3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(12):1645-1652. doi:10.7150/jca.14192 This issue Cite
Research Paper
High Stoichiometry Phosphorylation of Talin at T144/T150 or S446 Produces Contrasting Effects on Calpain-mediated Talin Cleavage and Cell Migration
1. Laboratory of Molecular and Cellular Biology, College of Life Sciences, Hunan Normal University, Changsha 410081, China
2. Laboratory of Medical Molecular and Immunological Diagnostics, College of Medicine, Hunan Normal University, Changsha 410013, China
Received 2015-10-21; Accepted 2016-3-13; Published 2016-7-25
Abstract
Focal adhesions are large multi-protein complexes that serve as the linkage between extracellular matrix (ECM) and actin cytoskeleton and control the network of signaling cascades underlying cell migration. Talin plays a key role in focal adhesion turnover, and calpain-mediated proteolysis of talin is central to focal adhesion disassembly, but its regulation is not well elucidated. Here we demonstrate that talin phosphorylation at three high stoichiometry sites on its head domain, T144 and T150, or S446, have contrasting effects on calpain-mediated cleavage of talin and cell migration by using site-directed mutagenesis to inhibit phosphorylation. Expression of talinT144A+T150A stimulated calpain-mediated cleavage of talin and accelerated focal adhesion disassembly, whereas expression of talinS446A fully inhibited talin cleavage by calpain, preventing focal adhesion disassembly. A large decrease in phospho-threonine or phospho-serine levels was seen with talinT144A+T150A or talinS446A respectively, while more active ERK was present in talinT144A+T150A than in talinS446A. Cell adhesion and transwell assays using uniformly expressing cells showed that expression of talinT144A+T150A or talinS446A have opposing effects on cell adhesion and migration. These findings define and highlight the integral role of site-specific high stoichiometry phosphorylation of talin in regulating calpain-mediated cleavage of talin and focal adhesion disassembly, thus controlling adhesion stability, cell adhesion and ultimately, cell migration.
Keywords: Talin, High stoichiometry phosphorylation, Calpain-mediated cleavage, Focal adhesion disassembly
Introduction
Activated integrins recruit many kinds of cytoplasmic proteins on their β-cytoplasmic tails. The resulting protein complexes form nascent adhesions and mature into focal adhesions, thus establishing the linkage between actin cytoskeleton and extracellular matrix (ECM) and the network of signaling pathways underlying cell migration [1-4]. Among over 180 adhesion components [5, 6], talin is the most upstream regulator of nascent adhesion assembly other than FAK [7]. Knocking out talin genes in embryonic stem (ES) cells abolishes the formation of focal adhesions and stress fibers [8]. Proteolytic cleavage of β-integrins and many adhesion components is performed by the intracellular calcium-dependent protease calpain (m-calpain or calpain 2), whose activity is dependent, on its cleavage of talin [11]. Talin is cleaved into a N-terminal head domain and a C-terminal rod domain by calpain [12]. The cleaved head domain of talin is degraded by Smurf1-mediated ubiquitylation, resulting in extensive focal adhesion turnover [13]. The head domain contains a FERM (band protein 4.1, erzin, radixin, moesin homology) domain as well as binding sites for the cytoplasmic tails of β-integrins, F-actin, layilin and FAK. The rod domain has several vinculin-binding sites, a second integrin-binding site and two F-actin binding sites. Three sites of high stoichiometry phosphorylation were detected by tandem mass spectrometry analysis: Thr144 and Thr150, potentially phosphorylated by Protein Kinase C (PKC), and Ser446, predicted to be phosphorylated by Protein Kinase A (PKA) [14]. Phosphopeptide abundance at these three sites were greater than at other talin phosphorylation sites. Talin is regulated by phosphorylation and calpain cleavage, but little is known about how these two regulatory mechanisms are integrated. In the study, we created site-directed mutations at the three high stoichiometry phosphorylation sites of talin to generate the expression vectors, talinT144A+T150A and talinS446A. Our goals were to identify the specific contexts and consequences associated with preventing high stoichiometry phosphorylation in transfected cells, and to investigate the role of talin site-specific high stoichiometry phosphorylation in regulating calpain-mediated cleavage of talin and focal adhesion disassembly in the context of adhesion stability, cell adhesion and cell migration.
Materials and methods
Reagents and antibodies
Anti-Green Fluorescent Protein (MAB3836), anti-Erk1/2 (p44/p42) (clone MK12 05-1152), anti-Paxillin (clone 5H11), anti-phospho-serine (clone 4A4) and anti-phospho-threonine (clone 20H6.1) antibodies (Ab) and fibronectin, purified from human plasma, were purchased from Millipore (Bedford, MA). Anti-ACTIVE® MAPK pAb(pTEpY) was purchased from Promega (Madison, WI). Anti-calapin 2 antibody (E-10) was purchased from Santa Cruz (Santa Cruz, CA).
Talin expression constructs
A construct for the FERM domain of wild-type human talin1 cDNA (1362bp, 454aa) containing three high stoichiometry phosphorylation sites at positions 144 and 150 (threonine) and 446 (serine) and a calpain cleavage site at position 432 [11], was synthesized by Shanghai Sangon Biotech. The mutant cDNAs, talinT144A+T150A, talinS446A and talinL432G, were created using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using the following primers: 5'-AAATAACAGGGGCCTTAAGAAAGGACAAGGCATTGCTGCGAGATGAAAAG-3' and 5'-TCGCAGCAATGCCTTGTCCTTTCTTAAGGCCCCTGTTATTTCCTCCTTTT-3'; 5'-CAAAAAGTCAACAGTCGGGCAGCAGCAATACAACCGGGTG-3' and 5'-GTTGTATTGCTGCTGCCCGACTGTTGACTTTTTGGGGGAC-3'; 5'-GAATTCATGGTTGCACTTTCACTGAAGATCAGCATTGGGAATGTGGTGAAGACGA-3' and 5'-GCGCATGATGGCAGGCAGGGCCACAGCGCCATGCTCCACTTTCCCCAC-3' respectively. The wild-type and mutant cDNAs were cloned into XhoI-EcoRI sites of pEGFP-N2 (Clontech, Palo Alto, CA) to generate EGFP-talin (WT), EGFP-talinT144A+T150A (T144A+T150A), EGFP-talinS446A (S446A) and EGFP- talinL432G (L432G). All the constructs were confirmed by DNA sequencing to ensure accuracy.
Cell culture and transfection
Fibroblast CHO-K1 cells, which do not express the epidermal growth factor receptor (EGFR), were from American Type Culture Collection(ATCC) and maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS). The cells, plated in 35-mm dishes and grown to confluence, were transfected with 2 μg of DNA and 4 μl of X-treme Gene HP (Roche, Indianapolis, IN) following the manufacturer's instructions. The cells were used 18-36 h post transfection.
Flow cytometry
At 12h after transfection, the cells expressing EGFP-talin or its mutants were collected and counted by sorting EGFP-positive cells in a BD FACSAria III cell sorter with 488-nm laser.
Co-immunoprecipitation and western blotting
At 24 h after transfection, the cells were lysed in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 plus halt protease inhibitor cocktail and halt phosphatase inhibitor cocktail). Expressed wild-type talin or its mutants were incubated with 2 μg of monoclonal anti-GFP Ab at 4°C with shaking for 2 h. The immunoprecipitates were captured with 20 μl of protein G-Sepharose beads (Millipore) on ice overnight. The beads were washed three times with lysis buffer and clarified by centrifugation. The proteins were denatured in SDS sample buffer by boiling water, and protein concentrations were determined using Bradford method to ensure equal amounts of total protein (30 ng) were loaded in each well. The protein samples were resolved on 10% SDS-PAGE and transferred to PVDF. Western blots were visualized and imaged with monoclonal anti-GFP Ab using Super-Signal West Pico (Pierce) under ChemiDoc TM XRS (Bio-rad), and quantified as integrated density to represent the amount of proteins with Image J software.
siRNA Calpain 2 siRNA duplex (5'-UGAGAACACUGGAACGAUAUU-3' and 5'-UAUCGUUCCAGUGUUCUCAUU-3') were designed and synthesized to target the sequence TGAGAACACTGGAACGATA (2172-2190bp) to knockdown calpain 2 activity. The control or siRNA was co-transfected into CHO cells with talinT144A+T150A using TurboFect siRNA transfection reagent (Thermos scientific) according to the manufacturer's instructions. At 36 h after transfection, the cells were lysed, and the protein was immunoprecipitated and analyzed by western blots with the anti-GFP Ab. Detection of phospho-threonine, phospho-serine and active ERK/MAPK
Equal amounts of the immunoprecipitates (30 ng) collected with anti-GFP Ab were separated by SDS-PAGE and transferred to PVDF. The proteins were analyzed by western blotting with monoclonal anti-phospho-threonine or anti-phospho-serineAbs. The homogenous cells expressing EGFP-talin or its mutants collected by flow cytometry were serum-starved overnight, then lysed and immunoprecipitated with a monoclonal anti-ERK1/2 Ab. Equal amounts of the proteins (30 ng) were resolved by SDS-PAGE and analyzed by western blotting with anti-ERK1/2 Ab or a polyclonal anti-ACTIVE MAPK Ab, which recognizes a dually phosphorylated peptide sequence representing the catalytic core of the active ERK2 (pT183EpY185).
Immuno-cytochemistry and live fluorescence microscopy
For immuno-cytochemistry, the cells were transfected, and at 18h after transfection the cells were plated on fibronectin-coated MatTek dishes and allowed to adhere for 1h.The cells were then fixed with 4% paraformaldehyde in PBS and stained with monoclonal anti-paxillin Ab and TRITC-conjugated goat-anti-mouse IgG as standard procedure, and viewed on a Nikon Eclipse Ti inverted microscope to image EGFP and TRITC fluorescence. For live fluorescence microscopy, at 18 h after transfection,the cells were plated on MatTek dishes coated with fibronectin (5 μg/ml) and grown for 4 h in serum-free DMEM. The dish was maintained in the chamber with 5% CO2 at 37 °C equipped with the microscope. Fluorescent images of focal adhesion disassembly were taken at 5-min intervals for a 60-min period.
Cell adhesion and cell migration
An equal number of transfected cells (5×104 cells) obtained by flow cytometry were plated on fibronectin-coated (5 μg /ml) MatTek dishes and were photographed after 4 h incubation. Cell adhesion assay was performed using a 48-well fibronectin-coated adhesion plate (Cell Biolabs) following the manufacturer's manual. Briefly, 150 μl of the counted cells expressing EGFP-talin or its mutants in serum-free DMEM were added to each well and incubated in the incubator for 90 min. The medium was aspirated and the wells were washed 4 times with PBS. The remaining cells in the well were collected with extraction solution and were counted by flow cytometry.
Transwell assays were performed using 24 well transwell permeable supports with 8 μm pores (Corning, Fig. S1) coated with fibronectin (5μg /ml) and placed over a bottom chamber containing 10% FBS DMEM. The various EGFP-tagged cells were added to the filter, serum-starved for 12 h and placed over the bottom chamber. After 12h incubation at 37 °C, the cells that migrated to the bottom chamber were collected and counted. For the cell adhesion and transwell assays, the samples were assayed in triplicate.
Results
Removal of site-specific high stoichiometry phosphorylation sites leads to large decreases of phospho-threonine or phospho-serine levels in talinT144A+T150A or talinS446A respectively, with more active ERK in talinT144A+T150A than in talinS446A
The talin head domain contains 7 phosphorylation sites at threonines, of which 2 high stoichiometry sites (T144, T150) had the highest level of phosphopeptide abundance, and 5 phosphorylation sites at serines, of which 1 high stoichiometry site (S446) had the highest level of phosphopeptide abundance [14]. To test the specificity of monoclonal anti-phospho-threonine or anti-phospho-serine Abs, we used western blotting to detect phospho-threonine or phospho-serine in transfected cells with wild-type talin and the vector EGFP-N2. The EGFP protein exhibited very weak phospho-threonine or phospho-serine signals, while EGFP-tagged wild-type talin possessed higher phospho-threonine or phospho-serine levels (Fig. S2), suggesting that anti-phospho-threonine or anti-phospho-serine Abs can differentiate the level of phospho-threonine or phospho-serine in talin proteins over controls. To identify whether preventing high stoichiometry phosphorylation by site-directed mutations leads to reduced phosphorylation of talinT144A+T150A or talinS446A, we performed western blotting to specifically recognize phospho-threonine or phospho-serine in wild-type or mutant talin. As shown in Fig.1A, the phosphorylation of talinT144A+T150A or talinS446A was significantly reduced, leading to the lower phospho-threonine or phospho-serine level respectively. Since no point mutation occurred at the high stoichiometry sites, wild-type talin exhibited higher levels of both phospho-threonine and phospho-serine. The result revealed that the phosphopeptides at three high stoichiometry sites were indeed highly abundant, and site-directed mutations at the three high stoichiometry sites to abolish phosphorylation led to significant reductions of phospho-threonine or phospho-serine levels in talinT144A+T150A or talinS446A respectively.
Since intracellular calcium levels fail to reach the near-millimolar concentrations required for calpain activation, epidermal growth factor (EGF) triggers the MAPK cascade and eventually activates ERK, which phosphorylates calpain at Ser50 and leads to calpain activation [15,16]. To ascertain whether the active amount of ERK varies in cells expressing wild-type or mutant talin, we used flow cytometry to obtain homogenous cells expressing various talin constructs and performed western blotting with a polyclonal anti-ACTIVE MAPK Ab to specifically detect active ERK. Fig.1B shows that the same amount of total ERK1/2 was detected in all of the serum-starved controls (non-transfected cells) and the 3 transfected cells with wild-type or mutant talin. Fig.1C demonstrates that no active ERK (ERK2) was detected in the serum-starved control cells. Active ERK was detected in the serum-starved cells expressing wild-type talin or its mutants, but its abundance significantly differed. Normalized to wild-type talin, active ERK activity was much higher in the cells expressing talinT144A+T150A (147%) than in the cells expressing talinS446A (62%, Fig.1D).
Expression of talinT144A+T150A stimulates calpain-mediated cleavage of talin, but expression of talinS446A fully inhibits the cleavage
To determine whether calpain-mediated proteolysis was affected by our talin mutants, CHO-K1 cells were transfected with wild-type and mutant talin.
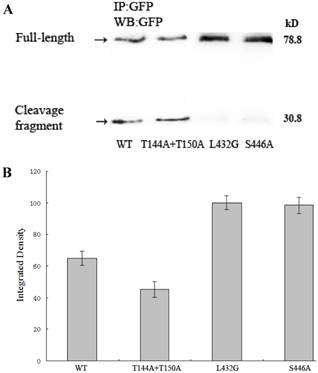
The recombinant EGFP-tagged talins were collected by immunoprecipitating with monoclonal anti-GFP Ab. Fig. 2A clearly shows that only wild-type talin and talinT144A+T150A were cleaved by calpain, producing one 30.8 kD cleavage fragment and one 78.8 kD full-length protein (uncleaved), but talinT144A+T150A was cleaved more efficiently and produced more cleavage fragments than in wild-type. As expected, a mutant resistant to calpain cleavage, talinL432G, was not cleaved and only yielded full-length protein. TalinS446A was also not cleaved, exhibiting a similar result to talinL432G.
Abolishing site-specific high stoichiometry phosphorylation leads to significant decreases in phospho-threonine or phospho-serine levels in talinT144A+T150A or talinS446A respectively, with more active ERK present in talinT144A+T150A than in talinS446A. (A), Wild-type and mutant talins were analyzed by western blots with monoclonal anti-phospho-threonine and anti-phospho-serine antibodies. The phosphopeptide abundance of talinT144A+T150A and talinS446A were significantly reduced. (B), The same amount of total ERK1/2 was detected in the serum-starved controls and 3 cell types transfected with mutant talins using a monoclonal anti-ERK1/2 antibody. (C), No active ERK (ERK2) was detected in the serum-starved control cells. Active ERK activity differed in 3 serum-starved cell populations transfected with talin mutants. (D), Active ERK was quantified as integrated density to represent the amount of protein. The vertical axis shows the quantified data as normalized to wild-type talin (P<0.01).
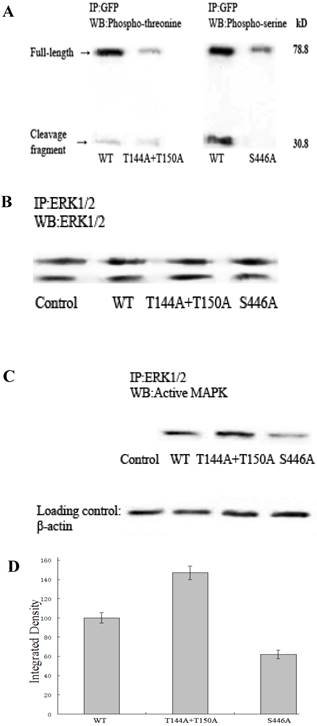
Fig. 2B demonstrates that the amount of full-length protein in talinS446A, wild-type talin and talinT144A+T150A varied significantly with abundances of 98.5%, 65.1% and 45.3% normalized to talinL432G respectively. To identify the effect of decreased calpain 2 activity on talin cleavage dynamics, we performed a siRNA to knockdown calpain 2 activity in cells transfected with talinT144A+T150A. As shown in Fig. S.3, calpain 2 activity was greatly inhibited by calpain 2 siRNA (top right), and the cleavage fragment of talin was greatly diminished when calpain 2 activity was knocked down (bottom right). This result indicates that expression of talinT144A+T150A stimulates calpain-mediated cleavage of talin, while expression of talinS446A fully inhibits the cleavage.
Calpain-mediated cleavage of talin greatly varies between wild-type and mutant talin. (A), Wild-type talin and its mutants were analyzed by western blotting with a monoclonal anti-GFP antibody. (B), Full-length protein (not cleaved by calpain) was quantified as integrated density to represent the amount of protein. The vertical axis shows the quantified data normalized to talinL432G (P<0.01).
Focal adhesions containing talinS446A resist calpain-mediated cleavageand disassemble much more slowly than adhesions containing talinT144A+T150A
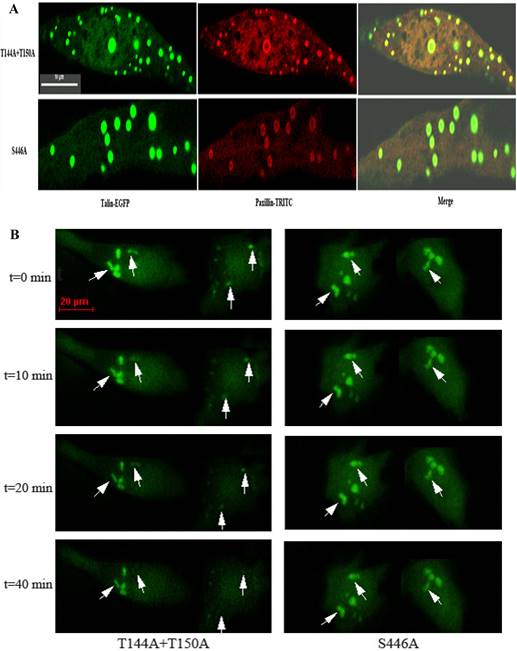
Differences in calpain-mediated proteolysis between talinT144A+T150A and talinS446A should reflect differences in the rate of focal adhesion disassembly. To monitor the dynamics of individual focal adhesion disassembly, we observed and imaged distinctly organized focal adhesions in EGFP-tagged cells over 1 h. Focal adhesions are typically elongated or dot-like and around 2 μm wide and 3-10 μm long in living cells [3]. As multi-protein complexes, focal adhesions are composed of many kinds of cytoplasmic proteins. Paxillin is a known marker localized to focal adhesions. As shown in Fig.3A, talin-EGFP and paxillin-TRITC were co-localized in transfected cells with talinT144A+T150A or talinS446A, indicating that the structures containing EGFP-tagged talin were focal adhesions. No focal adhesions were observed in cells expressing pEGFP-N2 (Fig. S.4). Fig.3B shows that organized focal adhesions in talinT144A+T150A were gradually diffused before 20 min and were completely dispersed and broken down before 40 min. In contrast, the focal adhesions observed in talinS446A cells maintained their structure and had no observable changes during this period, suggesting that focal adhesions containing talinS446A have the ability to resist calpain-mediated cleavage and disassemble much more slowly than those containing talinT144A+T150A. Quantification of focal adhesions lifespan by measuring average time before dissociation in groups of 20 individual focal adhesions revealed that the focal adhesions containing talinS446A persist for much longer (60-80 min) than those containing talinT144A+T150A (30-40 min).
Expression of talinT144A+T150A or talinS446A have contrasting effects on cell adhesion and migration
To avoid interference from non-expressing cells and identify the true effect of site-specific high stoichiometry phosphorylation on cell adhesion and migration, homogenous cells expressing wild-type or mutant talin and collected by flow cytometry were used in cell adhesion and transwell assays.
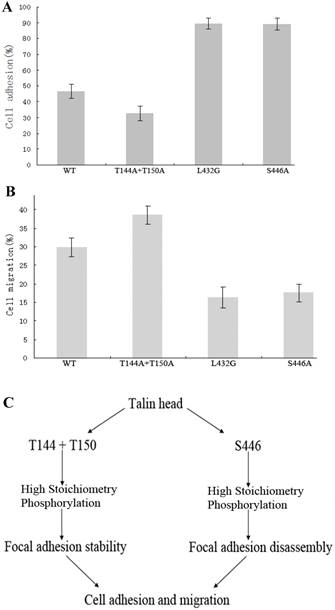
The results show that cell adhesion and growth were greatly varied in cells expressing talinT144A+T150A or talinS446A after the same number of both cell types were plated on fibronectin-coated MatTek dishes for 4 h. The cells expressing talinS446A adhered well, and became larger and elongated, whereas most of the cells expressing talinT144A+T150A did not adhere during this period, and the cells that did adhere were smaller and round. 76.9% (100/130) of the cells expressing talinS446A were elongated, compared to 14.3% (9/63) of the cells expressing talinT144A+T150A (Fig. S.5). The rate of cell adhesion in the cells expressing talinT144A+T150A or talinS446A as determined from cell adhesion assays was 32.48 and 88.97%, respectively (Fig.4A). In a transwell assay with noninvasive CHO-K1 cells, the proportion of cells that migrated to the bottom chamber were 38.54% and 17.58% respectively (Fig.4B). These results reveal that talinT144A+T150A stimulates cell migration by inhibiting cell adhesion while talinS446A inhibits cell migration by stimulating cell adhesion.
Immuno-cytochemistry and live fluorescence microscopy. (A), Talin-EGFP and paxillin-TRITC were co-localized to focal adhesions in cells transfected with talinT144A+T150A or talinS446A. Scale bars, 10μm. (B), At 18h after transfection, individual focal adhesions were imaged at 5-min intervals for a 60-min period. Arrowheads point to disassembling focal adhesions. Scale bars, 20μm.
Discussion
Focal adhesion assembly refers to the incorporation of adhesion components into adhesions, whereas disassembly is the detachment of these proteins from adhesions [2]. Focal adhesion assembly and disassembly occur in a common cytoplasm, but it is not simply a common reversible mechanism [1,17]. Focal adhesion disassembly enables adhesion release, rear retraction and cell body translocation, but the molecular mechanism remains to be elucidated. It has been clear that disassembly of other adhesion components by calpain depends on the ability of calpain to cleave talin [11], suggesting that calpain cleavage of talin plays a pivotal role in regulating calpain-mediated focal adhesion disassembly.
Our data demonstrate that calpain-mediated cleavage of the talin head domain is sufficient to promote focal adhesion disassembly, verifying that the FERM domain has an essential role in performing the functions of talin [18, 19]. In the study, the transfection efficiency of wild-type talin and the mutants were almost the same. The protein concentrations were equal between mutants and the cells assayed were homogenous expressing cells obtained by flow cytometry. Wild-type talin or talinL432G were used as positive and negative controls for talin cleavage by calpain. Expression of two site-specific phosphorylation-deficient mutants, talinT144A+T150A or talinS446A, regulates calpain-mediated cleavage of talin and focal adhesion disassembly in a contrasting manner: talinT144A+T150A triggers calpain cleavage, 30% more than wild-type, but talinS446A almost completely abolishes cleavage like the talinL432G mutant. Further study is needed to investigate how the site-specific high stoichiometry phosphorylation of talin leads to up- or down-regulation of calpain activation through active ERK and influences cell adhesion and migration. Taken together, our findings define and highlight the prominent role of talin site-specific high stoichiometry phosphorylation at T144/T150 and S446 in regulating calpain-mediated cleavage of talin and focal adhesion disassembly, thus controlling adhesion stability, cell adhesion and ultimately, cell migration (Fig.4C).
Expression of two site-specific phosphorylation-deficient mutants, talinT144A+T150A or talinS446A have contrasting effects on cell adhesion and migration. (A), The vertical axis shows the rate of cell adhesion (P<0.01). (B), The vertical axis shows the cells that migrated to the bottom chamber (P<0.01). (C), Proposed mechanism whereby talin site-specific high stoichiometry phosphorylation has opposing effects on focal adhesion stability, cell adhesion and migration.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank Dr. Chunyi Tong for the help in the work of flow cytometry, and the support of the colleagues of the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486). This work was supported in funds by the grants from National Natural Science Foundation of China (No.30871269 and No. 81172051), Hunan Provincial Natural Science Foundation of China (No. 10JJ2014) to X.G.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells — over and over and over again. Nature Cell Biol. 2002;4(4):E97-E100
2. Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biol. 2004;6(2):154-161
3. Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nature Rev. Mol. Cell Biol. 2010;11(9):633-643
4. Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nature Cell Biol. 2008;10(9):1039-1050
5. Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nature Cell Biol. 2007;9(8):858-867
6. Zaidel-Bar Z, Geiger B. The switchable integrin adhesome. J. Cell Sci. 2010;123(9):1385-1388
7. Lawson C, Lim ST, Uryu S, Chen X, Calderwood DA, Schlaepfer D. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J.Cell Biol. 2012;196(2):223-232
8. Priddle H, Hemmings L, Monkley S, Woods A, Patel B, Sutton D, Dunn GA, Zicha D, Critchley DR. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol. 1998;142(4):1121-1133
9. Glading A, Lauffenburger DA, Wells A Cutting to the chase. calpain proteases in cell motility. Trends Cell Biol. 2002;12(1):46-54
10. Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J. Cell Sci. 2005;118(17):3829-3838
11. Franco SJ, Rodgers MA, Benjamin J, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nature Cell Biol. 2004;6(10):977-983
12. Critchley DR, Gingras AR. Talin at a glance. J Cell Sci. 2008;121(9):1345-1347
13. Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nature Cell Biol. 2009;11(5):624-630
14. Ratnikov B, Ptak C, Han J, Shabanowitz J, Hunt DF, Ginsberg MH. Talin phosphorylation sites mapped by mass spectrometry. J.Cell Sci. 2005;118(21):4921-4923
15. Glading A, Chang P, Lauffenburger DA, Wells A. Epidermal growth factor receptor activation of calpain is required for fibroblast motility and occurs via an ERK/MAP kinase signaling pathway. J. Biol. Chem. 2000;275(4):2390-2398
16. Glading A, Bodnar RJ, Reynolds IJ, Shiraha H, Satish L, Potter DA, Blair HC, Wells A. Epidermal growth factor activates m-calpain (calpain II), at least in part, by extracellular signal-regulated kinase-mediated phosphorylation. Mol. Cell Biol. 2004;24(6):2499-2512
17. Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nature Cell Biol. 2005;7(7):581-590
18. Calderwood DA, Zent R, Grant DJG, Hynes RO, Ginsberg MH. The talin head domain binds to integrin subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 1999;274(40):28071-28074
19. Paolo GD, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, Camilli PD. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 " by the FERM domain of talin. Nature. 2002;420(6911):85-89
Author contact
![]() Corresponding authors: Rushi Liu: liurushiedu.cn, Xiangrong Guo: xrguoedu.cn.
Corresponding authors: Rushi Liu: liurushiedu.cn, Xiangrong Guo: xrguoedu.cn.

 Global reach, higher impact
Global reach, higher impact