Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(12):1680-1693. doi:10.7150/jca.15494 This issue Cite
Research Paper
GSTT1 Null Genotype Significantly Increases the Susceptibility to Urinary System Cancer: Evidences from 63,876 Subjects
1. Department of Cardiovascular Surgery of the First Affiliated Hospital& Institute for Cardiovascular Science, Soochow University, Suzhou, Jiangsu, China
2. Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Department of Experimental Research, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, Guangdong, China.
3. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China
4. Department of Internal Medicine, Harbin Medical University, Harbin 150081, Heilongjiang, China
5. Department of Urinary Surgery, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China
#These authors contributed equally to this work.
Received 2016-3-9; Accepted 2016-5-18; Published 2016-7-26
Abstract
GSTT1 gene plays an important role in detoxification and clearance of reactive oxygen species(ROS). A null variant in this gene has been demonstrated to confer cancer susceptibility. Although many studies have demonstrated the association between GSTT1 null polymorphism and urinary system cancer susceptibility, several publications reported opposite conclusions. For better understanding the effects of this polymorphism on the risk of urinary system cancer, a updated meta-analysis was performed with a total of 26,666 cases and 37,210 controls extracted from 117 studies, by following the latest meta-analysis guidelines (PRISMA). The results suggested that the GSTT1 null genotype was significantly associated with an increased risk of urinary system cancer (OR=1.13, 95%CI=1.05-1.22). Furthermore, stratified analyses by the type of cancer, ethnicity, source of control and quality score presented a significantly increased risk associated with GSTT1 null genotype in bladder and prostate cancer subgroup, Caucasians and Indians subgroup, population-based(PB) subgroup, medium quality and low quality subgroup. Overall, our meta-analysis suggested that GSTT1 null genotype is a potential cancer susceptibility variant. Well-designed and large-cohort studies are needed to confirm the association between GSTT1 null genotype and urinary system cancer risk.
Keywords: GSTT1, meta-analysis, oxidative stress, polymorphism, susceptibility, urinary system cancer
Introduction
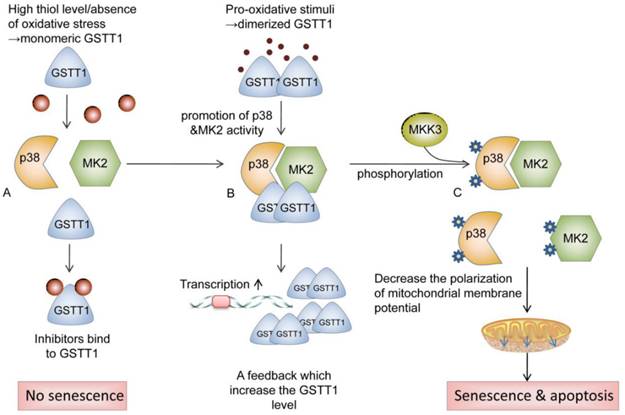
Cancer is one of the leading causes of morbidity and mortality worldwide, with approximately 14 million new cases per year according to the World Cancer Report 2014[1]. Exposure to carcinogens and harmful metabolites is one of etiologic factors for urinary system cancer[2, 3]. Cytosolic glutathione S-transferases (GSTs) are members of a phase II metabolic isozymes super family, which protect cells against electrophilic damage by catalyzing the conjugation between carcinogens and glutathione. This catalytic reaction decreases the toxicity of carcinogens and facilitates their excretion from the body. Additionally, certain GSTs also protect tissues against injury by attenuating oxidative stress or modulating the signaling pathways of cell proliferation, apoptosis and differentiation[4, 5]. The glutathione S-transferase theta 1 (GSTT1) is located on the long arm of chromosome 22 (22q11.23)[6]. Some studies reported that GSTT1 protein was involved in catalyzing the conjugation of ethylene oxide and halogenated metabolites[7]. Moreover, GSTT1 is also involved in the activation of p38/MAPKAP kinase 2(MK2) signal pathway. GSTT1 may prevent cells from tumorigenesis via promoting p38/MK2-mediated apoptosis and senescence. High thiol levels and the absence of oxidative stress keep the GSTT1 in monomeric form. In the presence of pro-oxidative stimuli, dimerized GSTT1 bind to p38 and MK2 to facilitate the activation of these kinases, which in turn elevate the expression of GSTT1. After interacting with MKK3, Phosphorylated p38 and MK2 reduce the polarization of mitochondrial membrane potential, which activates apoptosis and senescence[8] (as shown in Fig 1). Dysfunction of GSTs has been implicated in the development of bladder cancer, prostate cancer, or renal cell carcinoma, due to the defective detoxification capacity [9, 10]. The GSTT1 polymorphism (GSTT1 presence and GSTT1 null) has been proved in several studies to be associated with an increased risk of urinary system cancer[11-13]. However, the results from different groups were to some extent divergent[14, 15], which might be owing to the limitations of individual studies, including the heterogeneity in the sample source, study design or disagreements among the investigations. A comprehensive meta-analysis might provide sufficient statistical power to draw a more reliable conclusion on the association between GSTT1 null genotype and urinary system cancer risk. Therefore, we conducted the current meta-analysis using genotype data from 117 eligible studies to assess such association.
Materials and Methods
The strategy of literature screening, data collection, and studies inclusion in this meta-analysis was according to the latest meta-analysis guidelines-Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)[16].
Search Strategy
A systematic literature retrieval was performed using the following key words: "glutathione S-transferase T1 or GSTT1", "variant or variation or polymorphism ” and “bladder or prostate or renal cell carcinoma or urinary system cancer” through PubMed and EMBASE databases without any restriction on language (the last search update was performed on Nov 24, 2015). We further searched articles from Chinese Biomedical (CBM) and China National Knowledge Infrastructure (CNKI) databases to ensure more studies were include in this research. We also searched the references of the relevant literatures manually. All the eligible studies were listed in the reference.
Schematic of the potential roles of GSTT1 in preventing cells from tumorigenesis. GSTT1 prevent cells from tumorigenesis via promoting p38/MK2 mediated apoptosis and senescence. High thiol levels and the absence of oxidative stress keep the GSTT1 in monomeric form. (A) Monomeric GSTT1 dissociates from p38 and MK2, and binds to its inhibitor. (B) In the presence of pro-oxidative stimuli, dimerized GSTT1 binds to p38 and MK2, promoting the activity of these kinases, which in turn elevate the expression of GSTT1. (C) After interacting with MKK3, Phosphorylated p38 and MK2 initiate the reduction of polarization of mitochondrial membrane potential, which activates apoptosis and senescence. Abbreviations: GSTT1, glutathione S‑transferase theta 1; MK2, mitogen-activated protein kinase-activated protein kinase 2; MKK3, mitogen-activated protein kinase 3.
Inclusion Criteria
The inclusion criteria were as follows: (a) evaluated the association between GSTT1 null genotype and urinary system cancer susceptibility; (b) only case-control studies were taken into account; (c) with details for calculating odds ratios (ORs) and corresponding 95% confidence intervals (CIs); (d) presented in English or Chinese. If the study had overlapping subjects with others, only the most complete and latest one was included.
The exclusion criteria were: (a) case only studies; (b)case reports; (c) conference abstracts; review articles; (d) non-cancer subjects only studies; (e)duplicate publications.
Data Extraction
Two of the authors (Y.W. and J.H.) independently extracted the information from all the eligible studies. While there was a disagreement between them, they made a discussion about the issue. If they could not reach a consensus, another author would participate to resolve the dispute, and authors would vote to make a final decision. Data extracted from each study were as follows: the surname of first author, year of publication, country and ethnicity of the subjects, case number and control number with the GSTT1 null, number of total subjects, source of control, genotyping method, and case confirmation approach. The types of cancer were consist of bladder cancer, prostate cancer and renal cell carcinoma. Ethnicities were categorized into Africans, Asians, Caucasians, Indians and Mixed (containing more than one ethnic group). The sources of control were classified as population-based(PB) and hospital-based(HB). The minimum number of patients was not defined for inclusion in this meta-analysis. Publications were divided into different categories by the type of cancer, ethnicity, source of control or quality score.
Quality Score Assessment
The quality of the study was independently assessed by two investigators (Y.W. and J.H.) via the quality assessment criteria (Table S1)[17,18]. The evaluation factors were as follows: representativeness of case, representativeness of control, ascertainment of urinary system cancer, control selection, genotyping examination, and total sample size. Each research was evaluated on a scale from 0-15. If the score of one study was ≤5, it was classified as "low quality"; 5<scores≤10, "medium quality"; scores>10, it was categorized as "high quality".
Statistical Methods
Crude ORs and the corresponding 95% CIs were used to evaluate the strength of association between GSTT1 null genotype and urinary system cancer susceptibility. The statistical significance of an association was determined by Z test. The between-studies heterogeneity was assessed and qualified using cochran Q-test and I2 statistic. For Q test, when P<0.10, the heterogeneity was considered statistically significant. I2 statistic represented the proportion of variants caused by heterogeneity across studies but not sampling error. In this meta-analysis, if heterogeneity was P>0.10 for Q-test, a fixed-effect model was applied; otherwise, a random-effect model was conducted. The leave-one-out sensitivity analysis was performed by excluding a study in the meta-analysis each time and recalculating ORs. Moreover, the publication bias was detected by Begg's and Egger's linear regression tests. The symmetry of the funnel plot suggested null publication bias; otherwise, the publication bias was present. Finally, a meta-regression was conducted to detect the main heterogeneity source among the studies included in our analysis.
All statistical analyses were performed using the meta software package of STATA version 12.0 (STATA Corporation, College Station, TX). All the P values were two-side tests. P<0.05 was considered statistically significant.
Results
Study characteristics
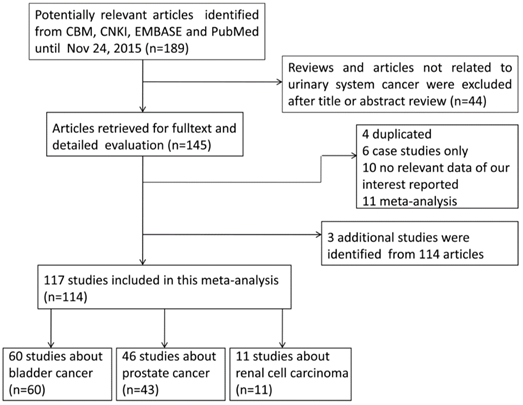
A total of 189 relative articles were obtained from CBM, CNKI, EMBASE and PubMed using search terms according to the search strategy in methods section. After screening the title or abstract, 44 articles that were either not related to urinary system cancer or reviews were excluded. The remaining 145 publications were carefully evaluated according to the eligibility criteria. Among them, 31 articles were removed because they were duplicated studies (n=4), not case-control studies (n=6), useless in GSTT1 genotyping data extraction (n=10) or meta-analysis (n=11). Therefore, 114 publications were subjected to further analysis. Among them, 3 additional studies were extracted from publications containing two studies[9, 17, 18]. For example, Steinhoff and his colleagues investigated the association of GSTT1 null genotype with both bladder and prostate cancer risk[9]. Finally, 117 eligible case-control studies with 26,666 cases and 37,210 controls meeting the criteria were included in our analysis[9-15, 17-89] [90-123]. The flowchart of the identification of eligible studies was shown in Fig 2.
Table 1 summarized all the eligible studies and main characteristics. The sample size ranged from 46 to 4,537 in the 117 studies, including 60, 46 and 11 studies focusing on bladder cancer, prostate cancer, and renal cell carcinoma, respectively. According to the race, these studies could be classified into 5 subgroups, Africans(68 studies), Asians(20 studies) , Caucasians(5 studies), Indians(10 studies) and mixed populations(14 studies). The majority of the patients(94%) with urinary system cancer were histologically confirmed, and 6% of the patients were determined by the medical records. Controls were frequency-matched to patients by age, gender and ethnicity in most of studies. Of all the studies, 49 studies were PB, while 68 were HB. Additionally, 19 studies were considered as low quality(score≤5), 79 studies were considered as medium quality(5<score≤10), and 19 studies were considered as high quality(score>10).
Flowchart of included studies for the meta-analysis of the association between GSTT1 null genotype and urinary system cancer risk.
Characteristics of the 117 studies included in the meta-analysis for the association between GSTT1 null genotype and risk of urinary system cancer
| Surname | Year | Country | Ethnicity | Type of cancer | Sourceof control | Genotyping method | Cases | Controls | MAF(T) | Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Kempkes | 1996 | Germany | Caucasians | Bladder cancer | PB | PCR | 113 | 170 | 0.18 | 6 |
| Bruning | 1997 | Germany | Caucasians | Renal cell caicinoma | PB | PCR | 45 | 48 | 0.23 | 6 |
| Salagovic | 1998 | Slovak | Caucasians | Bladder cancer | PB | PCR | 67 | 248 | 0.17 | 5 |
| Kim | 1998 | Korea | Asians | Bladder cancer | HB | PCR | 67 | 67 | 0.43 | 6 |
| Abdel-Rahman | 1998 | Egypt | Caucasians | Bladder cancer | PB | PCR | 37 | 34 | 0.15 | 8 |
| Katoh | 1998 | Japan | Asians | Bladder cancer | PB | PCR | 112 | 112 | 0.47 | 9 |
| Salagovic | 1999 | Slovak | Caucasians | Bladder cancer | PB | PCR | 76 | 248 | 0.17 | 5 |
| Autrup | 1999 | Denmark | Caucasians | Prostate cancer | PB | PCR | 153 | 288 | 0.15 | 7 |
| Lee | 1999 | Korea | Asians | Bladder cancer | HB | PCR | 158 | 131 | 0.50 | 7 |
| Rebbeck | 1999 | USA | Mixed | Prostate cancer | PB | PCR | 232 | 231 | 0.69 | 8 |
| Longuemaux | 1999 | France | Caucasians | Renal cell caicinoma | HB | PCR | 173 | 211 | 0.19 | 8 |
| Schnakenberg | 2000 | Germany | Caucasians | Bladder cancer | PB | PCR | 157 | 223 | 0.22 | 5 |
| Steinhoff | 2000 | Germany | Caucasians | Bladder cancer | HB | PCR | 135 | 127 | 0.13 | 5 |
| Steinhoff | 2000 | Germany | Caucasians | Prostate cancer | HB | PCR | 91 | 127 | 0.13 | 5 |
| Peluso | 2000 | Italy | Caucasians | Bladder cancer | HB | PCR | 122 | 54 | 0.11 | 6 |
| Kim | 2000 | Korea | Asians | Bladder cancer | HB | PCR | 112 | 220 | 0.46 | 7 |
| Kelada | 2000 | USA | Mixed | Prostate cancer | HB | PCR | 256 | 469 | 0.33 | 7 |
| Sweeney | 2000 | USA | Mixed | Renal cell caicinoma | PB | PCR | 126 | 504 | 0.18 | 9 |
| Murata | 2001 | Japan | Asians | Prostate cancer | PB | PCR | 115 | 200 | 0.48 | 6 |
| Toruner | 2001 | Turkey | Caucasians | Bladder cancer | HB | PCR | 121 | 121 | 0.17 | 6 |
| Gsur | 2001 | Austria | Caucasians | Prostate cancer | HB | PCR | 166 | 166 | 0.20 | 8 |
| Kote-Jarai | 2001 | UK | Caucasians | Prostate cancer | PB | PCR | 273 | 278 | 0.24 | 10 |
| Ma | 2002 | China | Asians | Bladder cancer | PB | PCR | 61 | 182 | 0.48 | 5 |
| Lee | 2002 | Korea | Asians | Bladder cancer | HB | PCR | 232 | 165 | 0.52 | 6 |
| Giannakopoulos | 2002 | Greece | Caucasians | Bladder cancer | HB | PCR | 89 | 147 | 0.11 | 6 |
| Beer | 2002 | USA | Caucasians | Prostate cancer | PB | PCR | 111 | 146 | 0.23 | 7 |
| Kim | 2002 | Korea | Asians | Bladder cancer | PB | PCR | 216 | 449 | 0.51 | 10 |
| Jong Jeong | 2003 | Korea | Asians | Bladder cancer | HB | PCR | 126 | 204 | 0.55 | 6 |
| Nakazato | 2003 | Japan | Asians | Prostate cancer | PB | PCR | 81 | 105 | 0.42 | 7 |
| Buzio | 2003 | Italy | Caucasians | Renal cell caicinoma | HB | PCR | 100 | 200 | 0.18 | 8 |
| Kidd | 2003 | Finland | Caucasians | Prostate cancer | PB | PCR | 202 | 189 | 0.15 | 10 |
| Gago-Dominguez | 2003 | USA | Mixed | Bladder cancer | PB | PCR | 196 | 176 | 0.19 | 10 |
| Srivastava | 2004 | India | Indians | Bladder cancer | HB | PCR | 106 | 182 | 0.16 | 4 |
| Mittal | 2004 | India | Indians | Prostate cancer | PB | PCR | 103 | 117 | 0.11 | 5 |
| Chen | 2004 | China | Asians | Bladder cancer | PB | PCR | 62 | 81 | 0.63 | 6 |
| Hung | 2004 | Italy | Caucasians | Bladder cancer | HB | PCR | 201 | 214 | 0.15 | 6 |
| Sanyal | 2004 | Sweden | Caucasians | Bladder cancer | PB | PCR | 270 | 122 | 0.10 | 8 |
| Medeiros | 2004 | Portugal | Caucasians | Prostate cancer | PB | PCR | 145 | 184 | 0.24 | 9 |
| Joseph | 2004 | USA | Caucasians | Prostate cancer | PB | PCR-RFLP | 177 | 265 | 0.23 | 9 |
| Moore | 2004 | Argentina | Caucasians | Bladder cancer | PB | PCR | 106 | 109 | 0.11 | 10 |
| Wang | 2005 | China | Asians | Prostate cancer | HB | PCR | 81 | 90 | 0.53 | 3 |
| Sobti | 2005 | India | Indians | Bladder cancer | PB | PCR | 100 | 76 | 0.14 | 5 |
| Golka | 2005 | Germany | Caucasians | Bladder cancer | HB | PCR | 136 | 163 | 0.23 | 5 |
| Saad | 2005 | Egypt | Caucasians | Bladder cancer | PB | PCR | 72 | 81 | 0.17 | 6 |
| Komiya | 2005 | Japan | Asians | Prostate cancer | PB | PCR | 186 | 288 | 0.52 | 8 |
| Srivastava | 2005 | India | Indians | Bladder cancer | PB | PCR | 106 | 370 | 0.21 | 8 |
| Srivastava | 2005 | India | Indians | Prostate cancer | PB | PCR | 127 | 144 | 0.20 | 8 |
| Kim | 2005 | Korea | Asians | Bladder cancer | HB | PCR | 153 | 153 | 0.58 | 8 |
| Broberg | 2005 | Sweden | Caucasians | Bladder cancer | PB | PCR | 61 | 154 | 0.14 | 9 |
| Karagas | 2005 | USA | Mixed | Bladder cancer | PB | PCR | 354 | 541 | 0.15 | 10 |
| Caceres | 2005 | Chile | Mixed | Prostate cancer | PB | PCR-RFLP | 100 | 129 | 0.11 | 10 |
| Garcia-Closas | 2005 | Spain | Caucasians | Bladder cancer | HB | TaqMan assay | 1146 | 1147 | 0.22 | 11 |
| Nam | 2005 | Canada | Mixed | Prostate cancer | PB | PCR-RFLP | 996 | 1092 | 0.23 | 13 |
| Ouerhani | 2006 | Tunisia | Africans | Bladder cancer | PB | PCR | 62 | 79 | 0.44 | 3 |
| Kogevinas | 2006 | Spain | Caucasians | Bladder cancer | HB | TaqMan assay | 99 | 91 | 0.19 | 7 |
| Silig | 2006 | Turkey | Caucasians | Prostate cancer | HB | PCR | 152 | 169 | 0.18 | 7 |
| Mittal | 2006 | India | Indians | Prostate cancer | HB | PCR | 54 | 105 | 0.29 | 7 |
| Yang | 2006 | China | Asians | Prostate cancer | HB | PCR | 163 | 202 | 0.47 | 9 |
| Nock | 2006 | USA | Caucasians | Prostate cancer | HB | PCR | 438 | 479 | 0.17 | 9 |
| Shao | 2006 | China | Asians | Bladder cancer | PB | PCR | 405 | 389 | 0.50 | 10 |
| Agalliu | 2006 | USA | Africans | Prostate cancer | PB | PCR | 31 | 15 | 0.47 | 11 |
| McGrath | 2006 | USA | Mixed | Bladder cancer | PB | PCR | 191 | 924 | 0.16 | 13 |
| Agalliu | 2006 | USA | Caucasians | Prostate cancer | PB | PCR | 558 | 522 | 0.17 | 14 |
| Lindstrom | 2006 | Sweden | Caucasians | Prostate cancer | PB | PCR | 1299 | 728 | 0.15 | 14 |
| Cengiz | 2007 | Turkey | Caucasians | Bladder cancer | HB | PCR | 51 | 53 | 0.21 | 5 |
| Cunningham | 2007 | USA | Mixed | Prostate cancer | PB | PCR | 499 | 493 | 0.43 | 8 |
| Wiesenbutter | 2007 | Germany | Caucasians | Renal cell caicinoma | HB | PCR | 98 | 324 | 0.18 | 8 |
| Mallick | 2007 | Guadeloupe | Mixed | Prostate cancer | HB | PCR | 134 | 134 | 0.37 | 8 |
| Kellen | 2007 | Belgium | Caucasians | Bladder cancer | PB | PCR | 194 | 380 | 0.16 | 9 |
| Zhao | 2007 | USA | Mixed | Bladder cancer | PB | PCR | 623 | 634 | 0.18 | 12 |
| Moore | 2007 | Europe | Caucasians | Renal cell caicinoma | HB | PCR | 861 | 1199 | 0.17 | 12 |
| Lima | 2008 | Brasil | Caucasians | Prostate cancer | HB | PCR | 125 | 100 | 0.22 | 6 |
| Covolo | 2008 | Italy | Caucasians | Bladder cancer | PB | PCR | 197 | 211 | 0.16 | 7 |
| Davydova | 2008 | Russia | Caucasians | Prostate cancer | PB | PCR | 61 | 100 | 0.22 | 7 |
| Song | 2008 | China | Asians | Bladder cancer | HB | PCR | 108 | 112 | 0.52 | 7 |
| Grando | 2008 | Brasil | Mixed | Bladder cancer | PB | PCR | 100 | 100 | 0.37 | 8 |
| Karami | 2008 | Europe | Caucasians | Renal cell caicinoma | HB | PCR | 628 | 913 | 0.18 | 9 |
| Yuan | 2008 | USA | Caucasians | Bladder cancer | PB | PCR | 658 | 680 | 0.18 | 12 |
| Altayli | 2009 | Turkey | Caucasians | Bladder cancer | HB | PCR | 135 | 128 | 0.07 | 7 |
| Sivonova | 2009 | Slovakia | Caucasians | Prostate cancer | PB | PCR | 129 | 228 | 0.20 | 8 |
| Song | 2009 | China | Asians | Bladder cancer | HB | PCR | 208 | 212 | 0.50 | 8 |
| Lavender | 2009 | USA | Africans | Prostate cancer | PB | TaqMan assay | 189 | 584 | 0.17 | 10 |
| Souiden | 2010 | Tunisia | Caucasians | Prostate cancer | HB | PCR | 110 | 122 | 0.15 | 6 |
| Coric | 2010 | Serbia | Caucasians | Renal cell caicinoma | HB | PCR | 76 | 182 | 0.29 | 8 |
| De Martino | 2010 | Austria | Caucasians | Renal cell caicinoma | HB | PCR | 147 | 112 | 0.21 | 8 |
| Steinbrecher | 2010 | Germany | Caucasians | Prostate cancer | PB | RT-PCR | 248 | 492 | 0.16 | 12 |
| Cantor | 2010 | Spain | Caucasians | Bladder cancer | HB | TaqMan assay | 678 | 710 | 0.23 | 12 |
| Moore | 2010 | USA | Caucasians | Bladder cancer | PB | Melt curve/copy number assays | 1004 | 1179 | 0.20 | 14 |
| Rouissi | 2011 | Tunisia | Africans | Bladder cancer | PB | PCR | 125 | 125 | 0.30 | 4 |
| Kwon | 2011 | Korea | Asians | Prostate cancer | PB | PCR | 166 | 327 | 0.50 | 6 |
| Thakur | 2011 | India | Indians | Prostate cancer | PB | PCR | 150 | 172 | 0.13 | 6 |
| Kumar | 2011 | India | Caucasians | Prostate cancer | HB | PCR | 57 | 46 | 0.48 | 6 |
| Rodrigues | 2011 | Brasil | Caucasians | Prostate cancer | PB | PCR | 154 | 154 | 0.26 | 7 |
| Salinas-Sanchez | 2011 | Spain | Caucasians | Bladder cancer | HB | PCR | 190 | 163 | 0.15 | 7 |
| Ashtiani | 2011 | Iran | Caucasians | Prostate cancer | HB | PCR | 110 | 100 | 0.47 | 7 |
| Safarinejad | 2011 | Iran | Caucasians | Prostate cancer | PB | PCR | 168 | 336 | 0.21 | 8 |
| Safarinejad | 2011 | Iran | Caucasians | Bladder cancer | PB | PCR | 332 | 166 | 0.21 | 9 |
| Norskov | 2011 | Denmark | Caucasians | Prostate cancer | PB | RT-PCR | 128 | 4409 | 0.15 | 10 |
| Koutros | 2011 | USA | Caucasians | Bladder cancer | PB | Melt curve/copy number assays | 215 | 289 | 0.20 | 13 |
| Goerlitz | 2011 | Egypt | Caucasians | Bladder cancer | PB | TaqMan assay | 617 | 620 | 0.25 | 13 |
| Henriquez-Hernandez | 2012 | Span | Caucasians | Bladder cancer | HB | PCR | 90 | 81 | 0.49 | 4 |
| Ovsiannikov | 2012 | Germany | Caucasians | Bladder cancer | HB | PCR | 196 | 235 | 0.20 | 4 |
| Salinas sanchez | 2012 | Spain | Caucasians | Renal cell caicinoma | HB | PCR | 132 | 163 | 0.15 | 6 |
| Hemelrijck | 2012 | Germany | Caucasians | Prostate cancer | PB | RT-PCR | 203 | 360 | 0.18 | 10 |
| Ahmad | 2012 | India | Indians | Renal cell caicinoma | PB | PCR | 196 | 250 | 0.42 | 11 |
| Catsburg | 2012 | USA | Mixed | Prostate cancer | PB | TaqMan assay | 909 | 736 | 0.21 | 12 |
| Catsburg | 2012 | USA | Mixed | Prostate cancer | PB | TaqMan assay | 491 | 736 | 0.21 | 12 |
| Lesseur | 2012 | USA | Caucasians | Bladder cancer | PB | TaqMan assay | 557 | 810 | 0.16 | 14 |
| Sharma | 2013 | India | Indians | Bladder cancer | HB | PCR | 50 | 50 | 0.36 | 4 |
| Choubey | 2013 | India | Indians | Prostate cancer | HB | PCR | 51 | 134 | 0.13 | 5 |
| Kang | 2013 | Korea | Asians | Bladder cancer | HB | PCR | 110 | 220 | 0.58 | 7 |
| Berber | 2013 | Turkey | Caucasians | Bladder cancer | HB | PCR | 114 | 114 | 0.14 | 7 |
| Matic | 2013 | Serbia | Caucasians | Bladder cancer | HB | PCR | 201 | 122 | 0.28 | 8 |
| Ceylan | 2014 | Turkey | Caucasians | Bladder cancer | PB | PCR | 65 | 70 | 0.13 | 5 |
| Sa | 2014 | Brazil | Caucasians | Prostate cancer | PB | PCR | 196 | 208 | 0.23 | 7 |
| Reszka | 2014 | Poland | Caucasians | Bladder cancer | PB | RT-PCR | 242 | 365 | 0.21 | 9 |
| Emeville | 2014 | Guadeloupe | Africans | Prostate cancer | PB | RT-PCR | 629 | 622 | 0.31 | 13 |
Meta-analysis of the association between GSTT1 null genotype and urinary system cancer risk
| Variables | No. of studies | Sample size Case/control | OR (95% CI) | P OR | I2 (%) | P heterogeneity |
|---|---|---|---|---|---|---|
| Alla | 117 | 26666/37210 | 1.13 (1.05-1.22) | 0.001 | 65.0 | 0.000 |
| Type of cancer | ||||||
| BC | 60 | 12887/15783 | 1.13 (1.03-1.24) | 0.012 | 55.5 | 0.000 |
| PC | 46 | 11197/17321 | 1.14 (1.01-1.29) | 0.042 | 72.6 | 0.000 |
| RCC | 11 | 2582/4106 | 1.10 (0.86-1.41) | 0.436 | 66.5 | 0.001 |
| Ethnicity | ||||||
| Caucasian | 68 | 16458/23377 | 1.16 (1.05-1.27) | 0.002 | 59.7 | 0.000 |
| Asian | 20 | 2922/3909 | 1.04 (0.90-1.19) | 0.614 | 44.1 | 0.019 |
| African | 5 | 1036/1425 | 0.80 (0.63-1.02) | 0.066 | 19.5 | 0.290 |
| Indians | 10 | 1043/1600 | 2.05 (1.70-2.48) | 0.000 | 1.7 | 0.423 |
| Mixed | 14 | 5207/6899 | 0.95 (0.80-1.14) | 0.596 | 71.0 | 0.000 |
| Source of control | ||||||
| HB | 49 | 9337/11133 | 1.10 (0.99-1.23) | 0.070 | 53.3 | 0.000 |
| PB | 68 | 17329/26077 | 1.15 (1.04-1.27) | 0.005 | 70.7 | 0.000 |
| Quality score | ||||||
| ≤5 (low) | 19 | 1803/2610 | 1.43 (1.13-1.79) | 0.002 | 57.6 | 0.001 |
| 5<score≤10 (medium) | 79 | 12956/21215 | 1.15 (1.04-1.26) | 0.005 | 64.0 | 0.000 |
| >10 (high) | 19 | 11907/13385 | 0.96 (0.86-1.07) | 0.438 | 61.7 | 0.000 |
BC, Bladder cancer; RCC, Renal cell carcinoma; PC, Prostate cancer; HB, Hospital based; PB, Population based.
Meta-analysis results
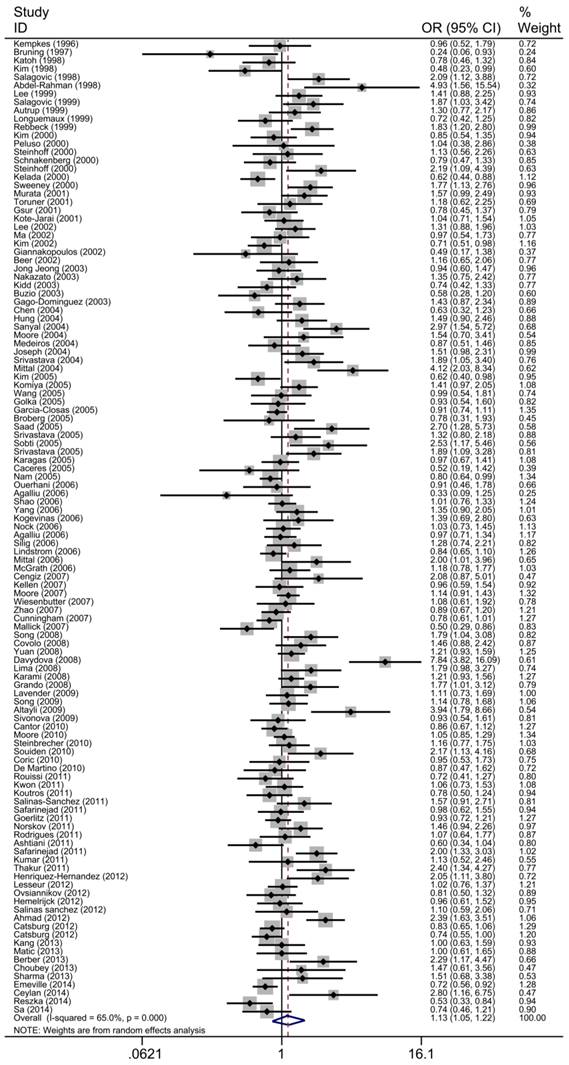
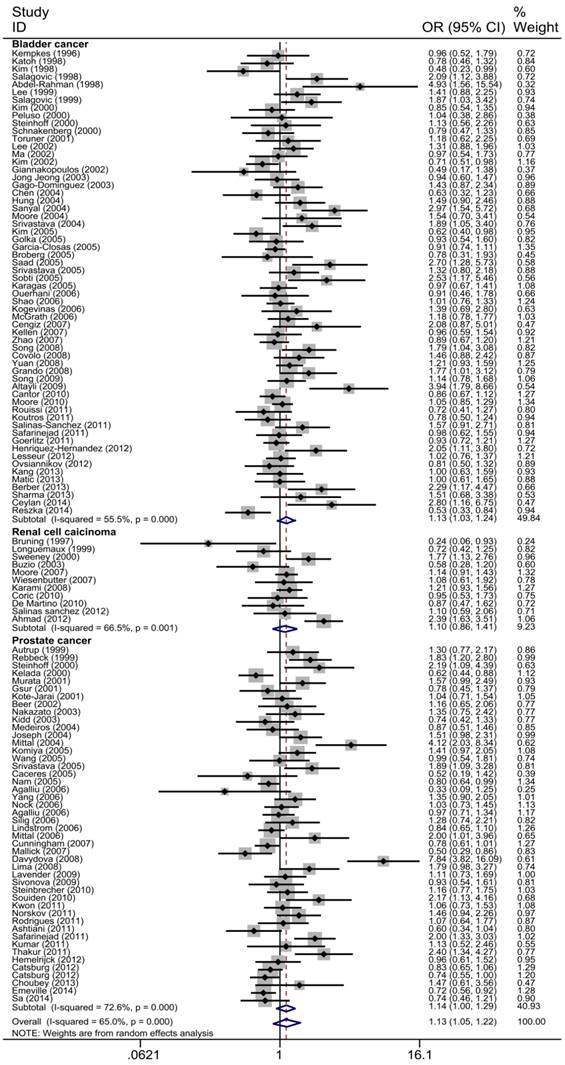
The main results of the meta-analysis were shown in Table 2, Fig 3 and Fig 4. Pooled analysis yielded a significant association between GSTT1 null genotype and urinary system risk (OR=1.13, 95%CI=1.05-1.22). In the stratification analysis by the type of cancer, significantly increased risk of bladder cancer and prostate cancer was observed with GSTT1 null genotype (bladder cancer: OR=1.13, 95%CI=1.03-1.24; prostate cancer: OR=1.14, 95%CI=1.01-1.29). Subgroup analysis by ethnicity revealed a significantly increased urinary system cancer risk for Caucasians and Indians (Caucasians: OR=1.16, 95%CI=1.05-1.27; Indians: OR=2.05, 95%CI=1.700-2.480). By source of control, a statistically significant association was confirmed in PB subgroup (OR=1.15, 95%CI=1.04-1.27). Moreover, when 117 studies were stratified by quality score, the presence of GSTT1 null genotype was associated with urinary system cancer risk in the low quality and medium quality subgroup (score≤5: OR=1.43, 95%CI=1.13-1.79; 5<score≤10: OR=1.15, 95%CI=1.40-1.26).
Q test and I2 statistic were applied to assess the heterogeneity between 117 studies. There were considerable heterogeneities for the GSTT1 genotype and urinary system cancer risk in the overall analysis(P=0.000, I2=65.0%). Thereafter, we applied meta-regression to investigate the sources of heterogeneity by the type of cancer, ethnicity, source of control and quality score. As shown in Table 3, we found the ethnicity and quality of the study contributing to the heterogeneity in this meta-analysis (ethnicity: P=0.003; quality score: P=0.002), but not the type of cancer and source of control (the type of cancer: P=0.703; source of control: P=0.175).
Forest plot of the association between GSTT1 null genotype and urinary system cancer risk. For each study, the estimates of ORs and corresponding 95% CI are plotted with a box and a horizontal line. Diamond, pooled ORs and its 95% CI.
Forest plot of the association between GSTT1 null genotype and urinary system cancer risk which is straitified by the type of cancer. For each study, the estimates of ORs and corresponding 95% CI are plotted with a box and a horizontal line. Diamond, pooled ORs and its 95% CI.
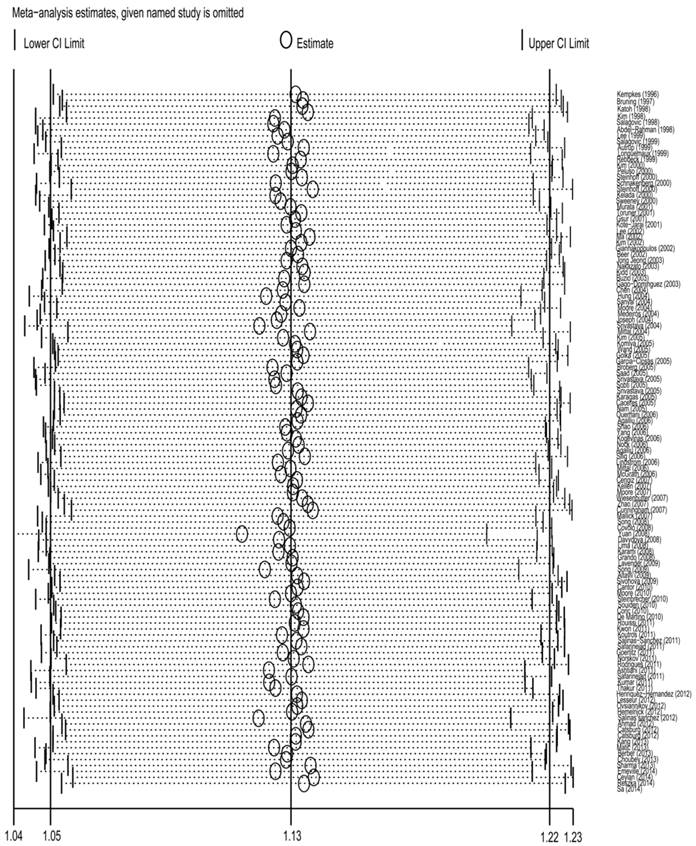
Sensitivity analysis
To detect the effect of individual data-set on the stability of the pooled analysis, we performed the sensitivity analysis by consecutively excluding a study at a time and recalculating ORs. As a result, there was no substantial change in the corresponding pooled ORs, suggesting that our analysis was statistically stable(Fig 5) .
Meta-regression analysis of the main characteristics of the 117 studies
| Study characteristics | Coef. | Std. Err. | t | P | 95%CI | |
|---|---|---|---|---|---|---|
| Type of cancer | -0.02 | 0.06 | -0.38 | 0.703 | -0.14 | 0.09 |
| Ethnicity | 0.14 | 0.05 | 3.03 | 0.003 | 0.05 | 0.12 |
| Source of control | 0.12 | 0.09 | 1.36 | 0.175 | -0.05 | 0.29 |
| Quality score | 0.28 | 0.09 | -3.24 | 0.002 | -0.45 | -0.11 |
Sensitivity analysis of the association between GSTT1 null genotype and urinary system cancer risk. Each point represents the recalculated OR after deleting a separate study.
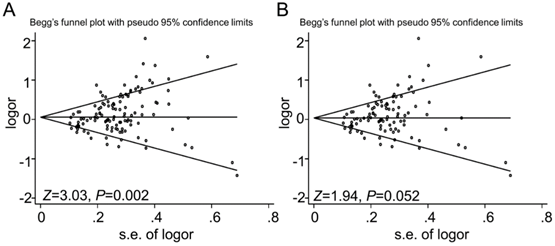
Funnel plot analysis to detect the publication bias for GSTT1 null genotype and urinary system cancer risk. Each point represents a separate study. Funnel plot of the Begg's test for (A) 117 studies, Z=3.03, P=0.002; and (B) for 98 studies after dropping 19 studies with low quality, Z=1.94, P=0.052.
Publication bias
Begg's (Fig 6) test and Egger's test were conducted to assess the publications bias of 117 studies. The results indicated a statistically significant evidence of publication bias in the meta-analysis (P=0.002). Interestingly, when we dropped the low quality studies with score≤5, publication bias disappeared (P=0.052). The data suggested that the bias might be caused by those studies with poor genotyping method or selectively reported positive results.
Discussion
This is a comprehensive meta-analysis for the relationship between the GSTT1 null genotype and the urinary system cancer risk. There were 117 studies with a total of 26,666 cases and 37,210 controls included. Pooled analysis suggested that GSTT1 null genotype was associated with a significantly increased risk of urinary system cancer. In the stratified analysis by the type of cancer , GSTT1 null genotype was shown to significantly increase the risk of bladder cancer and prostate cancer, but not renal cell carcinoma. Moreover, while stratified analysis were conducted by ethnicity, source of control and quality score, significant association were identified among Caucasians and Indians, PB subgroup and high score subgroup. So far, this is the first meta-analysis to evaluate the association between GSTT1 null genotype and urinary system cancer risk with the largest sample size of 63,876 subjects in this area.
The GST super family belongs to phase II detoxifying enzymes. They catalyze the reduction reactions between glutathione and electrophilic substrates, producing stable and harmless compounds. Then these compounds are excreted or compartmentalized[124]. GSTT1 gene is located on the long arm of chromosome 22 (22q11.23) and composed of 5 exons[6]. Apart from the well-established roles in detoxification, each GST has specific function. For instance, GSTT1 is involved in p38/MK2 mediated senescence[8], and conjugation with small molecular-epoxides, which may attenuate the injury from inflammation to cancer progression[125]. There is a null variant allele of GSTT1 gene, which results in an absence of GST enzyme activity and an increased risk to malignancies. Many studies were performed to evaluate the association between GSTT1 null genotype and the susceptibility to urinary system cancer. However, the results from different laboratories were not consistent. Numerous reports suggested that there was an significant association between GSTT1 null genotype and urinary system cancer[9-13, 20, 21], while several other studies failed to detect any association between renal cell carcinoma and GSTT1 null variant[114, 117, 118]. Some meta-analysis were performed to evaluate the association between GSTT1 null genotype and bladder cancer, prostate cancer or renal cell carcinoma, respectively, overall or in a certain population[126-129]. Our analysis included all the eligible studies regardless of the types of cancer like bladder cancer, prostate cancer or renal cell carcinoma. The results of the current meta-analysis confirmed a significant association between GSTT1 null genotype and the susceptibility of urinary system cancer. However, this variant was proven to significantly increase the predisposition to bladder cancer and prostate cancer in stratified analysis by the type of cancer, but not renal cell carcinoma. SNPs in a gene were typically cancer-specific. The discrepancy might owe to the inherent heterogeneity of oncogenic progression in different cancer. The stratified analysis by ethnicity verified that GSTT1 null variant significantly increased the urinary system cancer predisposition in Caucasian and Indian subgroups.
Moreover, a significant increased risk was indicated in PB subgroup, but not in HB subgroup. This was probably due to that the controls collected from hospital were not capable of representing the general population, which might decrease the risk value. Stratification analysis also showed the heterogeneities of between-studies in the overall analysis. We further conducted the meta-regression to assess the factors contributing to the strong heterogeneities among overall analysis. The results suggested that the ethnicity and quality of the study were the main factors associated with the heterogeneities. First, cancer is a complicated disease induced by the gene-environment interaction. People born in different geographic areas has different genetic background and life style. There are different linkage disequilibrium patterns in different populations. Supposed that GSTT1 polymorphism is in close linkage with a causal mutation in Caucasians and Indians but not other ethnicity, the discrepancy can be explained. Besides, the effects from clinical features and live surroundings of different population should be well-contemplated. The second, studies with poor genotyping method and selectively reported positive results also contributed to the heterogeneities. Therefore, more studies are needed to investigate the heterogeneity in further research.
Compared to the former meta-analysis, the current one might possess the following merits: (a) This meta-analysis included the latest studies till Nov, 2015 to make it more comprehensive and minimize the selection bias; (b) The study drew a more complete conclusion about GSTT1 null variant and urinary system cancer risk, but not only bladder cancer or prostate cancer; (c)This investigation conducted a systematical stratification and meta-regression analysis, which provided a more precise conclusion; (d)The sample size and study number were relatively larger, and the statistical power was more potent. However, there were some limitations in this meta-analysis which should be taken into consideration. First, the study number about renal cell carcinoma (n=11) or Africans (n=5) was small, compared to the studies about bladder cancer(n=60) or Caucasians(n=68). Because of the relatively small sample size, the stratified studies regarding renal cell carcinoma and Africans might have a lower statistical power to detect the substantial effects of GSTT1 null genotype on the risk. Second, not all the studies provided enough data about lifestyles (example, smoking, drinking alcohol or tea) or environmental exposure which were reported to be important for the development of urinary system cancer. Third, the selection bias was exist. When we dropped the low quality studies with score≤5, publication bias was not present. There might be a language bias, inflated evaluations or selectively reported positive result in studies of low quality.
To summarize, this systematic meta-analysis regarding the relationship between GSTT1 null genotype and urinary system cancer indicated that this variant significantly increased the risk of urinary system cancer. Moreover, the significant association was found in bladder cancer, prostate, Caucasians, Indians, PB subgroup, but not in renal cell carcinoma, Africans, Asians, Mixed and HB subgroup. The well-designed, large-cohort and multi-center studies are needed to present more rigorous data to confirm our findings.
Supplementary Material
Table S1.
Acknowledgements
This work is supported by the National Natural Science Foundation of China (No.81402090 to Y.W.), the Colleges and Universities Natural Science Foundation of Jiangsu Province (General Program, 15KJB180017 to W.L.) and the Suzhou Science and Technology Program (SYS201540 to W.L.).
Author contribution
All authors contributed significantly to this work, Y.W., J.H., T.M. and Z.S. performed the research design and data collection; W.L. and H.S. assessed the studies quality and conducted the stratified analysis; H.S., T.M. and F.L. did the sensitive analysis and publication bias test. Y.W. and J.H. wrote the paper and prepared Figure 1-6 and Table 1-3. All authors reviewed the manuscript. Moreover, all authors confirmed the final edition.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stewart BW, Wild CP. World Cancer Report 2014. Lyon, France: International Agency for Research on Cancer. 2014
2. Moore LE, Wilson RT, Campleman SL. Lifestyle factors, exposures, genetic susceptibility, and renal cell cancer risk: a review. Cancer Invest. 2005;23:240-55
3. Thier R, Golka K, Bruning T. et al. Genetic susceptibility to environmental toxicants: the interface between human and experimental studies in the development of new toxicological concepts. Toxicol Lett. 2002;127:321-7
4. Strange RC, Spiteri MA, Ramachandran S. et al. Glutathione-S-transferase family of enzymes. Mutat Res. 2001;482:21-6
5. Dusinska M, Staruchova M, Horska A. et al. Are glutathione S transferases involved in DNA damage signalling? Interactions with DNA damage and repair revealed from molecular epidemiology studies. Mutat Res. 2012;736:130-7
6. Webb G, Vaska V, Coggan M. et al. Chromosomal localization of the gene for the human theta class glutathione transferase (GSTT1). Genomics. 1996;33:121-3
7. Minelli C, Wei I, Sagoo G. et al. Interactive effects of antioxidant genes and air pollution on respiratory function and airway disease: a HuGE review. Am J Epidemiol. 2011;173:603-20
8. Ito M, Imai M, Muraki M. et al. GSTT1 is upregulated by oxidative stress through p38-MK2 signaling pathway in human granulosa cells: possible association with mitochondrial activity. Aging (Albany NY). 2011;3:1213-23
9. Steinhoff C, Franke KH, Golka K. et al. Glutathione transferase isozyme genotypes in patients with prostate and bladder carcinoma. Arch Toxicol. 2000;74:521-6
10. Sweeney C, Farrow DC, Schwartz SM. et al. Glutathione S-transferase M1, T1, and P1 polymorphisms as risk factors for renal cell carcinoma: a case-control study. Cancer Epidemiol Biomarkers Prev. 2000;9:449-54
11. Mittal RD, Mishra DK, Mandhani A. Evaluating polymorphic status of glutathione-S-transferase genes in blood and tissue samples of prostate cancer patients. Asian Pac J Cancer Prev. 2006;7:444-6
12. Lindstrom S, Zheng SL, Wiklund F. et al. Systematic replication study of reported genetic associations in prostate cancer: Strong support for genetic variation in the androgen pathway. Prostate. 2006;66:1729-43
13. Srivastava D, Kumar A, Mittal B. et al. Polymorphism of GSTM1 and GSTT1 genes in bladder cancer: a study from North India. Archives of Toxicology. 2004:78
14. Longuemaux S, Delomenie C, Gallou C. et al. Candidate genetic modifiers of individual susceptibility to renal cell carcinoma: a study of polymorphic human xenobiotic-metabolizing enzymes. Cancer Res. 1999;59:2903-8
15. Toruner GA, Akyerli C, Ucar A. et al. Polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and bladder cancer susceptibility in the Turkish population. Arch Toxicol. 2001;75:459-64
16. Liberati A, Altman DG, Tetzlaff J. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700
17. Agalliu I, Langeberg WJ, Lampe JW. et al. Glutathione S-transferase M1, T1, and P1 polymorphisms and prostate cancer risk in middle-aged men. The Prostate. 2006;66:146-56
18. Catsburg C, Joshi AD, Corral R. et al. Polymorphisms in carcinogen metabolism enzymes, fish intake, and risk of prostate cancer. Carcinogenesis. 2012;33:1352-9
19. Abdel-Rahman SZ, Anwar WA, Abdel-Aal WE. et al. GSTM1 and GSTT1 genes are potential risk modifiers for bladder cancer. Cancer Detect Prev. 1998;22:129-38
20. Salagovic J, Kalina I, Stubna J. et al. Genetic polymorphism of glutathione S-transferases M1 and T1 as a risk factor in lung and bladder cancers. Neoplasma. 1998;45:312-7
21. Salagovic J, Kalina I, Habalova V. et al. The role of human glutathione S-transferases M1 and T1 in individual susceptibility to bladder cancer. Physiol Res. 1999;48:465-71
22. Peluso M, Airoldi L, Magagnotti C. et al. White blood cell DNA adducts and fruit and vegetable consumption in bladder cancer. Carcinogenesis. 2000;21:183-7
23. Schnakenberg E, Lustig M, Breuer R. et al. Gender-specific effects of NAT2 and GSTM1 in bladder cancer. Clin Genet. 2000;57:270-7
24. Sanyal S, Festa F, Sakano S. et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729-34
25. Golka K, Seidel T, Dietrich H. et al. [Occupational and non-occupational risk factors in bladder cancer patients in an industrialized area located in former East-Germany]. Aktuelle Urol. 2005;36:417-22
26. Saad AA, O'Connor PJ, Mostafa MH. et al. Glutathione S-transferase M1, T1 and P1 polymorphisms and bladder cancer risk in Egyptians. Int J Biol Markers. 2005;20:69-72
27. Altayli E, Gunes S, Yilmaz AF. et al. CYP1A2, CYP2D6, GSTM1, GSTP1, and GSTT1 gene polymorphisms in patients with bladder cancer in a Turkish population. International Urology and Nephrology. 2008;41:259-66
28. Broberg K. Constitutional short telomeres are strong genetic susceptibility markers for bladder cancer. Carcinogenesis. 2004;26:1263-71
29. Cantor KP, Villanueva CM, Silverman DT. et al. Polymorphisms in GSTT1, GSTZ1, and CYP2E1, Disinfection By-products, and Risk of Bladder Cancer in Spain. Environmental Health Perspectives. 2010;118:1545-50
30. Cengiz M, Ozaydin A, Ozkilic AC. et al. The investıgatıon of GSTT1, GSTM1 and SOD polymorphism in bladder cancer patıents. International Urology and Nephrology. 2007;39:1043-8
31. Ceylan GG, Ceylan C, Taşdemir S. et al. The effect of Glutathione-S-transferases in the susceptibility to bladder cancer. Irish Journal of Medical Science (1971 -). 2014
32. Covolo L, Placidi D, Gelatti U. et al. Bladder cancer, GSTs, NAT1, NAT2, SULT1A1, XRCC1, XRCC3, XPD genetic polymorphisms and coffee consumption: a case-control study. European Journal of Epidemiology. 2008;23:355-62
33. García-Closas M, Malats N, Silverman D. et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. The Lancet. 2005;366:649-59
34. Goerlitz D, El Daly M, Abdel-Hamid M. et al. GSTM1, GSTT1 Null Variants, and GPX1 Single Nucleotide Polymorphism Are Not Associated with Bladder Cancer Risk in Egypt. Cancer Epidemiology Biomarkers & Prevention. 2011;20:1552-4
35. Henríquez-Hernández LA, Navarro P, Luzardo OP. et al. Polymorphisms of glutathione S-transferase μ and θ, MDR1 and VEGF genes as risk factors of bladder cancer: A case-control study. Urologic Oncology: Seminars and Original Investigations. 2012;30:660-5
36. Hung RJ, Boffetta P, Brennan P. et al. GST, NAT, SULT1A1, CYP1B1 genetic polymorphisms, interactions with environmental exposures and bladder cancer risk in a high-risk population. International Journal of Cancer. 2004;110:598-604
37. Kellen E, Zeegers M, Paulussen A. et al. Does occupational exposure to PAHs, diesel and aromatic amines interact with smoking and metabolic genetic polymorphisms to increase the risk on bladder cancer?; The Belgian case control study on bladder cancer risk. Cancer Letters. 2007;245:51-60
38. Kogevinas M, Fernandez F, Garcia-Closas M. et al. Hair dye use is not associated with risk for bladder cancer: Evidence from a case-control study in Spain. European Journal of Cancer. 2006;42:1448-54
39. Koutros S, Silverman DT, Baris D. et al. Hair dye use and risk of bladder cancer in the New England bladder cancer study. International Journal of Cancer. 2011;129:2894-904
40. Lesseur C, Gilbert-Diamond D, Andrew AS. et al. A case-control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicology Letters. 2012;210:100-6
41. Matic M, Pekmezovic T, Djukic T. et al. GSTA1, GSTM1, GSTP1, and GSTT1 polymorphisms and susceptibility to smoking-related bladder cancer: A case-control study. Urologic Oncology: Seminars and Original Investigations. 2013;31:1184-92
42. Moore LE, Baris DR, Figueroa JD. et al. GSTM1 null and NAT2 slow acetylation genotypes, smoking intensity and bladder cancer risk: results from the New England bladder cancer study and NAT2 meta-analysis. Carcinogenesis. 2010;32:182-9
43. Moore LE, Wiencke JK, Bates MN. et al. Investigation of genetic polymorphisms and smoking in a bladder cancer case-control study in Argentina. Cancer Letters. 2004;211:199-207
44. Ovsiannikov D, Selinski S, Lehmann M-L. et al. Polymorphic Enzymes, Urinary Bladder Cancer Risk, and Structural Change in the Local Industry. Journal of Toxicology and Environmental Health, Part A. 2012;75:557-65
45. Reszka E, Jablonowski Z, Wieczorek E. et al. Polymorphisms of NRF2 and NRF2 target genes in urinary bladder cancer patients. Journal of Cancer Research and Clinical Oncology. 2014;140:1723-31
46. Safarinejad MR, Safarinejad S, Shafiei N. et al. Association of genetic polymorphism of glutathione S-transferase (GSTM1, GSTT1, GSTP1) with bladder cancer susceptibility. Urologic Oncology: Seminars and Original Investigations. 2013;31:1193-203
47. Yuan JM, Chan KK, Coetzee GA. et al. Genetic determinants in the metabolism of bladder carcinogens in relation to risk of bladder cancer. Carcinogenesis. 2008;29:1386-93
48. Katoh T, Inatomi H, Kim H. et al. Effects of glutathione S-transferase (GST) M1 and GSTT1 genotypes on urothelial cancer risk. Cancer Lett. 1998;132:147-52
49. Kim WJ, Lee HL, Lee SC. et al. Polymorphisms of N-acetyltransferase 2, glutathione S-transferase mu and theta genes as risk factors of bladder cancer in relation to asthma and tuberculosis. J Urol. 2000;164:209-13
50. Kim WJ, Kim H, Kim CH. et al. GSTT1-null genotype is a protective factor against bladder cancer. Urology. 2002;60:913-8
51. Kim HK WJ, Lee HL, Lee MS. et al. A case-control study on the effects of the genetic polymorphisms of N-acetyltransferase 2 and glutathione S-transferase mu and theta on the risk of bladder cancer. Korean J Prev. 1998;31:275-84
52. Kim EJ, Jeong P, Quan C. et al. Genotypes of TNF-alpha, VEGF, hOGG1, GSTM1, and GSTT1: useful determinants for clinical outcome of bladder cancer. Urology. 2005;65:70-5
53. Lee SJ, Cho SH, Park SK. et al. Combined effect of glutathione S-transferase M1 and T1 genotypes on bladder cancer risk. Cancer Lett. 2002;177:173-9
54. Ma QW, Lin GF, Chen JG. et al. Polymorphism of glutathione S-transferase T1, M1 and P1 genes in a Shanghai population: patients with occupational or non-occupational bladder cancer. Biomed Environ Sci. 2002;15:253-60
55. Lee SJ, Kang D, Cho SH. et al. Association of genetic polymorphism of glutathione s-transferase M1 and T1 and bladder cancer. J korean Cancer Assoc. 1999;31:548-55
56. Shao CX, Xiang YB, Zhang W. et al. Polymorphism of GSTM1 and GSTT1 with smoking and bladder cancer risk: a population-based case control study. Tumor. 2006;26:346-51
57. Song DK, Xing DL, Zhang LR. et al. Relationship between polymorphism of glutathione S-transferase and genetic susceptibility to bladder cancer. Chi J Urol. 2008;29:80-3
58. Song DK, Xing DL, Zhang LR. et al. Association of NAT2, GSTM1, GSTT1, CYP2A6, and CYP2A13 gene polymorphisms with susceptibility and clinicopathologic characteristics of bladder cancer in Central China. Cancer Detection and Prevention. 2009;32:416-23
59. Jong Jeong H, Jin Kim H, Young Seo I. et al. Association between glutathione S-transferase M1 and T1 polymorphisms and increased risk for bladder cancer in Korean smokers. Cancer Letters. 2003;202:193-9
60. Chen Yc, Xu L, Guo YlL. et al. Polymorphisms inGSTT1andp53and urinary transitional cell carcinoma in south-western Taiwan: A preliminary study. Biomarkers. 2004;9:386-94
61. Kang Hw, Song PH, Ha Y-S. et al. Glutathione S-transferase M1 and T1 polymorphisms: Susceptibility and outcomes in muscle invasive bladder cancer patients. European Journal of Cancer. 2013;49:3010-9
62. Ouerhani S, Tebourski F, Slama MRB. et al. The role of glutathione transferases M1 and T1 in individual susceptibility to bladder cancer in a Tunisian population. Annals of Human Biology. 2006;33:529-35
63. Rouissi K, Ouerhani S, Hamrita B. et al. Smoking and Polymorphisms in Xenobiotic Metabolism and DNA Repair Genes are Additive Risk Factors Affecting Bladder Cancer in Northern Tunisia. Pathology & Oncology Research. 2011;17:879-86
64. Gago-Dominguez M, Bell DA, Watson MA. et al. Permanent hair dyes and bladder cancer: risk modification by cytochrome P4501A2 and N-acetyltransferases 1 and 2. Carcinogenesis. 2003;24:483-9
65. Karagas M, Park S, Warren A. et al. Gender, smoking, glutathione-transferase variants and bladder cancer incidence: a population-based study. Cancer Letters. 2005;219:63-9
66. McGrath M, Michaud D, De Vivo I. Polymorphisms in GSTT1, GSTM1, NAT1 and NAT2 genes and bladder cancer risk in men and women. BMC Cancer. 2006;6:239
67. Zhao H, Lin J, Grossman HB, Hernandez LM. et al. Dietary isothiocyanates, GSTM1, GSTT1, NAT2 polymorphisms and bladder cancer risk. International Journal of Cancer. 2007;120:2208-13
68. Grando JPS, Kuasne H, Losi-Guembarovski R. et al. Association between polymorphisms in the biometabolism genes CYP1A1, GSTM1, GSTT1 and GSTP1 in bladder cancer. Clinical and Experimental Medicine. 2008;9:21-8
69. Autrup JL, Thomassen LH, Olsen JH. et al. Glutathione S-transferases as risk factors in prostate cancer. Eur J Cancer Prev. 1999;8:525-32
70. Rebbeck TR, Walker AH, Jaffe JM. et al. Glutathione S-transferase-mu (GSTM1) and -theta (GSTT1) genotypes in the etiology of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:283-7
71. Kelada SN, Kardia SL, Walker AH. et al. The glutathione S-transferase-mu and -theta genotypes in the etiology of prostate cancer: genotype-environment interactions with smoking. Cancer Epidemiol Biomarkers Prev. 2000;9:1329-34
72. Gsur A, Haidinger G, Hinteregger S. et al. Polymorphisms of glutathione-S-transferase genes (GSTP1, GSTM1 and GSTT1) and prostate-cancer risk. Int J Cancer. 2001;95:152-5
73. Kote-Jarai Z, Easton D, Edwards SM. et al. Relationship between glutathione S-transferase M1, P1 and T1 polymorphisms and early onset prostate cancer. Pharmacogenetics. 2001;11:325-30
74. Murata M, Watanabe M, Yamanaka M. et al. Genetic polymorphisms in cytochrome P450 (CYP) 1A1, CYP1A2, CYP2E1, glutathione S-transferase (GST) M1 and GSTT1 and susceptibility to prostate cancer in the Japanese population. Cancer Lett. 2001;165:171-7
75. Beer TM, Evans AJ, Hough KM. et al. Polymorphisms of GSTP1 and related genes and prostate cancer risk. Prostate Cancer and Prostatic Diseases. 2002;5:22-7
76. Nakazato H, Suzuki K, Matsui H. et al. Association of genetic polymorphisms of glutathione-S-transferase genes (GSTM1, GSTT1 and GSTP1) with familial prostate cancer risk in a Japanese population. Anticancer Res. 2003;23:2897-902
77. Kidd LC, Woodson K, Taylor PR. et al. Polymorphisms in glutathione-S-transferase genes (GST-M1, GST-T1 and GST-P1) and susceptibility to prostate cancer among male smokers of the ATBC cancer prevention study. Eur J Cancer Prev. 2003;12:317-20
78. Joseph MA, Moysich KB, Freudenheim JL. et al. Cruciferous vegetables, genetic polymorphisms in glutathione S-transferases M1 and T1, and prostate cancer risk. Nutr Cancer. 2004;50:206-13
79. Medeiros R, Vasconcelos A, Costa S. et al. Metabolic susceptibility genes and prostate cancer risk in a southern European population: The role of glutathione S-transferases GSTM1, GSTM3, and GSTT1 genetic polymorphisms. The Prostate. 2004;58:414-20
80. Mittal RD, Srivastava DS, Mandhani A. et al. Polymorphism of GSTM1 and GSTT1 genes in prostate cancer: a study from North India. Indian J Cancer. 2004;41:115-9
81. Komiya Y, Tsukino H, Nakao H. et al. Human glutathione S-transferase A1, T1, M1, and P1 polymorphisms and susceptibility to prostate cancer in the Japanese population. Journal of Cancer Research and Clinical Oncology. 2004;131:238-42
82. Wang YL, Jiang J, Wang LF. et al. Polymorphisms of Glutathione S-transferase genes GSTM1 and GSTT1 and prostate cancer risk in Chinese population. Acta Academiae Medicinae Militaris Tertiae. 2005;27:1039-41
83. Srivastava DSL, Mandhani A, Mittal B. et al. Genetic polymorphism of glutathione S-transferase genes (GSTM1, GSTT1 and GSTP1) and susceptibility to prostate cancer in Northern India. BJU International. 2005;95:170-3
84. Caceres DD, Iturrieta J, Acevedo C. et al. Relationship among metabolizing genes, smoking and alcohol used as modifier factors on prostate cancer risk: exploring some gene-gene and gene-environment interactions. Eur J Epidemiol. 2005;20:79-88
85. Nam RK. The Use of Genetic Markers to Determine Risk for Prostate Cancer at Prostate Biopsy. Clinical Cancer Research. 2005;11:8391-7
86. Kempkes M, Golka K, Reich S. et al. Glutathione S-transferase GSTM1 and GSTT1 null genotypes as potential risk factors for urothelial cancer of the bladder. Arch Toxicol. 1996;71:123-6
87. Sharma T, Jain S, Verma A. et al. Gene environment interaction in urinary bladder cancer with special reference to organochlorine pesticide: a case control study. Cancer Biomark. 2013;13:243-51
88. Henriquez-Hernandez LA, Navarro P, Luzardo OP. et al. Polymorphisms of glutathione S-transferase mu and theta, MDR1 and VEGF genes as risk factors of bladder cancer: a case-control study. Urol Oncol. 2012;30:660-5
89. Salinas-Sanchez AS, Sanchez-Sanchez F, Donate-Moreno MJ. et al. Polymorphic deletions of the GSTT1 and GSTM1 genes and susceptibility to bladder cancer. BJU Int. 2011;107:1825-32
90. Berber U, Yilmaz I, Yilmaz O. et al. CYP1A1 (Ile462Val), CYP1B1 (Ala119Ser and Val432Leu), GSTM1 (null), and GSTT1 (null) polymorphisms and bladder cancer risk in a Turkish population. Asian Pac J Cancer Prev. 2013;14:3925-9
91. Yang J, Wu HF, Zhang W. et al. Polymorphisms of metabolic enzyme genes, living habits and prostate cancer susceptibility. Front Biosci. 2006;11:2052-60
92. Nock NL. Polymorphisms in Polycyclic Aromatic Hydrocarbon Metabolism and Conjugation Genes, Interactions with Smoking and Prostate Cancer Risk. Cancer Epidemiology Biomarkers & Prevention. 2006;15:756-61
93. Silig Y, Pinarbasi H, Günes S. et al. Cetinkaya O. Polymorphisms of CYP1A1, GSTM1, GSTT1, and Prostate Cancer Risk in Turkish Population. Cancer Investigation. 2006;24:41-5
94. Cunningham JM, Hebbring SJ, McDonnell SK. et al. Evaluation of Genetic Variations in the Androgen and Estrogen Metabolic Pathways as Risk Factors for Sporadic and Familial Prostate Cancer. Cancer Epidemiology Biomarkers & Prevention. 2007;16:969-78
95. Mallick S, Romana M, Blanchet P. et al. GSTM1 and GSTT1 Polymorphisms and the Risk of Prostate Cancer in a Caribbean Population of African Descent. Urology. 2007;69:1165-9
96. Davydova NA, Dmitrieva AI, Selivanov SP. et al. [Polymorphism of glutathion-S-transferase genes in patients with prostatic cancer]. Urologiia. 2008:26-9
97. Lima MM Jr, Oliveira MN, Granja F. et al. Lack of association of GSTT1, GSTM1, GSTO1, GSTP1 and CYP1A1 polymorphisms for susceptibility and outcome in Brazilian prostate cancer patients. Folia Biol (Praha). 2008;54:102-8
98. Lavender NA, Benford ML, VanCleave TT. et al. Examination of polymorphic glutathione S-transferase (GST) genes, tobacco smoking and prostate cancer risk among Men of African Descent: A case-control study. BMC Cancer. 2009;9:397
99. Sivoňová M, Waczulíková I, Dobrota D. et al. Polymorphisms of glutathione-S-transferase M1, T1, P1 and the risk of prostate cancer: a case-control study. Journal of Experimental & Clinical Cancer Research. 2009;28:32
100. Souiden Y, Mahdouani M, Chaieb K. et al. Polymorphisms of glutathione-S-transferase M1 and T1 and prostate cancer risk in a Tunisian population. Cancer Epidemiology. 2010;34:598-603
101. Steinbrecher A, Rohrmann S, Timofeeva M. et al. Dietary Glucosinolate Intake, Polymorphisms in Selected Biotransformation Enzymes, and Risk of Prostate Cancer. Cancer Epidemiology Biomarkers & Prevention. 2010;19:135-43
102. Kwon DD, Lee JW, Han DY. et al. Relationship between theGlutathione-S-TransferaseP1,M1, andT1Genotypes and Prostate Cancer Risk in Korean Subjects. Korean Journal of Urology. 2011;52:247
103. Ashtiani ZO, Hasheminasab S-M, Ayati M. et al. Are GSTM1, GSTT1 and CAG Repeat Length of Androgen Receptor Gene Polymorphisms Associated with Risk of Prostate Cancer in Iranian Patients? Pathology & Oncology Research. 2010;17:269-75
104. Kumar V, Yadav CS, Datta SK. et al. Association of GSTM1 and GSTT1 polymorphism with lipid peroxidation in benign prostate hyperplasia and prostate cancer: a pilot study. Dis Markers. 2011;30:163-9
105. Norskov MS, Frikke-Schmidt R, Bojesen SE. et al. Copy number variation in glutathione-S-transferase T1 and M1 predicts incidence and 5-year survival from prostate and bladder cancer, and incidence of corpus uteri cancer in the general population. Pharmacogenomics J. 2011;11:292-9
106. Rodrigues IS, Kuasne H, Losi-Guembarovski R. et al. Evaluation of the influence of polymorphic variants CYP1A1*2B, CYP1B1*2, CYP3A4*1B, GSTM1*0, and GSTT1*0 in prostate cancer. Urologic Oncology: Seminars and Original Investigations. 2011;29:654-63
107. Safarinejad MR, Shafiei N, Safarinejad SH. Glutathione S-transferase gene polymorphisms (GSTM1, GSTT1, GSTP1) and prostate cancer: a case-control study in Tehran, Iran. Prostate Cancer and Prostatic Diseases. 2011;14:105-13
108. Thakur H, Gupta L, Sobti RC. et al. Association of GSTM1T1 genes with COPD and prostate cancer in north Indian population. Molecular Biology Reports. 2010;38:1733-9
109. Hemelrijck MV, Rohrmann S, Steinbrecher A. et al. Heterocyclic Aromatic Amine [HCA] Intake and Prostate Cancer Risk: Effect Modification by Genetic Variants. Nutrition and Cancer. 2012;64:704-13
110. Choubey VK, Sankhwar SN, Tewari R. et al. Null genotypes at theGSTM1andGSTT1genes and the risk of benign prostatic hyperplasia: A case-control study and a meta-analysis. The Prostate. 2013;73:146-52
111. Emeville E, Broquere C, Brureau L. et al. Copy number variation of GSTT1 and GSTM1 and the risk of prostate cancer in a Caribbean population of African descent. PLoS ONE. 2014;9:e107275
112. Sa RA, Moreira Ados S, Cabello PH. et al. Human glutathione S-transferase polymorphisms associated with prostate cancer in the Brazilian population. Int Braz J Urol. 2014;40:463-73
113. Bruning T, Lammert M, Kempkes M. et al. Influence of polymorphisms of GSTM1 and GSTT1 for risk of renal cell cancer in workers with long-term high occupational exposure to trichloroethene. Arch Toxicol. 1997;71:596-9
114. Buzio L, De Palma G, Mozzoni P. et al. Glutathione S-transferases M1-1 and T1-1 as risk modifiers for renal cell cancer associated with occupational exposure to chemicals. Occup Environ Med. 2003;60:789-93
115. Moore LE, Brennan P, Karami S. et al. Glutathione S-transferase polymorphisms, cruciferous vegetable intake and cancer risk in the Central and Eastern European Kidney Cancer Study. Carcinogenesis. 2007;28:1960-4
116. Wiesenhütter B, Selinski S, Golka K. et al. Re-assessment of the influence of polymorphisms of phase-II metabolic enzymes on renal cell cancer risk of trichloroethylene-exposed workers. International Archives of Occupational and Environmental Health. 2007;81:247-51
117. Karami S, Boffetta P, Rothman N. et al. Renal cell carcinoma, occupational pesticide exposure and modification by glutathione S-transferase polymorphisms. Carcinogenesis. 2008;29:1567-71
118. Ćorić V, Plješa-Ercegovac M, Matić M. et al. The Role of GSTM1 and GSTT1 Polymorphism in Patients with Renal Cell Carcinoma. Journal of Medical Biochemistry. 2010:29
119. De Martino M, Klatte T, Schatzl G. et al. Renal cell carcinoma Fuhrman grade and histological subtype correlate with complete polymorphic deletion of glutathione S-transferase M1 gene. J Urol. 2010;183:878-83
120. Salinas-Sánchez AS, Sánchez-Sánchez F, Donate-Moreno MJ. et al. GSTT1, GSTM1, and CYP1B1 gene polymorphisms and susceptibility to sporadic renal cell cancer. Urologic Oncology: Seminars and Original Investigations. 2012;30:864-70
121. Ahmad ST, Arjumand W, Seth A. et al. Impact of Glutathione TransferaseM1, T1,andP1Gene Polymorphisms in the Genetic Susceptibility of North Indian Population to Renal Cell Carcinoma. DNA and Cell Biology. 2012;31:636-43
122. Wiesenhutter B, Selinski S, Golka K. et al. Re-assessment of the influence of polymorphisms of phase-II metabolic enzymes on renal cell cancer risk of trichloroethylene-exposed workers. Int Arch Occup Environ Health. 2007;81:247-51
123. Sivonova M, Waczulikova I, Dobrota D. et al. Polymorphisms of glutathione-S-transferase M1, T1, P1 and the risk of prostate cancer: a case-control study. J Exp Clin Cancer Res. 2009;28:32
124. Higgins LG, Hayes JD. Mechanisms of induction of cytosolic and microsomal glutathione transferase (GST) genes by xenobiotics and pro-inflammatory agents. Drug Metab Rev. 2011;43:92-137
125. Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51-88
126. Gong M, Dong W, Shi Z. et al. Genetic polymorphisms of GSTM1, GSTT1, and GSTP1 with prostate cancer risk: a meta-analysis of 57 studies. PLoS One. 2012;7:e50587
127. Liu D, Liu Y, Ran L. et al. GSTT1 and GSTM1 polymorphisms and prostate cancer risk in Asians: a systematic review and meta-analysis. Tumour Biol. 2013;34:2539-44
128. Yang X, Long S, Deng J. et al. Glutathione S-transferase polymorphisms (GSTM1, GSTT1 and GSTP1) and their susceptibility to renal cell carcinoma: an evidence-based meta-analysis. PLoS One. 2013;8:e63827
129. Gong M, Dong W, An R. Glutathione S-transferase T1 polymorphism contributes to bladder cancer risk: a meta-analysis involving 50 studies. DNA Cell Biol. 2012;31:1187-97
Author contact
![]() Corresponding author: Zhen-Ya Shen, Department of Cardiovascular Surgery & Institute of Cardiovascular Science, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China, email: uuzyshencom.
Corresponding author: Zhen-Ya Shen, Department of Cardiovascular Surgery & Institute of Cardiovascular Science, First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, China, email: uuzyshencom.

 Global reach, higher impact
Global reach, higher impact