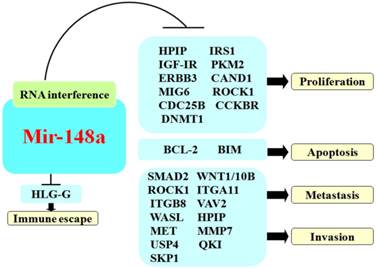
Regulation of tumor cell migration and invasion
The transfer of solid tumor cells from primary foci to distant foci is a result of epithelial-mesenchymal transition (EMT), a process that polarizes the transformation of the epithelium into stromal cells that subsequently acquire invasion and metastatic abilities. The pathological features are characterized by decreased expression of the epithelial marker E-cadherin, increased mesenchymocyte markers (e.g., N-cadherin, fibronectin, and vimentin), changed cellular morphology, loss of cell adhesion and increased motility. EMT is an important mechanism in tumor invasiveness and metastasis, which involves transforming growth factor-β (TGF-β), the WNT signaling pathway, the Snail signaling pathway, and E-cadherin expression.88-90 SMAD2 is the downstream effector of the TGF-β signaling pathway. The highly active TGF-β-smad complex can enhance both tumor cell metastasis and invasion.91 Wang et al.92 confirmed that mir-148a could suppress EMT, and target the inhibition of SMAD2 to ultimately depress GC metastasis and invasion. Several studies showed that mir-148a could target the regulation of WNT1 and WNT10B in endometrial carcinoma (EC) and HCC respectively, wherein EMT was inhibited to realize both tumor metastasis and invasiveness.39,93 Li et al.94 detected enhanced expression of mir-148a could inhibit both growth and metastasis of tumor by controlling the ACVR1/BMP/Wnt signaling pathway in the tumor stem cell-line HCC. Mir-148a could inhibit EMT to weaken the invasive capacity of cancer cells by targeted regulation of ROCK1 expression in non-small cell lung cancer47 and MET in HCC.95
Integrins are members of the heterodimeric transmembrane receptor family, which mainly mediate cell to cell, as well as cell to extracellular matrix interactions, and subsequently improve cell motility as well as tumor cells metastasis.96,97 Mir-148a could also depress the motility and metastasis of NC cells by targeting the regulation of ITGA11, ITGB8, VAV2, ROCK1 and WASL.71
Mir-148a could suppress tumor metastasis and invasion by regulating specific targeted genes including HPIP,70 matrix metalloproteinase-7 (MMP-7),98 ubiquitin specific protease 4 (USP4),37 HOX transcript antisense RNA (HOTAIR),99 OKI58 and S-Phase Kinase-Associated Protein 1 (SKP1) (Fig 2).58
Regulation of tumor cell apoptosis
Bcl-2 (B-cell lymphoma 2 gene) is an important molecule that can inhibit apoptosis as well as being able to promote oncogenesis.100 MiR-148a can induce tumor cell apoptosis by the targeted inhibition of Bcl-2 in CC and PC.68,86 Bcl-2-like protein 11 (BIM) is a pro-apoptotic factor that belongs to the Bcl-2 protein family BH3 group. BIM has been identified as a critical pro-apoptotic factor in solid tumors.101 Kim et al. found that upregulated mir-148a could suppress BIM, which led to inhibition of glioma apoptosis, while downregulated mir-148a could increase BIM expression to induce apoptosis (Fig 2).57
Conclusion
Accumulating studies have shown that mir-148a is aberrantly expressed in various tumors, which has been linked to tumor size, stage of development, metastasis and prognosis. However, the biological function and mechanism of mir-148a remains unsolved. Current studies of mir-148a function are limited in terms of cellular and in vitro xenograft experiments; however, the mir-148a gene knockout mouse model is necessary for future studies. Besides the methylation-mediated regulation of mir-148a expression, mir-148a might also be regulated by histone modification and SNPs changes; however, this remains to be verified. Additionally, research results of the relationship between mir-148a and tumors are inconsistent, which requires further study to determine its mechanism in tumors. Thus, mir-148a is predicted to be a potential biological marker and novel therapeutic target in diagnostic and prognostic applications.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Grant No. 81472860) and the Construct Program of the Key Discipline of Basic Medicine in Hunan Province.
Conflict of Interests
All authors declare that they have no any conflict of interests.
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854
2. Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70:6401-6406
3. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297
4. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20
5. Wei R, Yang J, Liu GQ. et al. Dynamic expression of microRNAs during the differentiation of human embryonic stem cells into insulin-producing cells. Gene. 2013;518:246-255
6. Ono K, Horie T, Nishino T. et al. MicroRNA-33a/b in lipid metabolism - novel "thrifty" models. Circ J. 2015;79:278-284
7. Calin GA, Sevignani C, Dumitru CD. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999-3004
8. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-688
9. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1-20
10. Barad O, Meiri E, Avniel A. et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486-2494
11. Landgraf P, Rusu M, Sheridan R. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-1414
12. Manaster I, Goldman-Wohl D, Greenfield C. et al. MiRNA-mediated control of HLA-G expression and function. PLoS One. 2012;7:e33395
13. Fu H, Tie Y, Xu C. et al. Identification of human fetal liver miRNAs by a novel method. FEBS Lett. 2005;579:3849-3854
14. Ribeiro-dos-Santos A, Khayat AS, Silva A. et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010;5:e13205
15. Nielsen S, Akerstrom T, Rinnov A. et al. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One. 2014;9:e87308
16. Porstner M, Winkelmann R, Daum P. et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur J Immunol. 2015;45:1206-1215
17. Palmieri A, Pezzetti F, Brunelli G. et al. Peptide-15 changes miRNA expression in osteoblast-like cells. Implant Dent. 2008;17:100-108
18. van Wijnen AJ, van de Peppel J, van Leeuwen JP. et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr Osteoporos Rep. 2013;11:72-82
19. Gailhouste L, Gomez-Santos L, Hagiwara K. et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153-1165
20. Ross SE, Hemati N, Longo KA. et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950-953
21. Qin L, Chen Y, Niu Y. et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11:320
22. Hu CW, Tseng CW, Chien CW. et al. Quantitative proteomics reveals diverse roles of miR-148a from gastric cancer progression to neurological development. J Proteome Res. 2013;12:3993-4004
23. Merkerova M, Vasikova A, Belickova M, Bruchova H. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010;19:17-26
24. Giraud-Triboult K, Rochon-Beaucourt C, Nissan X, Champon B, Aubert S, Pietu G. Combined mRNA and microRNA profiling reveals that miR-148a and miR-20b control human mesenchymal stem cell phenotype via EPAS1. Physiol Genomics. 2011;43:77-86
25. Zhang J, Zhang XH, Wang CX. et al. Dysregulation of microRNA biosynthesis enzyme Dicer plays an important role in gastric cancer progression. Int J Clin Exp Pathol. 2014;7:1702-1707
26. Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of microRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668-1674
27. Chen Z, Saad R, Jia P. et al. Gastric adenocarcinoma has a unique microRNA signature not present in esophageal adenocarcinoma. Cancer. 2013;119:1985-1993
28. Zheng G, Xiong Y, Xu W. et al. A two-microRNA signature as a potential biomarker for early gastric cancer. Oncol Lett. 2014;7:679-684
29. Chen Y, Song Y, Wang Z. et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170-1179
30. Sun J, Song Y, Wang Z. et al. Clinical significance of promoter region hypermethylation of microRNA-148a in gastrointestinal cancers. Onco Targets Ther. 2014;7:853-863
31. Song P, Zhu H, Zhang D. et al. A genetic variant of miR-148a binding site in the SCRN1 3'-UTR is associated with susceptibility and prognosis of gastric cancer. Sci Rep. 2014;4:7080
32. Takahashi M, Cuatrecasas M, Balaguer F. et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7:e46684
33. Tsai HL, Yang IP, Huang CW. et al. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl Res. 2013;162:258-268
34. Szafranska AE, Davison TS, John J. et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452
35. Hanoun N, Delpu Y, Suriawinata AA. et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56:1107-1118
36. Ladeiro Y, Couchy G, Balabaud C. et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955-1963
37. Heo MJ, Kim YM, Koo JH. et al. microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget. 2014;5:2792-2806
38. Pan L, Huang S, He R, Rong M, Dang Y, Chen G. Decreased expression and clinical significance of miR-148a in hepatocellular carcinoma tissues. Eur J Med Res. 2014;19:68
39. Yan H, Dong X, Zhong X. et al. Inhibitions of epithelial to mesenchymal transition and cancer stem cells-like properties are involved in miR-148a-mediated anti-metastasis of hepatocellular carcinoma. Mol Carcinog. 2014;53:960-969
40. Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853-861
41. Hummel R, Hussey DJ, Michael MZ. et al. MiRNAs and their association with locoregional staging and survival following surgery for esophageal carcinoma. Ann Surg Oncol. 2011;18:253-260
42. Hummel R, Watson DI, Smith C. et al. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15:429-438
43. Yu J, Li Q, Xu Q, Liu L, Jiang B. MiR-148a inhibits angiogenesis by targeting ERBB3. J Biomed Res. 2011;25:170-177
44. Xu Q, Jiang Y, Yin Y. et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3-13
45. Xu Q, Liu LZ, Yin Y. et al. Regulatory circuit of PKM2/NF-kappaB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene. 2015;34:5482-5493
46. Tao S, He H, Chen Q, Yue W. GPER mediated estradiol reduces miR-148a to promote HLA-G expression in breast cancer. Biochem Biophys Res Commun. 2014;451:74-78
47. Li J, Song Y, Wang Y, Luo J, Yu W. MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem. 2013;380:277-282
48. Li L, Chen YY, Li SQ, Huang C, Qin YZ. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 2015;21:1155-1161
49. Yang JS, Li BJ, Lu HW. et al. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035-3042
50. Chen Y, Min L, Zhang X. et al. Decreased miRNA-148a is associated with lymph node metastasis and poor clinical outcomes and functions as a suppressor of tumor metastasis in non-small cell lung cancer. Oncol Rep. 2013;30:1832-1840
51. Fujita Y, Kojima K, Ohhashi R. et al. MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J Biol Chem. 2010;285:19076-19084
52. Lombard AP, Mooso BA, Libertini SJ. et al. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol Carcinog. 2015;55(5):757-67
53. Zhou X, Zhao F, Wang ZN. et al. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep. 2012;27:447-454
54. Gu Y, Zhang M, Peng F. et al. The BRCA1/2-directed miRNA signature predicts a good prognosis in ovarian cancer patients with wild-type BRCA1/2. Oncotarget. 2015;6:2397-2406
55. Roth P, Wischhusen J, Happold C. et al. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449-457
56. Hua D, Mo F, Ding D. et al. A catalogue of glioblastoma and brain MicroRNAs identified by deep sequencing. Omics. 2012;16:690-699
57. Kim J, Zhang Y, Skalski M. et al. microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res. 2014;74:1541-1553
58. Wang H, Pan JQ, Luo L. et al. NF-κB induces miR-148a to sustain TGF-beta/Smad signaling activation in glioblastoma. Mol Cancer. 2015;14:2
59. Li S, Chowdhury R, Liu F. et al. Tumor-suppressive miR148a is silenced by CpG island hypermethylation in IDH1-mutant gliomas. Clin Cancer Res. 2014;20:5808-5822
60. Ma W, Zhang X, Chai J, Chen P, Ren P, Gong M. Circulating miR-148a is a significant diagnostic and prognostic biomarker for patients with osteosarcoma. Tumour Biol. 2014;35:12467-12472
61. Cimmino A, Calin GA, Fabbri M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944-13949
62. Nobusawa S, Lachuer J, Wierinckx A. et al. Intratumoral patterns of genomic imbalance in glioblastomas. Brain Pathol. 2010;20:936-944
63. Arslantas A, Artan S, Oner U. et al. Genomic alterations in low-grade, anaplastic astrocytomas and glioblastomas. Pathol Oncol Res. 2007;13:39-46
64. Brezinova J, Zemanova Z, Ransdorfova S. et al. Structural aberrations of chromosome 7 revealed by a combination of molecular cytogenetic techniques in myeloid malignancies. Cancer Genet Cytogenet. 2007;173:10-16
65. Kulkarni S, Qi Y, O'HUigin C. et al. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc Natl Acad Sci U S A. 2013;110:20705-20710
66. Miao RY, Drabsch Y, Cross RS. et al. MYB is essential for mammary tumorigenesis. Cancer Res. 2011;71:7029-7037
67. Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523-534
68. Zhang H, Li Y, Huang Q. et al. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011;18:1702-1710
69. Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS One. 2012;7:e35331
70. Xu X, Fan Z, Kang L. et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645
71. Li HP, Huang HY, Lai YR. et al. Silencing of miRNA-148a by hypermethylation activates the integrin-mediated signaling pathway in nasopharyngeal carcinoma. Oncotarget. 2014;5:7610-7624
72. Kalimutho M, Di Cecilia S, Del Vecchio Blanco G. et al. Epigenetically silenced miR-34b/c as a novel faecal-based screening marker for colorectal cancer. Br J Cancer. 2011;104:1770-1778
73. Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865-873
74. Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971-983
75. Pan W, Zhu S, Yuan M. et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773-6781
76. Zhu A, Xia J, Zuo J. et al. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol. 2012;29:2701-2709
77. Zuo J, Xia J, Ju F. et al. MicroRNA-148a can regulate runt-related transcription factor 3 gene expression via modulation of DNA methyltransferase 1 in gastric cancer. Mol Cells. 2013;35:313-319
78. Long XR, He Y, Huang C, Li J. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J Oncol. 2014;44:1915-1922
79. Manavathi B, Acconcia F, Rayala SK, Kumar R. An inherent role of microtubule network in the action of nuclear receptor. Proc Natl Acad Sci U S A. 2006;103:15981-15986
80. Wang X, Yang Z, Zhang H. et al. The estrogen receptor-interacting protein HPIP increases estrogen-responsive gene expression through activation of MAPK and AKT. Biochim Biophys Acta. 2008;1783:1220-1228
81. Berns K, Horlings HM, Hennessy BT. et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395-402
82. Murata T, Takayama K, Katayama S. et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010;13:356-361
83. Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741-744
84. Zheng B, Liang L, Wang C. et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574-7583
85. Liffers ST, Munding JB, Vogt M. et al. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. 2011;91:1472-1479
86. Zhang R, Li M, Zang W. et al. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2014;35:837-844
87. Zhan Q, Fang Y, Deng X. et al. The Interplay Between miR-148a and DNMT1 Might be Exploited for Pancreatic Cancer Therapy. Cancer Invest. 2015;33:267-275
88. Charpentier M, Martin S. Interplay of Stem Cell Characteristics, EMT, and Microtentacles in Circulating Breast Tumor Cells. Cancers (Basel). 2013;5:1545-1565
89. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428
90. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-454
91. Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987-2002
92. Wang SH, Li X, Zhou LS. et al. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013;34:3705-3712
93. Aprelikova O, Palla J, Hibler B. et al. Silencing of miR-148a in cancer-associated fibroblasts results in WNT10B-mediated stimulation of tumor cell motility. Oncogene. 2013;32:3246-3253
94. Li L, Liu Y, Guo Y. et al. Regulatory MiR-148a-ACVR1/BMP circuit defines a cancer stem cell-like aggressive subtype of hepatocellular carcinoma. Hepatology. 2015;61:574-584
95. Zhang JP, Zeng C, Xu L, Gong J, Fang JH, Zhuang SM. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2014;33:4069-4076
96. Missan DS, DiPersio M. Integrin control of tumor invasion. Crit Rev Eukaryot Gene Expr. 2012;22:309-324
97. Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816-826
98. Sakamoto N, Naito Y, Oue N. et al. MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci. 2014;105:236-243
99. Tao S, He H, Chen Q. Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J Transl Med. 2015;13:131
100. Sun CY, Wang BL, Hu CQ. et al. Expression of the bcl-2 gene and its significance in human pancreatic carcinoma. Hepatobiliary Pancreat Dis Int. 2002;1:306-308
101. Bean GR, Ganesan YT, Dong Y. et al. PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal. 2013;6:ra20
Author contact
![]() Corresponding authors: Xiaoning Peng, MD. Department of Pathology and Pathophysiology, Hunan Normal University Medical School, Changsha 410013, Hunan, China. Tel: +86-13786169507 Fax: +86-21-64085875 E-mail: pxiaoningedu.cn or Xiaomin Zeng, MS. Department of Statistics and Epidemiology, Public Health School, Central South University, Changsha, Hunan410078, China. Tel: +86-13467630366 Fax: +86-21-64085875 E-mail: zxiaominedu.cn.
Corresponding authors: Xiaoning Peng, MD. Department of Pathology and Pathophysiology, Hunan Normal University Medical School, Changsha 410013, Hunan, China. Tel: +86-13786169507 Fax: +86-21-64085875 E-mail: pxiaoningedu.cn or Xiaomin Zeng, MS. Department of Statistics and Epidemiology, Public Health School, Central South University, Changsha, Hunan410078, China. Tel: +86-13467630366 Fax: +86-21-64085875 E-mail: zxiaominedu.cn.

 Global reach, higher impact
Global reach, higher impact