Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(11):1534-1540. doi:10.7150/jca.15689 This issue Cite
Research Paper
SOX4 Promotes Progression in OLP-Associated Squamous Cell Carcinoma
1. Department of Oral Medicine, Sichuan Academy of Medical Sciences, Sichuan Provincial People's Hospital, Affiliated Hospital to University of Electronic Science and Technology of China, Chengdu 610072, Sichuan, China.
2. UCLA School of Dentistry and Jonsson Comprehensive Cancer Center, 10833 Le Conte Ave, Los Angeles, CA 90095, USA.
*Yi Liu and Li Cui contributed equally to this study
Received 2016-3-29; Accepted 2016-6-1; Published 2016-7-8
Abstract
Background: The development of oral squamous cell carcinoma (OSCC) is a multistep process that involves in both genetic alterations and epigenetic modifications. Previous studies suggest SOX4 might function as an oncogene or a tumor suppressor in different types of cancers. However, whether SOX4 involves in promoting the progression of oral precancer to cancer is unknown.
Methods: Liquid chromatography with tandem mass spectrometry (LC-MS/MS) was used to identify the proteins that may be differentially expressed between oral lichen planus (OLP) and OLP-associated OSCC (OLP-OSCC) formalin-fixed paraffin-embedded (FFPE) tissues. Immunohistochemistry (IHC) and Western blotting were performed to evaluate SOX4 expression between OLP and OLP-OSCC tissues and among oral cancer cell lines and normal human oral keratinocytes (NHOKs). SOX4 siRNA was used to knock down the expression of SOX4 in UM1 oral cancer cells. MTT, cell counting, migration and Matrigel invasion assays were utilized to examine the effect of SOX4 down-regulation on proliferation, migration and invasion capacity of UM1 cells.
Results: LC-MS/MS analysis showed that 88 proteins including SOX4 were only identified in OLP-OSCC FFPE tissues when compared to OLP FFPE tissues. IHC confirmed that SOX4 expression was significantly higher in OLP-OSCC than OLP and Western blot analysis indicated that SOX4 was over-expressed in UM1/UM2 cells when compared to NHOKs. Knockdown of SOX4 significantly inhibited the proliferation, migration and invasion of UM1 cells (P<0.01).
Conclusions: Our study indicated that SOX4 is significantly upregulated in OLP-OSCC versus OLP tissues. In addition, down-regulation of SOX4 led to significantly reduced proliferation, migration and invasion capability of oral cancer cells. These findings suggest that SOX4 might be actively involved in the progression of OLP to OSCC.
Keywords: Invasion, Migration, Oral lichen planus, Oral squamous cell carcinoma, SOX4
Introduction
Despite the advances in surgery, radiation therapy and chemotherapy, oral squamous cell carcinoma (OSCC) remains to be a challenging public health problem worldwide [1-2]. The 5-year overall survival rate following treatment for OSCC has been stagnant at 50% in the past few decades [3-4]. The main reasons for the poor prognosis is that early molecular events responsible for the progression of oral precancer to cancer are poorly understood, and most OSCC cases are diagnosed at the advanced stage. This highlights the importance of studying the underlying molecular mechanisms and developing molecular biomarkers for early detection of this devastating disease.
In human, SOX (sex-determining region Y-related high-mobility-group box transcription factor) gene family has at least 20 members [5]. SOX4, one of group-C SOX genes, plays an important role in the regulation of transcription during developmental processes such as embryonic cardiac development, nervous system development, osteoblastic differentiation, and thymocyte development [6]. Recently, multiple studies reported altered expression of SOX4 in human cancers. SOX4 was found to be overexpressed in prostate cancer (PCa) and its expression level was closely correlated with prognosis [7]. In fact, upregulation of SOX4 in PCa was induced by PTEN loss as a result of the activation of PI3K-AKT-mTOR signaling [8]. SOX4 was identified as a master regulator of epithelial-mesenchymal transition (EMT) by directly regulating the expression of the epigenetic modifier Ezh2 in breast cancer, indicating SOX4 might be indispensable for tumor progression [9]. However, studies also showed that SOX4 was significantly down-regulated in metastatic melanoma compared with dysplastic nevi and primary melanoma. In addition, knockdown of SOX4 promoted the migration and invasion of melanoma cells in an NF-κB p50-dependent manner, suggesting that SOX4 might function as a tumor suppressor gene in melanoma [10].
Oral lichen planus (OLP) is a chronic inflammatory disease which presents as white striations, white papules, white plaques, erythema, erosions or blisters affecting predominantly buccal mucosa, tongue and gingivae, although other oral sites may be occasionally involved [11]. The World Health Organization (WHO) classifies OLP as a “premalignant disorder” with unspecified malignant transformation risk and suggests that OLP patients should be under close monitoring. Clinically, OLP is characterized by T-cell-mediated chronic immune response and abnormal epithelial keratinization cycle. However, the etiology of OLP is poorly understood and the molecular mechanisms underlying OLP progression to OSCC remains largely unknown. Further investigation of the genetic/protein alterations in OLP and associated OSCC is warranted, which will lead to an improved understanding of the malignant potential of OLP and help diagnose and treat OLP-associated OSCC patients at early stage.
Although SOX4 seems to be a crucial regulator responsible for cancer initiation and progression, whether it involves in the progression of oral precancer to OSCC is unknown. Thus, in this study, we aim to investigate if SOX4 is differentially expressed between OLP and OLP-OSCC and whether SOX4 has a functional role in the proliferation, migration and invasion of oral cancer cells.
Materials and methods
Tissue samples and immunochemistry (IHC)
The study was approved by the UCLA Institutional IRB Committee. Archived FFPE tissues of OLPs and OLP-OSCCs were used for liquid chromatography with tandem mass spectrometry (LC-MS/MS) and IHC. To perform IHC analysis, FFPE tissue sections were deparaffinized by sequential washing with xylene, 100% ethanol, 95% ethanol, 80% ethanol and PBS. The sections were incubated with 0.3% H2O2 in methanol for 5 min to quench the endogenous peroxidase activity. The slides were blocked in PBS with 5% BSA for 30 min and then incubated overnight with a 1:100 dilution of anti-SOX4 primary antibody (Abcam, Cambridge, MA, USA) at 4 °C. After sections were rinsed with PBS, they were incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:2000; GE Healthcare, Piscataway, NJ, USA) for 2 h at room temperature.
Protein extraction for mass spectrometry
Intact proteins were isolated from the FFPE tissues of OLP (n=6) and OLP-OSCC (n=6) with the Qproteome FFPE Tissue kit (Qiagen, Valencia, CA, USA). In total, 5 μg of proteins from pooled OLP or OLP-OSCC sample were digested with 50 ng enzyme-grade trypsin (Promega, Madison, WI, USA) for overnight. LC-MS/MS of the resulting peptides was performed using a NanoLC system (Eksigent Technologies, Dublin, CA, USA) and a LTQ mass spectrometer (Thermo Fisher, Waltham, MA, USA). The peptides were concentrated and desalted on a C18 IntegraFrit Nano-Precolumn (New Objective, Woburn, MA, USA) for 10 min, then eluted and resolved using a C18 reversed-phase capillary column (New Objective). LC separation was performed at 400 nL/min with the following mobile phases: A, 5% acetonitrile/0.1%formic acid (v/v); B, 95% acetonitrile/0.1% formic acid (v/v). The chosen LC gradient was: from 5% to 15% B in 1 min, from 15% to 100% B in 40 min, and then maintained at 100%B for 15 min. Database searches were performed using the X! Tandem search engine against the SwissProt protein sequence database. The search criteria were set with a mass accuracy of 0.4 Da and semi-style cleavage by trypsin. Proteins with two unique peptides are considered as positively identified.
Cell culture
Normal human oral keratinocytes (NHOKs) were maintained in the EpiLife media supplemented with the human keratinocyte growth supplement (Invitrogen). UM1 and UM2 oral cancer cell lines were cultured in the Dulbecco's modified eagle medium (DMEM) plus 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). The cells were maintained at 37°C in a humidified 5% CO2 incubator and passaged when they reached 90-95% confluence.
siRNA knockdown of SOX4
UM1 cells were transfected with siRNA in 96-well or 6-well plates using the Lipofectamine 3000 transfection regent (Invitrogen, Carlsbad, CA, USA) according to the company's instruction. Validated double-stranded siRNAs of SOX4 (sc-38412) or non-target control siRNAs (Santa Cruz Biotech, Santa Cruz, CA, USA) were mixed with the transfection reagent and then added to the cell culture. After overnight incubation, the siRNAs were removed and the cells were further cultured in fresh media for 48 h.
Western blotting
Protein samples were separated with a 4-12% Bis-Tris NuPAGE gel (Invitrogen) and transferred onto nitrocellulose membrane by the Trans-blot SD semi-dry transfer cell (Bio-Rad, Brea, CA, USA). The membranes were blocked in TBST buffer containing 5% nonfat milk (Santa Cruz Biotech), and incubated with rabbit antibody against human SOX4 at a dilution of 1:500 (Abcam) overnight, followed by HRP linked goat anti-rabbit IgG (1:5000; GE Healthcare). The detection was performed with the ECL-Plus Western blotting reagent kit (GE Healthcare).
MTT assay
UM1 cells are highly invasive while UM2 cells have low invasive capacity. Therefore, we choose UM1 cell line as the in vitro model for functional analysis. After 24 h of serum starvation, the siRNA transfected cells were seeded into a 96-well plate at a density of 4000 cells/well in 100 μL medium for the MTT assay. At the indicated time points, 20μL of MTT (Sigma-Aldrich, St. Louis, MO, USA) dissolved in PBS at 5 mg/ml was added to each culture well. Following by incubation for 4 h at 37 °C, the supernatant was then discarded and the precipitate dissolved in 200 μl of dimethyl sulfoxide (DMSO, Sigma). The absorbance of each well was measured using a Synergy HT microplate reader (BioTek Instruments, Winooski, VT, USA) at 490 nm.
Cell counting assay
Cells were harvest from 10-cm petri dish with 0.25% trypsin treatment to prepare single-cell suspension, followed by counting the cells with a vi-cell® cell counter (Beckman Coulter, Inc. Brea, CA, USA). Then the cells were seeded into 12-well plates at a density of 1 x105 cells per well. At the indicated time points, cells were harvested and resuspended in 500 µL complete media and subjected to cell counting analysis.
Wound healing assay
Wound healing assay was used to evaluate the migratory ability of siRNA-transfected UM1 cells. Briefly, 5x105 cells were seeded into a 6-well plate and were allowed to grow to confluent cell monolayers. Subsequently, four vertical wounds were generated per well using a micropipette tip. The plate was then returned to the tissue culture incubator. For quantification, images at the beginning and after 8 h were captured and the distance between the edges of the wound were calculated and analyzed by the NIH Image J software.
Invasion assay
The invasion assays were performed with the Matrigel-coated transwells. Following 24 h serum starvation, cells were harvested and resuspended in the DMEM media containing 0.1% FBS, and 5 x104 cells were then added to the upper chamber of transwell inserts. DMEM containing 10% FBS was used as chemoattractant in the lower chamber. After 24 h, cells that had migrated through the membrane were fixed and stained with the HEMA 3 staining kit (Thermo Fisher Scientific, Pittsburgh, PA, USA). The invaded cells in four random fields were counted and expressed as the average number of cells per field under light microscopy (Eclipse TE2000, Nikon, Tokyo, Japan).
Statistical analysis
The data were expressed as the mean ± standard deviation, and analyzed by the independent samples t-test using the MedCalc (MedCalc Software, Ostend, Belgium). P values < 0.05 were considered as statistically significant.
Results
SOX4 was over-expressed in OLP-OSCC versus OLP
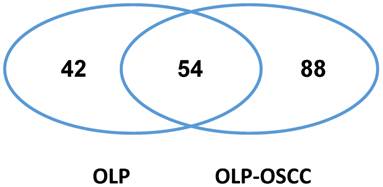
We isolated intact proteins from archived FFPE tissues of OLPs and OLP-OSCCs using the Qproteome FFPE Tissue kit and then analyzed the protein samples with LC-MS/MS and database searching. In total, 96 and 142 proteins were identified in the pooled OLP FFPE tissue sample (n=6) and the pooled OLP- OSCC FFPE tissue sample (n=6), respectively (Fig.1 and supplementary data). Fifty-four proteins were commonly detected between OLP and OLP-OSCC FFPE tissues, and 88 proteins including SOX4 were only identified in OLP-OSCC. A list of tumor promoting proteins that only detected in OLP-OSCC was summarized in Table 1.
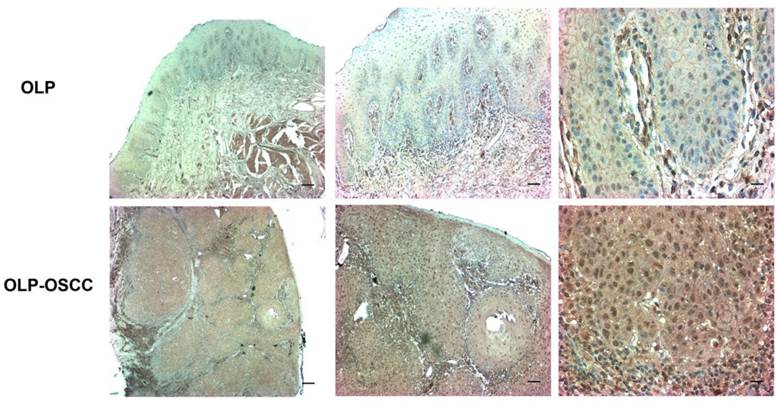
The expression level of SOX-4 in clinical samples was further assessed using IHC. The positive staining of cells was identified as yellow-brown granules. The intensity of SOX4 staining was significantly stronger in OLP-OSCC than OLP tissues (Fig. 2).
The commonly and differentially expressed proteins between the archived FFPE tissues of OLP and OLP-OSCC. OLP-OSCC were progressed from OLP of the same patients. Proteins were isolated from archived FFPE tissues with the Qproteome FFPE tissue kit and identified by LC-MS/MS and database searching. Forty-two proteins were only identified from the FFPE tissues of OLP while 88 proteins were only identified from FFPE tissues of OLP-OSCC.
A list of proteins that only detected in OLP-OSCC
| Accession No | # of Peptide | Protein name | Molecular function |
|---|---|---|---|
| Q06945 | 2 | SOX-4 | DNA binding |
| Q9NZT1 | 2 | Calmodulin-like protein 5 | Calcium binding |
| P17813 | 2 | Endoglin | Angiogenesis regulation |
| P15502 | 2 | Elastin | ECM structural constituent |
| P05164 | 4 | Myeloperoxidase | Peroxidase activity |
| P60174 | 3 | Triosephosphate isomerase | Ubiquitin protein ligase binding |
| P32119 | 2 | Peroxiredoxin-2 | Redox regulation |
| Q15084 | 2 | Protein disulfide-isomerase A6 | Cell redox homeostasis |
| P19971 | 2 | Thymidine phosphorylase | Angiogenesis regulation |
| P30044 | 2 | Peroxiredoxin-5, mitochondrial | Redox regulation |
SOX4 was over-expressed in OSCC cell lines
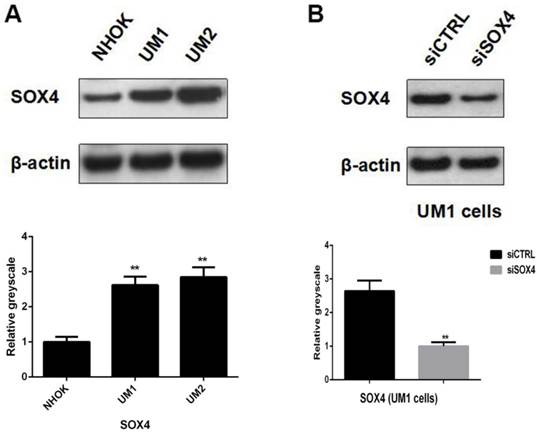
Western blot analysis showed that SOX4 was significantly over-expressed in both UM1 and UM2 cells compared to NHOKs, which is in line with IHC results and further confirms that SOX4 is overexpressed in oral cancer cells (Fig. 3A).
Knockdown of SOX4 in UM1 oral cancer cells
UM1 cells were successfully transfected with siSOX4 by using the Lipofectamine 3000 reagent. Western blotting results showed that the protein expression level of SOX4 was significantly lower in the UM1 cells transfected with siSOX4 when compared to the UM1 cells transfected with control siRNA (Fig. 3B).
The expression level of SOX4 in OLP and OLP-OSCC FFPE tissues. The IHC results showed that the distribution and intensity of SOX4 staining in OLP-OSCC was significantly broader and stronger than that in OLP tissues (From left to right, scale bar, 25 µm, 50 µm or 100µm)
(A) Significant over-expression of SOX4 in UM1 and UM2 oral cancer cells versus NHOKs (normal cells). (B) Knockdown of SOX4 in UM1 cells with siRNA.
Knockdown of SOX4 inhibited the proliferation of UM1 cells
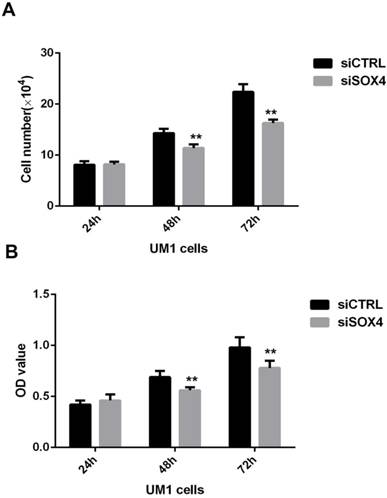
To assess the role of SOX4 in the proliferation of OSCC, MTT assay and cell counting assay were performed using oral cancer cells transfected with siRNA against SOX4 or control siRNA at different time points (24, 48, and 72h). The results showed that the number of oral cancer cells in the SOX4 suppression group was significantly lower in comparison with the negative control group at 48h and 72h (**P <0.01) (Fig.4).
Knockdown of SOX4 suppressed the migration capacity of UM1 cells
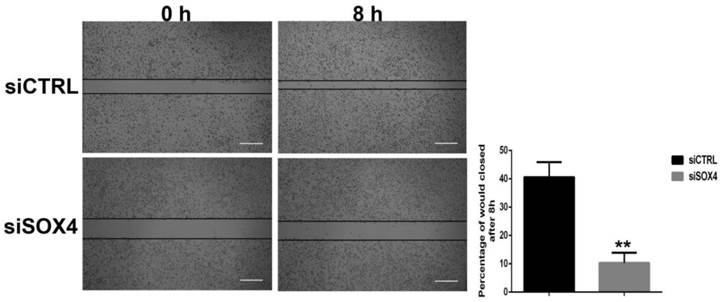
For wound healing assay, the effect of SOX4 inhibition on cell migration was determined at 8 h after the cells reached confluence. As shown in Figure 5, relative to control cells, cells treated with siSOX4 reduced the migration capacity significantly. The empty space between cell layers was significantly greater in siSOX4 treated cells as compared to the control cells (P<0.01) (Fig.5).
Knockdown of SOX4 suppressed the invasion capability of UM1 cells
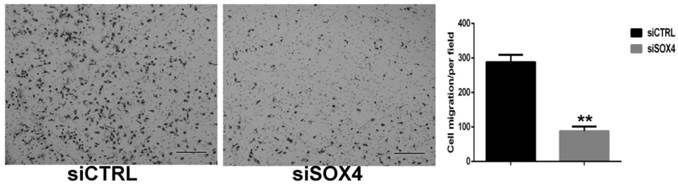
To investigate whether SOX4 involves in regulation invasion capability of oral cancer cells, Matrigel invasion assay was used to evaluate the motility of UM1 cells after SOX4 suppression. Compared with control cells, SOX4 knockdown UM1 cells showed about 68 % reduction in the number of cells crossing the membrane (P<0.01) (Fig. 6).
Effect of SOX4 inhibition on the proliferation of UM1 oral cancer cells. Both cell counting (A) and MTT (B) assay results showed that SOX4 downregulation inhibited the proliferation of UM1 cells at 48h and 72h (**, P < 0.01).
Effect of SOX4 inhibition on the migration of UM1 oral cancer cells. The percentage of wound closed in the siSOX4 transfected UM1 cells was significantly less than the cells transfected with siCTRL (**, P < 0.01) (Scale bar, 100µm).
Effect of SOX4 inhibition on the invasion of UM1 oral cancer cells. siSOX4 transfected UM1 cells showed about 68 % reduction in the number of cells invading through the membrane in comparison with those cells transfected with siCTRL (**, P < 0.01) (Scale bar, 100µm).
Discussion
In this study, we have clearly demonstrated that SOX4 is over-expressed in OLP-OSCC versus OLP. It should be noted that the OSCC in the study progressed from the OLP of the same patients. We have also shown that down-regulation of SOX4 in UM1 oral cancer cells significantly impairs the cell proliferation, migration and invasion, suggesting that SOX4 may have a potential role in promoting the progression of OLP-associated oral cancer.
Oral carcinogenesis is a multistep process characterized by genetic alterations that influence key cellular pathways of growth and development. To date, the underlying molecular mechanisms responsible for OSCC initiation and development, especially the progression of oral precancer to cancer, are inadequately understood. Multiple types of oral precancerous lesions including, but not limited to, oral leukoplakia, OLP, oral submucous fibrosis, oral erythroplakia and actinic cheilitis, may progress to OSCC at different conversion rate. In fact, OLP is defined by the WHO as a precancerous lesion which should be closely monitored for the risk of cancer development. It is a good clinical model for understanding the role of chronic inflammation in oral carcinogenesis. To the best of our knowledge, this is the first LC-MS/MS study revealing the differentially expressed proteins in the archived FFPE tissues between OLP and OLP-associated OSCC. It appeared to be challenging for us to use LC-MS/MS for the analysis of isolated proteins from archived FFPE tissues. We initially tested LC-MS/MS with iTRAQ (isobaric tagging for relative and absolute quantification) for quantification of intact proteins isolated from FFPE tissues. However, the iTRAQ labeling of peptides failed in our experiments presumably due to cross-linking of proteins in archived FFPE tissues. Therefore, we had to use a direct LC-MS/MS identification method to compare the proteins identified from FFPE tissues between OLP and OLP-OSCC. Even with the direct LC-MS/MS identification method, we were unable to identify a large number of proteins from archived FFPE tissues, which again might be due to that cross-linking altered the molecular weights of proteins and peptides. Nevertheless, a number of proteins that promote cancer progression were only detected in OLP-OSCC, and further investigation of these proteins may reveal their potential clinical significance in early stage of OSCC.
Our findings based on LC-MS/MS, IHC and functional analyses are in line with those from previous studies. Yoon et al. reported that SOX4 was significantly upregulated in OSCC tissues compared to adjacent normal mucosa and contributed to oncogenic phenotypes of oral cancer cells by promoting cell survival and elevating chemoradioresistance [12]. Watanabe et al. found that SOX4 expression levels were significantly correlated with various clinicopathological parameters of OSCC including gender, T status, and stage levels, and increased SOX4 expression was detected in poorly differentiated OSCC or in the metastatic focus [13]. In addition to OSCC, studies have demonstrated that SOX4 may play an oncogenic role in other solid malignant tumors [14-17]. Zhang et al. elucidated that SOX4 was a direct target of C/EBPα and downregulation of SOX4 suppressed the self-renewal of leukemic cells and restored their differentiation, indicating that SOX4 overexpression resulting from inactivation of C/EBPα promotes leukemia development [18]. Zhang et al. discovered that SOX4 positively regulated the epithelial-mesenchymal transition (EMT) process and may combine with oncogenic Ras together to promote tumorigenesis in vivo [19]. However, it should be noted that some of the published studies reported a tumor suppressor function of SOX4. For instance, Li et al. found that SOX4 was essential for p53 activation in response to DNA damage. In addition, SOX4 could interact with and stabilize p53 protein by inhibiting Mdm2-mediated p53 ubiquitination and degradation, suggesting that SOX4 might suppress the progression DNA damage response-associated cancer [20]. Wang et al. reported that over-expression of SOX4 in primary gallbladder carcinoma (PGC) was significantly associated with favorable clinical parameters such as low histologic grade, low pathologic T stage, and early clinical stage. In addition, SOX4 upregulation was an independent prognostic factor for better overall and disease-free survival in patients with PGC [21]. Taken together, these previous findings seem to suggest that the concrete role of SOX4 is closely associated with tumor microenvironment and might be tissue specific.
Conclusion
Our study demonstrated that SOX4 was overexpressed when OLP progressed to OSCC. In addition, SOX4 might have a functional role in OSCC by modulating the proliferation, migration and invasion of oral cancer cells. Further studies are needed to validate the clinical significance of SOX4 in the early stage OSCC so that this target protein may be applied to early detection of the malignancy.
Acknowledgements
This work was supported by the TRDRP grant (S.H.), UCLA School of Dentistry Faculty Grant (S.H.), National Natural Science Foundation of China (Grant no. 81502362, Y.L.), and Doctoral Foundation of Sichuan Academy of Medical Sciences & Sichuan Provincial People's Hospital, China (Grant no. 30305030568, Y.L.).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695-709
2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30
3. Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309-16
4. Casiglia J, Woo SB. A comprehensive review of oral cancer. Gen Dent. 2001;49:72-82
5. Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239-55
6. Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ. et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038-46
7. Wang L, Zhang J, Yang X, Chang YW, Qi M, Zhou Z. et al. SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis. 2013;16:301-7
8. Bilir B, Osunkoya AO, Wiles WG 4th, Sannigrahi S, Lefebvre V, Metzger D. et al. SOX4 is essential for prostate tumorigenesis initiated by PTEN ablation. Cancer Res. 2016;76:1112-21
9. Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M. et al. SOX4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768-83
10. Jafarnejad SM, Wani AA, Martinka M, Li G. Prognostic significance of SOX4 expression in human cutaneous melanoma and its role in cell migration and invasion. Am J Pathol. 2010;177:2741-52
11. Sugerman PB, Savage NW, Walsh LJ, Zhao ZZ, Zhou XJ, Khan A. et al. The pathogenesis of oral lichen planus. Crit Rev Oral Biol Med. 2002;13:350-65
12. Yoon TM, Kim SA, Cho WS, Lee DH, Lee JK, Park YL. et al. SOX4 expression is associated with treatment failure and chemoradioresistance in oral squamous cell carcinoma. BMC Cancer. 2015;15:888
13. Watanabe M, Ohnishi Y, Wato M, Tanaka A, Kakudo K. SOX4 expression is closely associated with differentiation and lymph node metastasis in oral squamous cell carcinoma. Med Mol Morphol. 2014;47:150-5
14. Fang CL, Hseu YC, Lin YF, Hung ST, Tai C, Uen YH. et al. Clinical and prognostic association of transcription factor SOX4 in gastric cancer. PLoS One. 2012;7:e52804
15. Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW. et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31:1298-307
16. Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ. et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038-46
17. Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sørensen FB, Verspaget HW. et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100:511-23
18. Zhang H, Alberich-Jorda M, Amabile G, Yang H, Staber PB, Di Ruscio A. et al. SOX4 is a key oncogenic target in C/EBPα mutant acute myeloid leukemia. Cancer Cell. 2013;24:575-88
19. Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X. et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597-608
20. Li AL, Zhang XM. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci U S A. 2009;106:3788-93
21. Wang C, Zhao H, Lu J, Yin J, Zang L, Song N. et al. Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn Pathol. 2012;7:41
Author contact
![]() Corresponding author: Shen Hu, PhD, MBA, CHS 63-070, 10833 Le Conte Ave, Los Angeles, CA 90095; E-mail: shenhuedu; Tel: (310) 206-8834; Fax: (310) 794-7109.
Corresponding author: Shen Hu, PhD, MBA, CHS 63-070, 10833 Le Conte Ave, Los Angeles, CA 90095; E-mail: shenhuedu; Tel: (310) 206-8834; Fax: (310) 794-7109.

 Global reach, higher impact
Global reach, higher impact