3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(13):1856-1860. doi:10.7150/jca.15605 This issue Cite
Short Research Communication
Low Proteolytic Clipping of Histone H3 in Cervical Cancer
1. Área de investigación clínica, Unidad de Innovación Clínica y Epidemiológica de la Secretaría de Salud del estado de Guerrero, Acapulco, Gro., México.
2. Laboratorio de Biología Molecular, Universidad Hipócrates, Acapulco, Gro., México.
3. Laboratorio de Biología Celular, Unidad Ciencias de la Salud, Facultad de Odontología, Universidad Autónoma de Baja California Mexicali, BC, México.
4. Laboratorio de Genética y Diagnóstico Molecular, Unidad de Investigación, Hospital Juárez de México, Ciudad de México, México.
5. Facultad de Estudios Superiores Zaragoza, Universidad Nacional Autónoma de México, Ciudad de México, México.
6. Unidad Académica de Ciencias Químico Biológicas, Universidad Autónoma de Guerrero, Chilpancingo, Gro., México.
7. Laboratorio de Bioquímica, Unidad Ciencias de la Salud, Facultad de Medicina Mexicali, Universidad Autónoma de Baja California Mexicali, BC, México.
8. Department of internal Medicine, Center for Arrhythmia Research, University of Michigan.
9. Centro Médico Nacional 20 de Noviembre, Instituto de Seguridad y Servicios Sociales de los Trabajadores del Estado, Ciudad de México, México.
10. Posgrado en Ciencias Genómicas, Universidad Autónoma de la Ciudad de México, Ciudad de México, México.
* Both authors have contributed equally to this work.
Received 2016-3-21; Accepted 2016-6-10; Published 2016-8-17
Abstract
Chromatin in cervical cancer (CC) undergoes chemical and structural changes that alter the expression pattern of genes. Recently, a potential mechanism, which regulates gene expression at transcriptional levels is the proteolytic clipping of histone H3. However, until now this process in CC has not been reported. Using HeLa cells as a model of CC and human samples from patients with CC, we identify that the H3 cleavage was lower in CC compared with control tissue. Additionally, the histone H3 clipping was performed by serine and aspartyl proteases in HeLa cells. These results suggest that histone H3 clipping operates as part of post-translational modification system in CC.
Keywords: Histone 3, Histone H3 clipping, Histone proteolysis, Histone modifications, Cervical cancer, HeLa.
Introduction
Cervical cancer (CC) is the third most common cancer worldwide with 530 000 new cases and 275 000 deaths each year. It is the sixth most common cancer in women in developed countries and the second most common cancer in underdeveloped countries [1]. Current treatment for CC is suboptimal and the identification of new carcinogenic pathways could provide new therapeutic targets.
During the different stages of CC development, accumulation of epigenetic alterations leading to changes in gene expression are observed. Altered mechanisms of epigenetic regulation in CC include DNA methylation, post-translational modifications (PTMs) of histones, and noncoding RNAs. Frequently in CC, hypermethylation within the promoter region of genes are involved in cell cycle, apoptosis, DNA repair, proliferation, cell adhesion, and metabolism leading to transcriptional repression [2]. DNA methylation in the promoter region of genes enables the recruitment of the histone deacetylases (HDACs) resulting in gene silencing in CC [3-5]. Additionally, it has been reported that histone modifying enzymes like HDAC1 and HDAC2 are over-expressed in cervical dysplasia and invasive carcinoma [6], suggesting that the dysregulation of enzymes that modify histones in CC is a mechanism by were gene silencing occurs. Finally, an association of different miRNAs to cervical carcinogenesis has been reported [2].
Recently an increasing number of publications have reported histone H3 clipping in human cells such as embryonic stem cells (hESCs) [7], senescent fibroblasts and melanocytes [8], peripheral blood mononuclear cells (PBMCs) [9], Raji cells [10], and hepatocytes [11]. Analysis of the proteolytic clipping of histone H3 in the differentiation of hESCs evidenced that the N-terminal tail of H3 can be cleaved in different sites and it was mediated by a serine protease [7]. In addition, proteolytic clipping of histone H3 was performed by a cysteine protease, cathepsin L, in models of oncogene-induced and replicative senescence. Notably, it was suggested that cleaved products of H3 could contribute to regulate genes implicated in cell cycle-promotion, DNA replication, cellular proliferation, apoptosis and migration in senescence [8].
Since histone H3 clipping only has been identify in a hepatocellular carcinoma cell line [11] and PTMs play an important role in cancer development, the aim of this study was to investigate if proteolytic clipping of histone H3 occurs in CC.
Material and Methods
Patients
In the present work 5 samples of patients diagnosed with CC and 1 patient non-cervical cancer (NCC) were used. NCC patient was negative to HPV presented normal pathology and was submitted to hysterectomy due to diagnosis of uterine myoma. The protocols were previously approved by the ethics committee of the Hospital Juarez de Mexico (HJM2231/13-B). After signing the informed consent, a biopsy was taken of each patient and divided into three parts; the first one for pathological confirmation of at least 80% of tumor cells, the second one for obtaining DNA and the remaining part was used to obtain total proteins extracts. All patients diagnosed with CC were classified according to the International Federation of Gynecology and Obstetrics (FIGO). However, the information concerning to FIGO classification was obtained only of 3 patients and it is included in the table 1.
Cell culture
HeLa cells were obtained from the American Type Culture Collection (ATCC) and cultivated in advanced DMEM (Gibco) supplemented with 8 % FBS (Gibco), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco) and 4 mM L-glutamine. Cells were grown at 37 °C in a humidified atmosphere containing 10% CO2. For inhibition experiments, cells were treated independently with 0.5 mM of 4-(2-Aminoethyl) benzen esulfonyl fluoride hydrochloride (AEBSF), 27 μM of trans-epoxysuccinyl-L-leucylamido-(4-guanidino) butane (E64) or 15 μM pepstatin A (dissolved in 10% (v/v) acetic acid in DMSO) (9:1 DMSO:acetic acid) for 12 hours. The inhibitors were purchased from (Sigma-Aldrich). AEBSF dose was used as manufacturer's instructions. Doses of E64 and pepstatin A inhibitors for HeLa cells are described in the literature [12].
Features of patients with CC.
| P1 | P2 | P3 | P4 | P5 | |
|---|---|---|---|---|---|
| Pathology | ADC | SCC | SCC | ADC | SCC |
| FIGO | IB1 | NI | IIIB | NI | IB2 |
| HPV genotype | 16, 31 | 16,18, 59 | 16, 31, 59 | 16, 31, 59, 70 | 16 |
| HPV copy number | 104, 102 | 102, 102, 102 | 104, 104, 102 | 102, 102, 102, 102 | 104 |
CC, cervical cancer; ADC, adenocarcinoma; SCC, squamous cell carcinoma; FIGO, International Federation of Gynecology and Obstetrics; NI, not identified.
Immunoblot analysis
Immunoblotting assays were performed using total proteins extracts from HeLa cells, NCC and CC. All samples were obtained with RSB buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1% NP40) supplemented with “complete” protease inhibitor cocktail. Briefly, samples were dissolved in 2X Laemmli-buffer with β-mercaptoethanol, heated to 95ºC for 8 min, and loaded onto a 15% SDS-polyacrylamide gel. Proteins were transferred to a PVDF membrane by damp blotting at 40 mA for 3 h. The PVDF membrane was blocked with 5% milk in TBS-T (10X TBS-T: 198 mM Tris, 1400 mM NaCl, 0.01% Tween 20, pH 7.6) for 1 h. Then the PVDF membrane was incubated overnight at 4ºC with the primary H3 antibody (Thermo Fisher Scientific) or HRP conjugated-GAPDH antibody (Cell Signaling Technology). We use a h3 primary antibody that recognizes the residues of the C-terminal of the human histone H3. H3 was used at a 1:500 dilution and GAPDH used at a 1:1000 dilution. Membrane was subsequently washed thrice in TBS-T. Incubation with HRP-conjugated secondary antibody (Jackson ImmunoResearch) was performed by 1 h at room temperature for only H3. Then, the membrane was washed thrice in TBS-T. Proteins were visualized using the SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and Gel Doc XR system (Bio-Rad). GAPDH was used as loading control.
Genotyping and load of HPV
DNA extraction from the patient's tissue was performed following manufactures instructions (Invitrogen Thermo Fisher Scientific). After this, genotyping and copy number determination of HPV was carried out through Anyplex II HPV28 detection kit (Seegene) following the manufactures instructions. Briefly, in 20 μl PCR reaction 1x of HPV28 primer mix A (14 high-risk HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68) or B (5 high risk HPV types: 26, 53, 69, 73, 82 and 9 low risk types: 6, 11, 40, 42, 43, 44, 54, 61, 70), 1x Anyplex master mix, and approximately 50 ng of DNA were added. The thermocycling conditions was as follow: an initial step at 50 °C for 4 minutes and a denaturation step at 95 °C for 15 minutes. The samples underwent 30 cycles at 95 °C for 30 seconds, 60 °C for 60 seconds and 72 °C for 30 seconds. After this, the reaction mix was followed by a 55 °C step for 30 seconds and a melting curve analysis at 55 to 85 °C (5s/0.5°C). The PCR reactions continue cycling at the same temperature for 10 cycles followed by melting curve analysis. This last step was repeated for another 10 cycles, so the melting curve analysis were obtained at 30, 40, and 50 cycles. The PCR reaction were done in a CFX96 Real Time PCR System (Bio-Rad) as recommended by the manufacturer.
As positive controls, three plasmid constructs with all HPV genotypes provided by the kit were used. The results were analyzed with the software Seegene Viewer version 2.0 (Seegene). This is a semi quantitative test where the results are indicated at 30 cycles (equivalent of 107 copies), 40 cycles (104 copies) or 50 cycles (102 copies).
Results and discussion
H3 truncated products generated by cleavage in distinct sites of H3 N-terminal tail has been reported in different cellular types (Fig. 1A) [7, 8]; nevertheless, proteolytic clipping of histone H3 has not been identified in CC.
Low proteolytic clipping of histone H3 in CC. A) The schematic drawing of Core Domain and H3 N-terminal tail shows the frequent cleavage sites (solid lines) reported in hESCs, senescent fibroblasts and melanocytes [7, 8]. Numbers designate the position of the amino acids in H3. B) Western blot analysis of H3 indicated low levels of cleavage product (asterisk) in HeLa cells as compared with sample of non-cervical cancer (NCC). Complete H3 (arrow) was only identified in HeLa cells. C) Immunodetection of H3 by western blot showed low levels of cleavage product (asterisk) in different samples of CC (lane 2 - 6; CC) as compared with sample of NCC (lane 1; NCC). GAPDH expression was analyzed in parallel as an internal control. The running position of protein markers is shown on the left. The result is representative of at least three separate experiments.

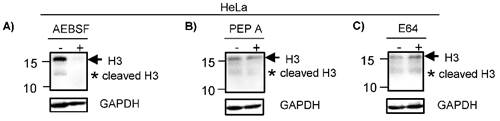
H3 is cleaved by serine and aspartyl proteases. HeLa cells were treated with different proteases inhibitors (+) or vehicle (-) as described in Material and Methods. A) Cells treated with AEBSF, an inhibitor of serine proteases, showed reduced levels of H3 complete (arrow) and cleavage (asterisk) as compared with vehicle. B) HeLa cells treated with Pepstatin A (Pep A) showed a reduction of H3 complete (arrow) and cleavage (asterisk). C) HeLa cells treated with E64 (an inhibitor of cysteine protease) did not showed any differences in the H3 proteolytic clipping as compared with vehicle. GAPDH expression was analyzed in parallel as an internal control. The running position of protein markers is shown on the left. The result is representative of at least three separate experiments.
Using HeLa cells, a cellular line of CC, we identified reduced levels of a cleaved H3 product in comparison to NCC, (Fig. 1B), suggesting that histone H3 is lower cleave in CC cells compared with control tissue. This observation is interesting because a recent study did not observe the cleavage of histone H3 in HeLa cells [11]. This discrepancy could be due to differences in the sample preparations in both protocols. We used RSB buffer, which contains 10 mM Tris-Cl pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 1 % NP40 supplemented with “complete” protease inhibitor cocktail. Tvardovskiy A, et al [11] used modified RIPA buffer, that contain: 50 mM Tris-Cl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 % sodium deoxycholate and 0.5% SDS supplemented with “complete” protease inhibitor cocktail and “PhosSTOP” phosphatase inhibitors. To last, a monoclonal antibody against C-termini of H3 histone (E.960.2, Thermo Fisher Scientific), was used in the present study compared to a polyclonal antibody (Anti-Histone H3 antibody, ab1791, Abcam) used in Tvardovskiy A, et al [11]. Monoclonal antibodies have been documented to possess higher specificity and it may explain why Tvardovskiy A, et al [11] did not inmunodetect the cleaved H3 product. Additionally, the truncated product of H3 (13 kDa) that we observed in HeLa cells had similar molecular weight as the H3 cleavage product reported in hESCs [7] and the proximal cleavage observed in senescent fibroblasts and melanocytes [8]. On the other hand, the band indicating complete histone H3 was not detected in NCC (Fig. 1B). Thus, these results could indicate that histone H3 is highly cleaved in NCC.
We also explore the proteolytic cleavage of the histone H3 in samples of patients with CC. The characteristics of the samples used in this study are indicated in table 1. In agreement with what we observed in HeLa cells, the western blot analysis showed a decreased in the cleavage product of H3 in CC samples of patients as compared with NCC (Fig. 1C). The intensity of the bands corresponding to complete and cleavage product of H3, was reduced in samples of adenocarcinoma (Fig.1C). The H3 cleavage could be reduced independently of the CC type (adenocarcinoma or squamous cell carcinoma), genotype or copy number of HPV. We do recognize that the number of human samples used here are low and the interpretation of this data should take cautiously.
To identify the enzymes participating in the histone H3 clipping, HeLa cells were treated with inhibitors of different protease classes as described in material and methods. Western blot indicated that the cleavage of H3 was inhibited when HeLa cells were pre-treated with AEBSF, a serine protease inhibitor, since the lower weight band was not observed. Of note, the intensity of the band indicating complete H3 was also reduced in treated cells (Fig. 2A). Consistent with this, it has been suggested that the peptide released during the histone H3 clipping participates regulating the expression of the H3 [20]. Similar to AEBSF, the pre-treatment with Pepstatin A (Pep A) reduced the levels of H3 cleavage product (Fig. 2B). In contrast, pre-treatment with E64 did not inhibit the cleavage of H3 (Fig. 2C). Together, these results suggest that the proteolytic clipping of histone H3 occurs in CC and this is exerted by serine and aspartyl proteases. In accordance, it has been reported that H3 cleavage was performed by a serine protease in hESC [7]. To our knowledge, the aspartyl protease activity participating in cleavage of H3 has not been reported heretofore and it may be a peculiarity in CC. It was previously showed in vitro that human recombinant legumain or purified bovine kidney legumain can cleave recombinant histone H3, additionally, even though this enzyme is expressed in nucleus of colon cancer cell lines, the proteolytic clipping of endogenous H3 histone has not been reported in these cell lines [13]. Others enzymes participating in this process are cathepsin L and D [8, 14, 15], granzyme A [10], glutamate dehydrogenase (GDH) [16], PRB1 [17], the viral protease 3C [18], and matrix metalloproteinase 9 (MMP-9) [19]. We did not investigate if these other enzymes classes are implicated in the cleavage of H3 in CC cells.
Histones modifications regulate gene expression and drastic changes in gene expression take place in CC [2]. Thus, it is not surprising that alterations in histone modifications occurs in different types of cancer [20]. Furthermore, different reports account the potential used of histone modifications as biomarkers or as a target for epigenetic therapy [2, 20, 21]. It has been suggested that H3 cleavage in humans regulates different biological process [7, 8]. In this work our results suggest that H3 cleavage was lower in CC compared with control tissue. Thus, it is valid to speculate that histone H3 clipping may be implicated in the genetic regulation during development of CC. More studies are needed to explore the functional outcomes of our findings.
Acknowledgements
We thank Fabiola Fragoso Sandoval, Xicoténcatl Jiménez Villanueva and Francisco Javier Hernández Aldana for all facilities with the patient recruitment, and Carlos Ugarte Briones for pathological characterization of all biological samples.
Competing Interests
We declare not conflict interests.
References
1. http://globocan.iarc.fr/old/FactSheets/cancers/cervix-new.asp. Globocan. Cervical cancer estimated incidence, mortality, and prevalence wordwide in. 2012
2. Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35:5083-5093
3. Zhang Z, Joh K, Yatsuki H. et al. Retinoic acid receptor beta2 is epigenetically silenced either by DNA methylation or repressive histone modifications at the promoter in cervical cancer cells. Cancer Lett. 2007;247:318-327
4. Lu TY, Kao CF, Lin CT. et al. DNA methylation and histone modification regulate silencing of OPG during tumor progression. J Cell Biochem. 2009;108:315-325
5. Liu D, Zhou P, Zhang L. et al. HDAC1/DNMT3A-containing complex is associated with suppression of Oct4 in cervical cancer cell. Biochemistry (Mosc). 2012;77:934-940
6. Huang BH, Laban M, Leung CH. et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395-404
7. Vossaert L, Meert P, Scheerlinck E. et al. Identification of histone h3 clipping activity in human embryonic stem cells. Stem Cell Res. 2014;13:123-134
8. Duarte LF, Young ARJ, Wang Z. et al. Histone h3.3 and its proteolytically processed form drive a cellular senescence programme. Nat Commun. 2014;5:5210
9. Howe CG, Gamble MV. Enzymatic cleavage of histone h3: a new consideration when measuring histone modifications in human samples. Clin Epigenetics. 2015;7:7
10. Lee PY, Park BC, Chi SW. et al. Histone H3 is digested by granzyme A during compromised cell death in the Raji Cell. J Microbiol Biotechnol. 2015;25:1578-1582
11. Tvardovskiy A, Wrzesinski K, Sidoli S. et al. Top-down and middle-down protein analysis reveals that intact and clipped human histones differ in post-translational modification patterns. Mol Cell Proteomics. 2015;14:3142-3153
12. Keng-Fu H, Chao-Liang W, Soon-Cen H. et al. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451-460
13. Haugen MH, Johansen HT, Pettersen SJ. et al. Nuclear legumain activity in colorectal cancer. Plos One. 2013;8:e52980
14. Duncan EM, Muratore-Schroeder TL, Cook RG. et al. Cathepsin L proteolytically processes histone h3 during mouse embryonic stem cell differentiation. Cell. 2008;135:284-294
15. Khalkhali Z, Goossens W, Margaryan NV. et al. Cleavage of histone 3 by cathepsin D in the involuting mammary gland. Plos One. 2014;9:e103230
16. Mandal P, Verma N, Chauhan S. et al. Unexpected Histone H3 Tail-clipping Activity of Glutamate Dehydrogenase. J Biol Chem. 2013;288:18743-18757
17. Xue Y, Vashisht AA, Tan Y. et al. PRB1 is required for clipping of the histone h3 n terminal tail in Saccharomyces cerevisiae. Plos One. 2014;9:e90496
18. Falk MM, Grigera PR, Bergmann IE. et al. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J Virol. 1990;64:748-756
19. Kim K, Punj V, Kim JM. et al. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016;30:208-219
20. Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391-3403
21. Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am j cancer res. 2015;2:589-597
Author contact
![]() Corresponding author: E-Mail: sofializethcom; Tel.: +52-55-5200-5003; Fax: +52-33-3632-6200.
Corresponding author: E-Mail: sofializethcom; Tel.: +52-55-5200-5003; Fax: +52-33-3632-6200.

 Global reach, higher impact
Global reach, higher impact