3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(10):1273-1280. doi:10.7150/jca.15035 This issue Cite
Research Paper
Association between the OGG1 Ser326Cys Polymorphism and Cancer Risk: Evidence from 152 Case-Control Studies
1. Cancer Center, Daping Hospital and Research Institute of Surgery, Third Military Medical University, Chongqing 400042, China;
2. Department of Diagnostic and Therapeutic Ultrasonography, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center of Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China;
3. Intensive care unit, Suining Central Hospital, Deshengxi Road 127, Chuanshan District, Suining, Sichuan 629000, China.
* These authors Hua Zou Qing Li and Wei Xia contributed equally to this work.
Received 2016-1-20; Accepted 2016-4-26; Published 2016-6-23
Abstract
Although it has been suggested that the 8-oxoguanine DNA glycosylase (OGG1) gene Ser326Cys polymorphism may be a risk factor for cancer, the conclusions from previous studies are inconsistent. Thus, we conducted an updated meta-analysis to estimate the effect of OGG1 variant genotypes on cancer susceptibility. We searched the PubMed for all eligible studies published in English for the period ending September 2014. We found the association between OGG1 Ser326Cys polymorphism and cancer susceptibility based on 152 case-control studies in different genetic model comparisons (dominant model: OR = 1.053, P = 0.018; recessive model: OR = 1.108, P < 0.001; homozygote: OR = 1.135, P < 0.001; additive model: OR = 1.059, P < 0.001). However, the results from the subgroup analyses based on types of cancer, health population as controls or studies with relatively large sample size did not support the conclusion. Although the overall results of this meta-analysis showed a positive association between OGG1 variant genotypes and cancer susceptibility, the subgroup analyses by cancer type, sample size, and source of controls presented inconsistent results. Therefore, the current evidence from the meta-analysis did not support the hypothesis of OGG1 Ser326Cys polymorphism as a risk factor of cancer.
Keywords: cancer, OGG1, polymorphism, meta-analysis.
Introduction
DNA damage can be induced by many factors, including mismatches during amplification, ultraviolet radiation, and chemical mutagens. Although endogenous DNA repair systems can repair the damages within a certain extent, the repair capability may be undermined by genetic variation in these systems. Even worse, the accumulation of errors can overwhelm the cell, resulting in permanent mutations in the gene and ultimately in the development of malignant tumors.[1] The 8-oxoguanine DNA glycosylase 1 (OGG1) gene, located on chromosome 3p26, plays a critical role in the base excision repair (BER) pathway.[2] The OGG1 promotes the hydroxylation of the glycosylic bond between the abnormal base and the sugar moiety, removing 8-oxoguanine from the oxidative lesion DNA, leaving a notch in the DNA strand, a so-called apurinic/apyrimidinic site, which could be subsequently filled by a normal base.[3, 4] The most studied OGG1 gene polymorphism located in exon 7 of this gene (rs1052133), an amino acid substitution from serine to cysteine at codon 326, has been proposed to modify OGG1 function and increase cancer susceptibility.[5, 6]
To date, though many studies have assessed the association between the OGG1 Ser326Cys polymorphism and cancer risk, there are still many inconsistencies among the findings from these studies. For this reason, we conducted a meta-analysis with 152 case-control studies to evaluate the linkage between this polymorphism and susceptibility for different types of cancer.[7-93]
Results
Study Characteristics
The database retrieval yielded 397 studies after duplicates were removed, of which 208 were irrelevant. The remaining 189 studies were further assessed, of which 37 were excluded (Figure 1). In total, 152 case-control studies involving 162 datasets were included in this meta-analysis. Table S1 in the supplementary materials provides the basic characteristics of the included studies. The datasets from the included studies were composed of various ethnicities: 74 datasets of Asian descendants, 75 datasets of Caucasian descendants, 12 datasets of mixed descendants, and 1 dataset of African descendants. The pattern of genotype distributions of the control groups in 30 studies were not in agreement with HWE.
Flowchart of selection of studies for inclusion in meta-analysis.
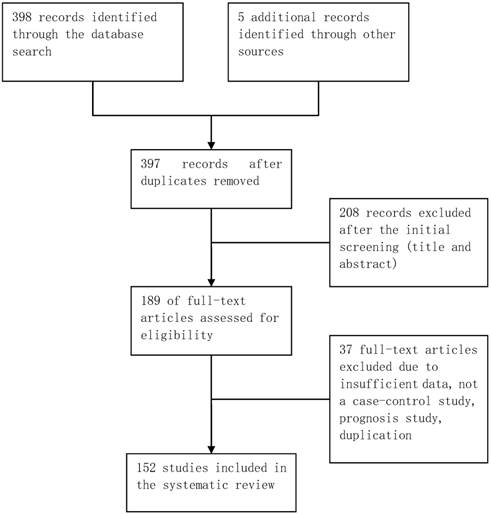
Quantitative synthesis
The frequencies of the 326Cys allele in control groups showed a statistically significant difference among different ethnicities. The 326Cys allele proportion in Asian controls was remarkably higher than in Caucasian controls (52.32% vs. 23.17%, P < 0.001). On the whole, the patients carried more SerCys or CysCys variant genotypes than controls in the dominant model comparison (OR = 1.053, 95%CI: 1.009~1.099, P = 0.018, Table 1), the recessive model comparison (OR = 1.108, 95%CI: 1.043~1.178, P < 0.001), the homozygote comparison (OR = 1.135, 95%CI: 1.055~1.220, P < 0.001), and additive model comparison (OR = 1.059, 95%CI: 1.024~1.095, P < 0.001).
Subgroup analyses
As displayed in Table 1, in the subset analysis by cancer type, we found that the association between OGG1 SerCys or CysCys polymorphism and cancer risk was not significantly different except for leukemia in the recessive model and additive model (P = 0.043 and P = 0.032, respectively). In the subset analysis stratified by ethnicity, the OGG1 variant genotypes were significantly associated with cancer susceptibility in Caucasian and Asian populations with the exception of Asian populations in the dominant model (OR = 1.060, 95%CI: 0.996~1.129, P = 0.068). In the subset analysis stratified by source of control, the association between OGG1 variant genotypes and cancer risk showed a statistical significance among studies with controls from hospitals in each genetic model comparison but not for controls from the population. In the subset analysis according to sample size, there was an obvious correlation between the OGG1 variant genotypes and cancer risk among studies with a sample size of less than 500 participants in each genetic model (P<0.05), but not for studies with a sample size larger than 500 participants in the dominant and additive models (P = 0.270 and P = 0.064, respectively). In addition, the subset analysis according to HWE revealed a statistically significant association between variant genotypes and cancer risk among studies in agreement with HWE, but not among studies not in agreement with HWE.
Stratification analyses of the hOGG1 Ser326Cys polymorphism with regard to cancer susceptibility.
| Variables | n | Dominant model (SerCys&CysCys versus SerSer) | Recessive model (CysCys versus SerSer&SerCys) | Homozygote comparison (CysCys versus SerSer) | Additive model (Cys allele versus Ser allele) | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI)a | Pb | OR (95% CI)a | Pb | OR (95% CI)a | Pb | OR (95% CI)a | Pb | ||
| Total | 162 | 1.053 (1.009, 1.099) | 0.018 | 1.108 (1.043, 1.178) | 0.001 | 1.135 (1.055, 1.220) | 0.001 | 1.059 (1.024, 1.095) | 0.001 |
| Cancer types | |||||||||
| biliary tract | 5 | 0.961 (0.776, 1.190) | 0.717 | 1.379 (0.920, 2.068) | 0.120 | 1.271 (0.867, 1.864) | 0.219 | 1.064 (0.909, 1.245) | 0.441 |
| bladder | 12 | 0.994 (0.874, 1.131) | 0.928 | 1.102 (0.811, 1.497) | 0.534 | 1.140 (0.872, 1.491) | 0.337 | 1.028 (0.920, 1.149) | 0.628 |
| breast | 16 | 1.006 (0.943, 1.072) | 0.867 | 1.078 (0.993, 1.171) | 0.074 | 1.080 (0.973, 1.198) | 0.146 | 1.025 (0.974, 1.078) | 0.347 |
| colorectal | 18 | 1.144 (0.992, 1.318) | 0.064 | 1.142 (0.950, 1.373) | 0.158 | 1.242 (0.985, 1.566) | 0.067 | 1.126 (1.008, 1.258) | 0.035 |
| endometrial | 5 | 1.270 (0.932, 1.732) | 0.131 | 1.079 (0.623, 1.870) | 0.785 | 1.132 (0.640, 2.004) | 0.670 | 1.195 (0.918, 1.554) | 0.185 |
| esophageal | 7 | 0.998 (0.867, 1.149) | 0.981 | 1.146 (0.802, 1.637) | 0.456 | 1.122 (0.797, 1.581) | 0.509 | 1.028 (0.915, 1.154) | 0.642 |
| gastric | 15 | 0.924 (0.799, 1.069) | 0.290 | 1.043 (0.813, 1.338) | 0.740 | 0.966 (0.754, 1.237) | 0.782 | 0.984 (0.862, 1.125) | 0.818 |
| head and neck | 15 | 1.055 (0.870, 1.280) | 0.584 | 1.156 (0.907, 1.472) | 0.241 | 1.219 (0.869, 1.711) | 0.251 | 1.060 (0.910, 1.234) | 0.455 |
| hepatocellular | 3 | 1.402 (0.756, 2.601) | 0.283 | 1.254 (0.572, 2.748) | 0.572 | 1.653 (0.581, 4.701) | 0.346 | 1.186 (0.754, 1.865) | 0.460 |
| leukemia | 4 | 1.385 (0.917, 2.090) | 0.121 | 1.884 (1.021, 3.476) | 0.043 | 2.044 (0.926, 4.515) | 0.077 | 1.444 (1.032, 2.021) | 0.032 |
| lung | 36 | 1.073 (0.984, 1.170) | 0.110 | 1.098 (0.983, 1.225) | 0.097 | 1.128 (0.989, 1.288) | 0.074 | 1.064 (0.994, 1.139) | 0.073 |
| pancreatic | 4 | 1.084 (0.929, 1.265) | 0.307 | 0.880 (0.683, 1.133) | 0.320 | 0.876 (0.666, 1.151) | 0.342 | 0.993 (0.889, 1.109) | 0.899 |
| prostate | 8 | 1.130 (0.891, 1.432) | 0.315 | 1.111 (0.666, 1.851) | 0.687 | 1.198 (0.669, 2.146) | 0.543 | 1.119 (0.889, 1.408) | 0.338 |
| others | 14 | 0.973 (0.839, 1.128) | 0.712 | 1.131 (0.909, 1.407) | 0.271 | 1.132 (0.885, 1.447) | 0.322 | 1.022 (0.913, 1.143) | 0.710 |
| Ethnicities | |||||||||
| Asian | 74 | 1.060 (0.996, 1.129) | 0.068 | 1.101 (1.023, 1.185) | 0.010 | 1.129 (1.032, 1.235) | 0.008 | 1.057 (1.012, 1.105) | 0.013 |
| Caucasian | 75 | 1.075 (1.003, 1.153) | 0.041 | 1.221 (1.075, 1.388) | 0.002 | 1.257 (1.090, 1.450) | 0.002 | 1.096 (1.030, 1.165) | 0.004 |
| mixed | 12 | 0.940 (0.875, 1.010) | 0.090 | 0.872 (0.740, 1.028) | 0.102 | 0.856 (0.719, 1.020) | 0.082 | 0.938 (0.882, 0.998) | 0.043 |
| African | 1 | 1.280 (0.884, 1.852) | 0.191 | 0.823 (0.281, 2.404) | 0.721 | 0.891 (0.303, 2.619) | 0.834 | 1.187 (0.859, 1.640) | 0.300 |
| Source of control | |||||||||
| population | 85 | 1.026 (0.968, 1.087) | 0.389 | 1.075 (0.995, 1.161) | 0.067 | 1.097 (0.996, 1.208) | 0.061 | 1.035 (0.990, 1.083) | 0.131 |
| hospital | 75 | 1.087 (1.018, 1.161) | 0.013 | 1.152 (1.043, 1.272) | 0.005 | 1.186 (1.060, 1.327) | 0.003 | 1.088 (1.032, 1.147) | 0.002 |
| family | 2 | 1.076 (0.879, 1.319) | 0.476 | 0.985 (0.645, 1.504) | 0.945 | 1.017 (0.655, 1.579) | 0.941 | 1.050 (0.884, 1.246) | 0.579 |
| Sample size | |||||||||
| large than 500 | 80 | 1.026 (0.980, 1.074) | 0.270 | 1.080 (1.014, 1.151) | 0.017 | 1.091 (1.007, 1.182) | 0.033 | 1.035 (0.998, 1.073) | 0.064 |
| less than 500 | 82 | 1.099 (1.005, 1.202) | 0.039 | 1.203 (1.046, 1.383) | 0.010 | 1.248 (1.073, 1.450) | 0.004 | 1.104 (1.028, 1.186) | 0.007 |
| HWEc | |||||||||
| yes | 132 | 1.073 (1.025, 1.123) | 0.002 | 1.118 (1.053, 1.187) | 0.000 | 1.153 (1.074, 1.239) | 0.000 | 1.070 (1.034, 1.109) | 0.000 |
| no | 30 | 0.962 (0.857, 1.080) | 0.513 | 1.103 (0.886, 1.373) | 0.380 | 1.080 (0.842, 1.385) | 0.544 | 1.002 (0.905, 1.108) | 0.973 |
Note: a Random-effect model; b P value of effect for genetic variants; C Hardy-Weinberg equilibrium.
Test of heterogeneity
Significant heterogeneity was found in the dominant model comparison (P = 0.018). In the subset analysis according to cancer type, significant heterogeneity was found in bladder cancer (P = 0.033), colorectal cancer (P<0.001), head and neck cancer (P<0.001), hepatocellular carcinoma (P = 0.002), leukemia (P = 0.005), lung cancer (P = 0.002), prostate cancer (P = 0.001), and other cancers (P = 0.018). We also found that there was significant heterogeneity in the subset analyses according to ethnicity, source of control, sample size, and HWE in dominant model comparisons (Supplementary Figures S1-S5). Similar heterogeneity was found in other genetic model comparisons.
Sensitivity analysis
In the sensitivity analysis, the effect of each dataset on the merged OR value was detected by repeating the meta-analysis by the leave-one-out method. In subset analyses according to cancer types, ethnicity, source of control, and sample size, we found that the overall result remained stable for each subgroup after completing sensitivity analysis (data not shown).
Publication bias
The shape of Begg's funnel plot seemed asymmetrical based on the dominant model, indicating that there was an obvious publication bias among studies evaluating the linkage between OGG1 Ser326Cys polymorphism and cancer susceptibility (Figure 2). Egger's regression test also indicated the existence of publication bias among included studies (dominant model: P = 0.041; recessive model: P = 0.003; homozygote comparison: P = 0.003; additive model: P = 0.017).
Discussion
The OGG1 gene encodes a repair enzyme for oxidative DNA damage.[2] Because of the critical role of OGG1 in base excision repair, the mutation in the OGG1 gene is considered to modify the susceptibility to cancer. In this meta-analysis, we obtained overall results similar to Wang et al. and Wei et al., respectively[94, 95] that the OGG1 variant genotypes might be a risk factor for developing malignant neoplasms.164, 165 However, the subgroup analyses according to cancer types, sample size, and source of controls did not support this conclusion.
Begg's funnel plot to assess publication bias for the dominant model.
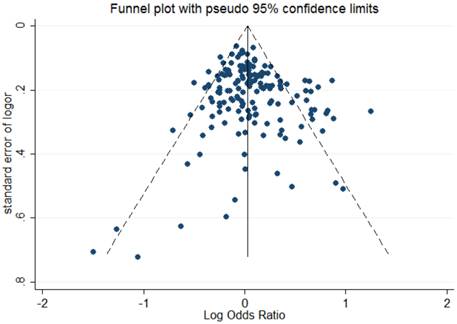
In the subset analysis according to cancer types, there was no obvious correlation between variant genotypes and cancer risk except for leukemia. The positive association between Ser326Cys polymorphism and leukemia might be a result of the relatively small sample size of leukemia studies and should be verified by future studies.
The association between OGG1 variant genotypes and cancer susceptibility was not found in Asian and Caucasian populations, except for Asian populations in the dominant model (P = 0.068). This conclusion is different from the previous meta-analysis result by Wang et al. and Wei et al. The possible reason for this difference might be that we treated the mixed population (mainly composed of Caucasian and African populations) and the African population as an individual subgroup, which did not exhibit cancer susceptibility for Ser/Cys and Cys/Cys genotypes.
When stratified by the source of controls, significant cancer susceptibility was found among studies using the hospital patients as controls but not for studies using healthy populations as controls. Moreover, in the subset analysis according to sample size, a significant association was found only among studies with a sample size less than 500 participants and not among studies with more than 500 participants. The results suggest that the correlation between OGG1 variant genotypes and cancer susceptibility might be weaker than we expected. Therefore, more large-scale studies with appropriate control subjects and strict methodologies are needed to assess genetic variants and cancer risk.
There are several limitations of our meta-analysis that need to be stated. First, only studies published in the PubMed database and written in English were included in the meta-analysis. Second, inadequate data regarding the gene-environment and gene-gene interactions in included studies prevented us from understanding in depth the magnitude of the effect of the variant gene on cancer susceptibility. Third, the significant publication bias due to non-publication of negative results and variability in small sample size studies in our meta-analysis might have led to false conclusions.
In general, the overall results of this meta-analysis showed a positive association between OGG1 variant genotypes and cancer susceptibility. However, it is more likely a false conclusion because most subgroup analyses by cancer type failed to support it. Furthermore, studies with different sample sizes or sources of controls with inconsistent results further weakened the reliability of these conclusions. In the future, additional well-designed and large-scale studies with representative controls are required to assess the relationship between OGG1 Ser326Cys polymorphism and cancer risk in single ethnicity populations, and information about gene-gene and gene-environment interactions is also necessary.
Materials and methods
Literature search and selection criteria
We retrieved relevant articles from the PubMed database with the following search terms: '8-oxoguanine DNA glycosylase / hOGG1 / OGG1 / OGG / Ser326Cys,' 'SNP / polymorphism' and 'carcinoma / cancer' (the latest retrieval was updated on September 20, 2014). We also examined the references of all included articles and previously published meta-analysis for potentially relevant articles. In order to avoid republication or overlapping data, only recently published studies with larger number of subjects were selected for our meta-analysis.[96, 97] Studies conforming to the following criteria were included: (1) has a case-control design or nested case-control design; (2) provides an evaluation of the linkage between the OGG1 Ser326Cys polymorphism and cancer risk; (3) provides available genotype frequency; and (4) was published in English.
Data extraction and qualitative assessment
Data were extracted by using a pre-designed data extraction form including first author, year of publication, research design, country, ethnicity, genotyping method, and genotype frequency. We categorized subjects as Asians, Caucasians, and Africans according to their ethnic descent. If a group of subjects comprised two or more than two ethnic descents, the group was termed ''mixed ethnicity''. Furthermore, if a study including subjects of different types of cancer or different ethnic groups, we treated each subgroup as an individual dataset (a total of 162 datasets from 152 articles). We evaluated the quality of included articles using the Newcastle-Ottawa quality assessment scale (NOS).[98] The data quality was quantitatively assessed based on a 'star system' that ranged from 0 to 9, including three aspects: the selection, comparability, and outcome of interest. Articles with 7 points or above were considered as high quality. Two investigators independently (L.Q. and Y.L.) extracted the data and evaluated the quality of studies. Disagreements between them were resolved by discussions with other authors.
Statistical analysis
We assessed the effect of OGG1 Ser326Cys polymorphism on cancer susceptibility using odds ratios (ORs) with 95% confidence intervals (CIs), with the following genetic models: dominant model, recessive model, homozygote comparison model, and additive model. Subset analysis was also carried out based on cancer types, ethnicity, and source of controls, sample size, and the Hardy-Weinberg equilibrium (HWE). Heterogeneity among the studies was determined by a Q test based on a chi-square test (P < 0.10 was regarded as a significant difference).[99, 100] Considering the clinical heterogeneity among included studies, a random effect model using the DerSimonian and Laird method was used to merge the datasets.[101] Sensitivity analysis was performed to evaluate the reliability of the results. Funnel plots and Egger's test were adopted to detect potential publication bias. The statistical analysis was completed by using STATA 13.0 (Stata Corp, College Station, TX.). The scheme of the meta-analysis was in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement [102].
Supplementary Material
Additional File 1Figures S1-S5.
Table S1.
Acknowledgements
We thank Meng-xia Li for his free assistance of language editing. We also thank Shi-Heng Zhang to provide professional opinions about OGG1 gene. We are especially grateful for strongly supporting to this study from Dong Wang. This study was supported by Grants from the National Natural Science Foundation of China (No. 30872975, 81401412).
Author contributions
D.W. X.W. H.Z. and L.Q. conceived and designed this study. L.Q. and X.W. were responsible for searching the articles in database. Q.L. X.W. and Z.H. were jointly involving in extracting data and writing the manuscript.
Competing Interests
The authors declare no competing financial interests.
References
1. Weiss JM, Goode EL, Ladiges WC, Ulrich CM. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Molecular carcinogenesis. 2005;42:127-41 doi:10.1002/mc.20067
2. Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Archives of biochemistry and biophysics. 2000;377:1-8 doi:10.1006/abbi.2000.1773
3. Dianov GL, Souza-Pinto N, Nyaga SG, Thybo T, Stevnsner T, Bohr VA. Base excision repair in nuclear and mitochondrial DNA. Progress in nucleic acid research and molecular biology. 2001;68:285-97
4. Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2002;16:1895-902 doi:10.1096/fj.02-0463com
5. Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic acids research. 1999;27:4001-7
6. Vodicka P, Stetina R, Polakova V, Tulupova E, Naccarati A, Vodickova L. et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28:657-64 doi:10.1093/carcin/bgl187
7. Arcand SL, Provencher D, Mes-Masson AM, Tonin PN. OGG1 Cys326 variant, allelic imbalance of chromosome band 3p25.3 and TP53 mutations in ovarian cancer. International journal of oncology. 2005;27:1315-20
8. Arizono K, Osada Y, Kuroda Y. DNA repair gene hOGG1 codon 326 and XRCC1 codon 399 polymorphisms and bladder cancer risk in a Japanese population. Japanese journal of clinical oncology. 2008;38:186-91 doi:10.1093/jjco/hym176
9. Bose S, Tripathi DM, Sukriti, Sakhuja P, Kazim SN, Sarin SK. Genetic polymorphisms of CYP2E1 and DNA repair genes HOGG1 and XRCC1: association with hepatitis B related advanced liver disease and cancer. Gene. 2013;519:231-7 doi:10.1016/j.gene.2013.02.025
10. Brevik A, Joshi AD, Corral R, Onland-Moret NC, Siegmund KD, Le Marchand L. et al. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:3167-73 doi:10.1158/1055-9965.epi-10-0606
11. Cai Q, Shu XO, Wen W, Courtney R, Dai Q, Gao YT. et al. Functional Ser326Cys polymorphism in the hOGG1 gene is not associated with breast cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2006;15:403-4 doi:10.1158/1055-9965.epi-05-0868
12. Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, Yamaner S. et al. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer research. 2010;30:1359-64
13. Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M. et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Current medical research and opinion. 2011;27:1295-302 doi:10.1185/03007995.2011.573544
14. Capella G, Pera G, Sala N, Agudo A, Rico F, Del Giudicce G. et al. DNA repair polymorphisms and the risk of stomach adenocarcinoma and severe chronic gastritis in the EPIC-EURGAST study. International journal of epidemiology. 2008;37:1316-25 doi:10.1093/ije/dyn145
15. Chang CH, Hsiao CF, Chang GC, Tsai YH, Chen YM, Huang MS. et al. Interactive effect of cigarette smoking with human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) polymorphisms on the risk of lung cancer: a case-control study in Taiwan. American journal of epidemiology. 2009;170:695-702 doi:10.1093/aje/kwp019
16. Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP Jr. et al. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis. 2009;30:78-87 doi:10.1093/carcin/bgn261
17. Chen L, Elahi A, Pow-Sang J, Lazarus P, Park J. Association between polymorphism of human oxoguanine glycosylase 1 and risk of prostate cancer. The Journal of urology. 2003;170:2471-4 doi:10.1097/01.ju.0000087498.23008.bb
18. Chen X, Liu X, Wang J, Guo W, Sun C, Cai Z. et al. Functional polymorphisms of the hOGG1 gene confer risk to type 2 epithelial ovarian cancer in Chinese. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2011;21:1407-13 doi:10.1097/IGC.0b013e31823122c6
19. Cho EY, Hildesheim A, Chen CJ, Hsu MM, Chen IH, Mittl BF. et al. Nasopharyngeal carcinoma and genetic polymorphisms of DNA repair enzymes XRCC1 and hOGG1. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2003;12:1100-4
20. Choi JY, Hamajima N, Tajima K, Yoo KY, Yoon KS, Park SK. et al. hOGG1 Ser326Cys polymorphism and breast cancer risk among Asian women. Breast cancer research and treatment. 2003;79:59-62
21. Cincin ZB, Iyibozkurt AC, Kuran SB, Cakmakoglu B. DNA repair gene variants in endometrial carcinoma. Medical oncology (Northwood, London, England). 2012;29:2949-54 doi:10.1007/s12032-012-0162-7
22. Curtin K, Samowitz WS, Wolff RK, Ulrich CM, Caan BJ, Potter JD. et al. Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:3384-8 doi:10.1158/1055-9965.epi-09-0955
23. De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K. et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutation research. 2007;631:101-10 doi:10.1016/j.mrgentox.2007.03.010
24. Dhillon VS, Yeoh E, Fenech M. DNA repair gene polymorphisms and prostate cancer risk in South Australia-results of a pilot study. Urologic oncology. 2011;29:641-6 doi:10.1016/j.urolonc.2009.08.013
25. Dianzani I, Gibello L, Biava A, Giordano M, Bertolotti M, Betti M. et al. Polymorphisms in DNA repair genes as risk factors for asbestos-related malignant mesothelioma in a general population study. Mutation research. 2006;599:124-34 doi:10.1016/j.mrfmmm.2006.02.005
26. Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23:1229-34
27. Engin AB, Karahalil B, Engin A, Karakaya AE. Oxidative stress, Helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genetic testing and molecular biomarkers. 2010;14:559-64 doi:10.1089/gtmb.2009.0195
28. Engin AB, Karahalil B, Engin A, Karakaya AE. DNA repair enzyme polymorphisms and oxidative stress in a Turkish population with gastric carcinoma. Molecular biology reports. 2011;38:5379-86 doi:10.1007/s11033-011-0690-9
29. Farinati F, Cardin R, Bortolami M, Nitti D, Basso D, de Bernard M. et al. Oxidative DNA damage in gastric cancer: CagA status and OGG1 gene polymorphism. International journal of cancer Journal international du cancer. 2008;123:51-5 doi:10.1002/ijc.23473
30. Farkasova T, Gurska S, Witkovsky V, Gabelova A. Significance of amino acid substitution variants of DNA repair genes in radiosusceptibility of cervical cancer patients; a pilot study. Neoplasma. 2008;55:330-7
31. Ferguson HR, Wild CP, Anderson LA, Murphy SJ, Johnston BT, Murray LJ. et al. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett's esophagus, or esophageal adenocarcinoma: results from the factors influencing the Barrett's adenocarcinoma relationship case-control study. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:736-9 doi:10.1158/1055-9965.epi-07-2832
32. Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S. et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Human genetics. 2007;121:233-42 doi:10.1007/s00439-006-0294-y
33. Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Do DNA repair genes OGG1, XRCC3 and XRCC7 have an impact on susceptibility to bladder cancer in the North Indian population? Mutation research. 2009;680:56-63 doi:10.1016/j.mrgentox.2009.09.008
34. Garcia-Quispes WA, Perez-Machado G, Akdi A, Pastor S, Galofre P, Biarnes F. et al. Association studies of OGG1, XRCC1, XRCC2 and XRCC3 polymorphisms with differentiated thyroid cancer. Mutation research. 2011;709-710:67-72 doi:10.1016/j.mrfmmm.2011.03.003
35. Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I. et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual's susceptibility to sporadic colorectal cancer. Molecular biology reports. 2012;39:527-34 doi:10.1007/s11033-011-0767-5
36. Gorgens H, Muller A, Kruger S, Kuhlisch E, Konig IR, Ziegler A. et al. Analysis of the base excision repair genes MTH1, OGG1 and MUTYH in patients with squamous oral carcinomas. Oral oncology. 2007;43:791-5 doi:10.1016/j.oraloncology.2006.10.004
37. Hall J, Hashibe M, Boffetta P, Gaborieau V, Moullan N, Chabrier A. et al. The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis. 2007;28:665-71 doi:10.1093/carcin/bgl160
38. Hanaoka T, Sugimura H, Nagura K, Ihara M, Li XJ, Hamada GS. et al. hOGG1 exon7 polymorphism and gastric cancer in case-control studies of Japanese Brazilians and non-Japanese Brazilians. Cancer letters. 2001;170:53-61
39. Hansen R, Saebo M, Skjelbred CF, Nexo BA, Hagen PC, Bock G. et al. GPX Pro198Leu and OGG1 Ser326Cys polymorphisms and risk of development of colorectal adenomas and colorectal cancer. Cancer letters. 2005;229:85-91 doi:10.1016/j.canlet.2005.04.019
40. Hansen RD, Krath BN, Frederiksen K, Tjonneland A, Overvad K, Roswall N. et al. GPX1 Pro(198)Leu polymorphism, erythrocyte GPX activity, interaction with alcohol consumption and smoking, and risk of colorectal cancer. Mutation research. 2009;664:13-9 doi:10.1016/j.mrfmmm.2009.01.009
41. Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G. et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer research. 2004;64:4378-84 doi:10.1158/0008-5472.can-04-0372
42. Hatt L, Loft S, Risom L, Moller P, Sorensen M, Raaschou-Nielsen O. et al. OGG1 expression and OGG1 Ser326Cys polymorphism and risk of lung cancer in a prospective study. Mutation research. 2008;639:45-54 doi:10.1016/j.mrfmmm.2007.11.002
43. Hosono S, Matsuo K, Ito H, Oze I, Hirose K, Watanabe M. et al. Polymorphisms in base excision repair genes are associated with endometrial cancer risk among postmenopausal Japanese women. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2013;23:1561-8 doi:10.1097/IGC.0b013e3182a80a7e
44. Huang M, Dinney CP, Lin X, Lin J, Grossman HB, Wu X. High-order interactions among genetic variants in DNA base excision repair pathway genes and smoking in bladder cancer susceptibility. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:84-91 doi:10.1158/1055-9965.epi-06-0712
45. Huang WY, Gao YT, Rashid A, Sakoda LC, Deng J, Shen MC. et al. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones: a population-based case-control study in China. Carcinogenesis. 2008;29:100-5 doi:10.1093/carcin/bgm247
46. Hung RJ, Brennan P, Canzian F, Szeszenia-Dabrowska N, Zaridze D, Lissowska J. et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. Journal of the National Cancer Institute. 2005;97:567-76 doi:10.1093/jnci/dji101
47. Ito H, Hamajima N, Takezaki T, Matsuo K, Tajima K, Hatooka S. et al. A limited association of OGG1 Ser326Cys polymorphism for adenocarcinoma of the lung. Journal of epidemiology / Japan Epidemiological Association. 2002;12:258-65
48. Janik J, Swoboda M, Janowska B, Ciesla JM, Gackowski D, Kowalewski J. et al. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutation research. 2011;709-710:21-31 doi:10.1016/j.mrfmmm.2011.02.009
49. Jankowska AM, Gondek LP, Szpurka H, Nearman ZP, Tiu RV, Maciejewski JP. Base excision repair dysfunction in a subgroup of patients with myelodysplastic syndrome. Leukemia. 2008;22:551-8 doi:10.1038/sj.leu.2405055
50. Jiao X, Huang J, Wu S, Lv M, Hu Y. et al. hOGG1 Ser326Cys polymorphism and susceptibility to gallbladder cancer in a Chinese population. International journal of cancer Journal international du cancer. 2007;121:501-5 doi:10.1002/ijc.22748
51. Karahalil B, Emerce E, Kocer B, Han S, Alkis N, Karakaya AE. The association of OGG1 Ser326Cys polymorphism and urinary 8-OHdG levels with lung cancer susceptibility: a hospital-based case-control study in Turkey. Arhiv za higijenu rada i toksikologiju. 2008;59:241-50 doi:10.2478/10004-1254-59-2008-1924
52. Karahalil B, Kocabas NA, Ozcelik T. DNA repair gene polymorphisms and bladder cancer susceptibility in a Turkish population. Anticancer research. 2006;26:4955-8
53. Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N. et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. Journal of experimental & clinical cancer research: CR. 2008;27:49. doi:10.1186/1756-9966-27-49
54. Kim EJ, Jeong P, Quan C, Kim J, Bae SC, Yoon SJ. et al. Genotypes of TNF-alpha, VEGF, hOGG1, GSTM1, and GSTT1: useful determinants for clinical outcome of bladder cancer. Urology. 2005;65:70-5 doi:10.1016/j.urology.2004.08.005
55. Kim HN, Kim NY, Yu L, Kim YK, Lee IK, Yang DH. et al. Polymorphisms in DNA repair genes and MDR1 and the risk for non-Hodgkin lymphoma. International journal of molecular sciences. 2014;15:6703-16 doi:10.3390/ijms15046703
56. Kim JI, Park YJ, Kim KH, Kim JI, Song BJ, Lee MS. et al. hOGG1 Ser326Cys polymorphism modifies the significance of the environmental risk factor for colon cancer. World journal of gastroenterology: WJG. 2003;9:956-60
57. Kim KY, Han W, Noh DY, Kang D, Kwack K. Impact of genetic polymorphisms in base excision repair genes on the risk of breast cancer in a Korean population. Gene. 2013;532:192-6 doi:10.1016/j.gene.2013.09.069
58. Klinchid J, Chewaskulyoung B, Saeteng S, Lertprasertsuke N, Kasinrerk W, Cressey R. Effect of combined genetic polymorphisms on lung cancer risk in northern Thai women. Cancer genetics and cytogenetics. 2009;195:143-9 doi:10.1016/j.cancergencyto.2009.08.011
59. Kohno T, Kunitoh H, Mimaki S, Shiraishi K, Kuchiba A, Yamamoto S. et al. Contribution of the TP53, OGG1, CHRNA3, and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:813-7 doi:10.1097/JTO.0b013e3181ee80ef
60. Kohno T, Kunitoh H, Toyama K, Yamamoto S, Kuchiba A, Saito D. et al. Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma risk. Cancer science. 2006;97:724-8 doi:10.1111/j.1349-7006.2006.00240.x
61. Kohno T, Shinmura K, Tosaka M, Tani M, Kim SR, Sugimura H. et al. Genetic polymorphisms and alternative splicing of the hOGG1 gene, that is involved in the repair of 8-hydroxyguanine in damaged DNA. Oncogene. 1998;16:3219-25 doi:10.1038/sj.onc.1201872
62. Kotnis A, Namkung J, Kannan S, Jayakrupakar N, Park T, Sarin R. et al. Multiple pathway-based genetic variations associated with tobacco related multiple primary neoplasms. PloS one. 2012;7:e30013. doi:10.1371/journal.pone.0030013
63. Krupa R, Sobczuk A, Poplawski T, Wozniak K, Blasiak J. DNA damage and repair in endometrial cancer in correlation with the hOGG1 and RAD51 genes polymorphism. Molecular biology reports. 2011;38:1163-70 doi:10.1007/s11033-010-0214-z
64. Kumar A, Pant MC, Singh HS, Khandelwal S. Role of OGG1 Ser326Cys polymorphism and 8-oxoguanine DNA damage in risk assessment of squamous cell carcinoma of head and neck in North Indian population. Mutation research. 2011;726:227-33 doi:10.1016/j.mrgentox.2011.09.015
65. Laantri N, Jalbout M, Khyatti M, Ayoub WB, Dahmoul S, Ayad M. et al. XRCC1 and hOGG1 genes and risk of nasopharyngeal carcinoma in North African countries. Molecular carcinogenesis. 2011;50:732-7 doi:10.1002/mc.20754
66. Lagadu S, Lechevrel M, Sichel F, Breton J, Pottier D, Couderc R. et al. 8-oxo-7,8-dihydro-2'-deoxyguanosine as a biomarker of oxidative damage in oesophageal cancer patients: lack of association with antioxidant vitamins and polymorphism of hOGG1 and GST. Journal of experimental & clinical cancer research: CR. 2010;29:157. doi:10.1186/1756-9966-29-157
67. Lan Q, Mumford JL, Shen M, Demarini DM, Bonner MR, He X. et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25:2177-81 doi:10.1093/carcin/bgh240
68. Le Marchand L, Donlon T, Lum-Jones A, Seifried A, Wilkens LR. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2002;11:409-12
69. Letkova L, Matakova T, Musak L, Sarlinova M, Krutakova M, Slovakova P. et al. DNA repair genes polymorphism and lung cancer risk with the emphasis to sex differences. Molecular biology reports. 2013;40:5261-73 doi:10.1007/s11033-013-2626-z
70. Li D, Suzuki H, Liu B, Morris J, Liu J, Okazaki T. et al. DNA repair gene polymorphisms and risk of pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:740-6 doi:10.1158/1078-0432.ccr-08-1607
71. Li Q, Huang L, Rong L, Xue Y, Lu Q, Rui Y. et al. hOGG1 Ser326Cys polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Cancer science. 2011;102:1123-7 doi:10.1111/j.1349-7006.2011.01928.x
72. Li Q, Wang JM, Peng Y, Zhang SH, Ren T, Luo H. et al. Association of DNA base-excision repair XRCC1, OGG1 and APE1 gene polymorphisms with nasopharyngeal carcinoma susceptibility in a Chinese population. Asian Pacific journal of cancer prevention: APJCP. 2013;14:5145-51
73. Li QD, Li H, Wang MS, Diao TY, Zhou ZY, Fang QX. et al. Multi-susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC gastroenterology. 2011;11:74. doi:10.1186/1471-230x-11-74
74. Li Z, Guan W, Li MX, Zhong ZY, Qian CY, Yang XQ. et al. Genetic polymorphism of DNA base-excision repair genes (APE1, OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Archives of medical research. 2011;42:226-34 doi:10.1016/j.arcmed.2011.04.005
75. Liang G, Pu Y, Yin L. Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer letters. 2005;223:265-74 doi:10.1016/j.canlet.2004.12.042
76. Liu CJ, Hsia TC, Tsai RY, Sun SS, Wang CH, Lin CC. et al. The joint effect of hOGG1 single nucleotide polymorphism and smoking habit on lung cancer in Taiwan. Anticancer research. 2010;30:4141-5
77. Liu X, Xiao N, Guo W, Wu Y, Cai Z, He Q. et al. The hOGG1 gene 5'-UTR variant c.-53G>C contributes to the risk of gastric cancer but not colorectal cancer in the Chinese population: the functional variation of hOGG1 for gastric cancer risk. Journal of cancer research and clinical oncology. 2011;137:1477-85 doi:10.1007/s00432-011-1022-0
78. Loft S, Svoboda P, Kasai H, Tjonneland A, Vogel U, Moller P. et al. Prospective study of 8-oxo-7,8-dihydro-2'-deoxyguanosine excretion and the risk of lung cancer. Carcinogenesis. 2006;27:1245-50 doi:10.1093/carcin/bgi313
79. Loizidou MA, Michael T, Neuhausen SL, Newbold RF, Marcou Y, Kakouri E. et al. DNA-repair genetic polymorphisms and risk of breast cancer in Cyprus. Breast cancer research and treatment. 2009;115:623-7 doi:10.1007/s10549-008-0084-4
80. Luo H, Li Z, Qing Y, Zhang SH, Peng Y, Li Q. et al. Single nucleotide polymorphisms of DNA base-excision repair genes (APE1, OGG1 and XRCC1) associated with breast cancer risk in a Chinese population. Asian Pacific journal of cancer prevention: APJCP. 2014;15:1133-40
81. Ma L, Chu H, Wang M, Shi D, Zhong D, Li P. et al. hOGG1 Ser326Cys polymorphism is associated with risk of bladder cancer in a Chinese population: a case-control study. Cancer science. 2012;103:1215-20 doi:10.1111/j.1349-7006.2012.02290.x
82. Malik MA, Zargar SA, Mittal B. Lack of influence of DNA repair gene OGG1 codon 326 polymorphisms of gastric cancer risk in the Kashmir valley. Asian Pacific journal of cancer prevention: APJCP. 2010;11:165-8
83. Marques CR, Da Silva TM, De Albuquerque DM, Chaves MS, Marques Filho MF, Oliveira JS. et al. NAT2, XRCC1 and hOGG1 polymorphisms, cigarette smoking, alcohol consumption and risk of upper aerodigestive tract cancer. Anticancer research. 2014;34:3217-24
84. Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S. et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997-1007 doi:10.1093/carcin/bgi280
85. McWilliams RR, Bamlet WR, Cunningham JM, Goode EL, de Andrade M, Boardman LA. et al. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer research. 2008;68:4928-35 doi:10.1158/0008-5472.can-07-5539
86. Ming-Shiean H, Yu JC, Wang HW, Chen ST, Hsiung CN, Ding SL. et al. Synergistic effects of polymorphisms in DNA repair genes and endogenous estrogen exposure on female breast cancer risk. Annals of surgical oncology. 2010;17:760-71
87. Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K. Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutation research. 2008;648:65-72 doi:10.1016/j.mrfmmm.2008.09.014
88. Takezaki T, Gao CM, Wu JZ, Li ZY, Wang JD, Ding JH. et al. hOGG1 Ser(326)Cys polymorphism and modification by environmental factors of stomach cancer risk in Chinese. International journal of cancer Journal international du cancer. 2002;99:624-7 doi:10.1002/ijc.10400
89. Tsai CW, Tsai MH, Tsou YA, Shih LC, Tseng HC, Chang WS. et al. The joint effect of smoking and hOGG1 genotype on oral cancer in Taiwan. Anticancer research. 2012;32:3799-803
90. Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L. et al. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer causes & control: CCC. 2008;19:1077-83 doi:10.1007/s10552-008-9171-4
91. Tsou YA, Hua CH, Tseng HC, Hsu CF, Tsai CW, Sun SS. et al. The joint effect of hOGG1 single nucleotide polymorphism and betel quid chewing on oral cancer in Taiwan. Anticancer research. 2010;30:4205-8
92. Tsukino H, Hanaoka T, Otani T, Iwasaki M, Kobayashi M, Hara M. et al. hOGG1 Ser326Cys polymorphism, interaction with environmental exposures, and gastric cancer risk in Japanese populations. Cancer science. 2004;95:977-83
93. Upadhyay R, Malik MA, Zargar SA, Mittal B. OGG1 Ser326Cys polymorphism and susceptibility to esophageal cancer in low and high at-risk populations of northern India. Journal of gastrointestinal cancer. 2010;41:110-5 doi:10.1007/s12029-009-9124-5
94. Wang W, Wang M, Chen Y, Zhang Z, Wang S, Xu M. et al. The hOGG1 Ser326Cys polymorphism contributes to cancer susceptibility: evidence from 83 case-control studies. Mutagenesis. 2012;27:329-36 doi:10.1093/mutage/ger083
95. Wei B, Zhou Y, Xu Z, Xi B, Cheng H, Ruan J. et al. The effect of hOGG1 Ser326Cys polymorphism on cancer risk: evidence from a meta-analysis. PloS one. 2011;6:e27545. doi:10.1371/journal.pone.0027545
96. Sterpone S, Cornetta T, Padua L, Mastellone V, Giammarino D, Testa A. et al. DNA repair capacity and acute radiotherapy adverse effects in Italian breast cancer patients. Mutation research. 2010;684:43-8 doi:10.1016/j.mrfmmm.2009.11.009
97. Srivastava K, Srivastava A, Mittal B. Polymorphisms in ERCC2, MSH2, and OGG1 DNA repair genes and gallbladder cancer risk in a population of Northern India. Cancer. 2010;116:3160-9 doi:10.1002/cncr.25063
98. Wells G, Shea B, O'Connell D, Peterson J, Welch V, Losos M. et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Ottawa Health Research Institute. 2011 wwwohrica/programs/clinical_epidemiology/oxfordasp. doi:10.1371/journal.pntd.0002195
99. Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Annals of internal medicine. 1997;127:820-6
100. Berman NG, Parker RA. Meta-analysis: neither quick nor easy. BMC medical research methodology. 2002;2:10
101. DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7:177-88
102. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International journal of surgery (London, England). 2010;8:336-41 doi:10.1016/j.ijsu.2010.02.007
Author contact
![]() Corresponding authors: Dong Wang, MD, PhD, Cancer Center, Daping Hospital and Research Institution of Surgery, Third Military Medical University, No. 10 Changjiang Zhi Rd, Yuzhong District, Chongqing 400042, China. dongwang64com or Xi Wei, MD, PhD, Department of Diagnostic and Therapeutic Ultrasonography, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center of Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China, weixiedu.cn.
Corresponding authors: Dong Wang, MD, PhD, Cancer Center, Daping Hospital and Research Institution of Surgery, Third Military Medical University, No. 10 Changjiang Zhi Rd, Yuzhong District, Chongqing 400042, China. dongwang64com or Xi Wei, MD, PhD, Department of Diagnostic and Therapeutic Ultrasonography, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center of Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin, China, weixiedu.cn.

 Global reach, higher impact
Global reach, higher impact