Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(13):1915-1925. doi:10.7150/jca.16390 This issue Cite
Research Paper
Screening of Pleural Mesotheliomas for DNA-damage Repair Players by Digital Gene Expression Analysis Can Enhance Clinical Management of Patients Receiving Platin-Based Chemotherapy
1. Ruhrlandklinik, West German Lung Centre, University Hospital Essen, University of Duisburg-Essen, Essen, Germany;
2. Institute of Pathology, University Hospital Essen, University of Duisburg-Essen, Essen, Germany;
3. Institute of Pathology, Division of Molecular Pathology, Charité, Berlin, Germany;
4. Department of Pathology, Helios Klinikum Emil von Behring, Berlin Germany;
5. Institute of Pathology, Ev.-Luth. Diakonissenkrankenhaus Flensburg, Flensburg, Germany;
6. Thoracic Surgery Department, Theagenio Cancer Hospital, Thessaloniki, Greece;
7. Pathology Department, “Theagenio” Cancer Hospital, Thessaloniki, Greece;
8. Pulmonary Department-Oncology Unit, “G. Papanikolaou” General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece;
9. Division of Pulmonology, Department of Clinical Therapeutics, National and Kapodistrian University of Athens School of Medicine, Alexandra Hospital, Athens, Greece;
10. Department of Medical Oncology, West German Cancer Centre, University Hospital Essen, University of Duisburg-Essen, Essen, Germany.
Received 2016-6-5; Accepted 2016-8-14; Published 2016-9-13
Abstract
Background: Malignant pleural mesothelioma (MPM) is a rare, predominantly asbestos-related and biologically highly aggressive tumour leading to a dismal prognosis. Multimodality therapy consisting of platinum-based chemotherapy is the treatment of choice. The reasons for the rather poor efficacy of platinum compounds remain largely unknown.
Material and Methods: For this exploratory mRNA study, 24 FFPE tumour specimens were screened by digital gene expression analysis. Based on data from preliminary experiments and recent literature, a total of 366 mRNAs were investigated using a Custom CodeSet from NanoString. All statistical analyses were calculated with the R i386 statistical programming environment.
Results: CDC25A and PARP1 gene expression were correlated with lymph node spread, BRCA1 and TP73 expression levels with higher IMIG stage. NTHL1 and XRCC3 expression was associated with TNM stage. CHECK1 as well as XRCC2 expression levels were correlated with tumour progression in the overall cohort of patients. CDKN2A and MLH1 gene expression influenced overall survival in this collective. In the adjuvant treated cohort only, CDKN2A, CHEK1 as well as ERCC1 were significantly associated with overall survival. Furthermore, TP73 expression was associated with progression in this subgroup.
Conclusion: DNA-damage response plays a crucial role in response to platin-based chemotherapeutic regimes. In particular, CHEK1, XRCC2 and TP73 are strongly associated with tumour progression. ERCC1, MLH1, CDKN2A and most promising CHEK1 are prognostic markers for OS in MPM. TP73, CDKN2A, CHEK1 and ERCC1 seem to be also predictive markers in adjuvant treated MPMs. After a prospective validation, these markers may improve clinical and pathological practice, finally leading to a patients' benefit by an enhanced clinical management.
Keywords: pleural mesothelioma, NanoString nCounter, digital gene expression analysis, DNA-damage repair, platin-based chemotherapy.
Introduction
Malignant pleural mesothelioma (MPM) is a rare, predominantly asbestos-related and biologically highly aggressive tumour leading to a dismal prognosis [1, 2]. In the US, approximately 2500 new cases of mesothelioma are diagnosed each year and the incidence of mesothelioma is expected to decline steadily [1, 3, 4]. In contrast, the incidence of mesothelioma in Europe continues to rise. Its peak is expected in the next two decades and may account for as many as 250,000 European deaths in the next 35 years [1, 2, 5-7].
Multimodality therapy consisting of chemotherapy, surgery and/or radiotherapy is centred on surgical resection in early stages. In clinical practice, the antifolate pemetrexed is used in combination with cisplatin [8] or carboplatin [9-12]. Cisplatin resulted in a response rate of merely 14% and a median survival of below 7 months [13]. Carboplatin resulted in similar response rates ranging from 6 to 16% [13, 14]. The reasons for the rather poor efficacy of platinum-compounds remain largely unknown.
Platinum cytotoxicity is based on forming bulky DNA adducts by chemically altering DNA bases by covalent binding of platinum [12], leading to both DNA inter- and (1,2 or 1,3)-intra-strand cross-linking [15-22]. Platinum-compounds prevent normal cell replication and trigger apoptosis [17, 21, 23], unless adducts from genomic DNA are repaired [20]. Even a single DNA cross-link, if not repairable, can be lethal [24]. There are also several local treatment modalities that have been used such as gene therapy, zoledronic acid and photodynamic therapy [25-30].
Nucleotide excision repair (NER) is capable of removing numerous types of DNA helix-distorting lesions, which are induced by platinum [15, 16, 31]. The endonuclease excision repair cross-complementing 1 (ERCC1) performs an essential late step in the NER process, where it nicks the damaged DNA strand at the 5' site of the helix-distorting lesion [31], and is the rate-limiting member of the NER pathway [15, 16, 20].
In contrast, mismatch repair (MMR) is one of the major DNA repair pathways, which is responsible for the repair of single-base or nucleotide mismatches and insertion-deletion loops that results from slippage during replication of repetitive sequences or during recombination [32-34]. The MutS homologue 2 (MSH2) protein belongs to the MMR pathway and is crucially involved in the repair of DNA cross-links. MSH2 also recognizes and binds to platinum-induced DNA interstrand cross-links, thereby initiating their excision and repair [24] whilst building a MSH2/MSH6 heterodimer [35, 36]. Defects in DNA MMR have been shown to be a mechanism of resistance to cisplatin both in vivo and in vitro [15, 37].
Base excision repair (BER) corrects small base alterations without distorting the DNA helix structure [38-40] by using two distinct ways. In the short patch pathway a single base replacement is performed by DNA polymerase b (POLB), DNA ligase3 and the X-ray cross complementing group 1 (XRCC1) protein, the latter as a scaffold protein to form the complex [41]. In the long patch pathway a DNA synthesis of multiple nucleotides (2-10 bases) occurs by the DNA pold/e, proliferating cell nuclear antigen (PCNA), the flap endonuclease 1 (FEN1) and DNA ligase1 [41]. Interestingly, BER components are also used to repair single-stranded DNA breaks (SSB), whereat XRCC1 and poly (ADP-ribose) polymerase (PARP1) serve as SSB sensors [38, 42]. As DNA lesions occurring in mesothelioma are in majority single strand breaks, the BER pathway plays a crucial role in survival of these cells [43, 44]. Further important pathways are the recombination repair system (Homologous recombination repair (HR), Non-homologous end-joining (NHEJ) and Fanconi anemia (FA) pathway) and the downstream target cascade that contributes to HR, including RAD52, RAD54, RPA and Breast Cancer 1 and 2 (BRCA1/2) [38, 45, 46]. These are of high interest for the understanding of the resistance mechanism to platin-compounds. Although impaired DNA repair could promote mutagenesis, an intact HR repair system may increase resistance to radiotherapy and chemotherapy [47].
In consideration of these aspects we aimed to investigate the impact of DNA-damage response and repair key players on platin-compound based therapy. Therefore, we analysed twenty-four MPM patients, whereof twelve underwent adjuvant and twelve neoadjuvant treatments. The selection of targets was based on recent literature and preliminary results of our group.
Material and Methods
Study Cohort and Experimental Design
For this exploratory mRNA study, twenty-four formalin-fixed paraffin-embedded (FFPE) tumour specimens were screened. Tumour classification was based on the WHO Classification Of Tumours guidelines (2004) [48] and TNM-staging was based on the UICC Classification of Malignant Tumours [49]. Those were confirmed by two experienced pathologists (JWO, KWS). The study included only MPM patients treated at the West German Cancer Centre or the West German Lung Centre between 2005 and 2009. All patients received platinum-based chemotherapy. Half of the collected samples were taken in an adjuvant and the other half in a neoadjuvant situation. The clinical stage was determined according to the criteria of the International Mesothelioma Interest Group (IMIG) tumour-node-metastasis staging system for MPM [50]. Radiologic response rate was assessed by modified Response Evaluation Criteria in Solid Tumors (modRECIST), which have been validated in MPM [51, 52]. Clinicopathological data including age, gender, histology and TNM are summarized in table 1. Surveillance for this study was stopped on August 31, 2014. Complete follow up was available for all patients with reported deaths in more than 96% (23/24). Progression under therapy was observed in approximately 80% (19/24) of patients. The retrospective study was approved by the Ethics Committee of the Medical Faculty of the University Duisburg-Essen (identifier: 14-5775-BO). The investigation conforms to the principles outlined in the declaration of Helsinki.
RNA Isolation and RNA Quantity Assessment
Three to five 10µm thick paraffin sections per sample were de-paraffinised using xylene prior to total RNA isolation including miRNAs. RNA was isolated by using the miRNeasy FFPE kit (Qiagen, Venlo, Netherlands) as recommended by the supplier except for proteinase K digestion performed overnight. Total RNA quantity was assessed using a Qubit flouormetric quantification system and Broad Range RNA Assay Kit (Thermo Fisher Scientific, MA, USA).
CodeSet Design
Based on preliminary experiments, recent literature and in silico predicted miRNA-targets, a total of 366 mRNAs (including reference genes) were investigated using a Custom CodeSet from NanoString. The nCounter standard chemistry was used and the preparation was carried out as recommended by the manufacturer. For each sample, 200 ng total RNA were processed. The high-sensitivity protocol was chosen; the cartridge was measurement at 555 fields of view.
NanoString Data Processing and Statistical Analysis
All statistical analyses were calculated using the R i386 statistical programming environment (v3.2.3). A technical normalization of the counts was done by subtracting the mean counts plus two-times standard deviation from the CodeSet inherent negative controls. Afterwards, a biological normalization using the included reference genes was performed [53-55]. In brief, the gene expression stability measures (M) were calculated by using the NormFinder algorithm and all sample counts were normalized against the geometric mean of the normalization gene counts. ACTB, MAPK14 and TCEB1 were identified as most stable reference genes.
Overview of therapeutic scheme, clinicopathological parameters as well as clinical outcome for each patient
| Therapy-Concept | Therapy-Regimen | Age (Years) | Gender | T-Stage | N-Status | M-Status | IMIG-Stage | Time to Death (Months) | Time to Progress (Months) | Progression | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| adjuvant | CARBO+PEM | 76 | m | 2 | 0 | 0 | 2 | 44,2 | 14,3 | Yes | Death |
| adjuvant | CARBO+PEM | 80 | m | 3 | 2 | 0 | 3 | 13,0 | 6,9 | Yes | Death |
| adjuvant | CIS+PEM | 51 | m | x | x | x | x | 8,0 | 6,2 | Yes | Death |
| adjuvant | CIS+PEM | 52 | m | 2 | 2 | 0 | 3 | 9,3 | 5,5 | Yes | Death |
| adjuvant | CIS+PEM | 56 | m | 3 | 0 | 1 | 4 | 43,2 | 5,5 | No | Death |
| adjuvant | CIS+PEM | 61 | m | 2 | 2 | 1 | 4 | 3,1 | 1,2 | Yes | Death |
| adjuvant | CIS+PEM | 65 | m | 2 | 2 | 0 | 3 | 8,8 | 4,9 | Yes | Death |
| adjuvant | CIS+PEM | 68 | f | 2 | 0 | 1 | 4 | 3,7 | 3,5 | No | Death |
| adjuvant | CIS+PEM | 70 | m | 1 | 2 | 0 | 3 | 14,5 | 6,7 | Yes | Death |
| adjuvant | CIS+PEM | 73 | m | 2 | 0 | 0 | 2 | 18,0 | 4,8 | Yes | Death |
| adjuvant | CIS+PEM | 75 | m | 3 | 0 | 0 | 3 | 21,7 | 6,4 | Yes | Death |
| adjuvant | CIS+PEM | 77 | m | 2 | 0 | 0 | 2 | 5,6 | 3,8 | Yes | Death |
| neoadjuvant | CIS+CARBO+PEM | 69 | m | 1 | 0 | 0 | 1 | 7,1 | 7,7 | No | Death |
| neoadjuvant | CIS+PEM | 54 | m | 3 | 2 | 1 | 4 | 13,8 | 4,1 | Yes | Alive |
| neoadjuvant | CIS+PEM | 56 | m | 2 | 0 | 0 | 2 | 37,0 | 23,1 | Yes | Death |
| neoadjuvant | CIS+PEM | 58 | m | 3 | 1 | 0 | 3 | 18,5 | 9,3 | Yes | Death |
| neoadjuvant | CIS+PEM | 58 | m | 2 | 0 | 0 | 2 | 6,7 | 2,8 | Yes | Death |
| neoadjuvant | CIS+PEM | 61 | m | 3 | 0 | 0 | 3 | 25,2 | 11,6 | Yes | Death |
| neoadjuvant | CIS+PEM | 62 | m | 2 | 2 | 0 | 3 | 41,5 | 34,7 | Yes | Death |
| neoadjuvant | CIS+PEM | 62 | m | 2 | 0 | 0 | 2 | 29,9 | 10,1 | Yes | Death |
| neoadjuvant | CIS+PEM | 62 | m | 2 | 2 | 0 | 3 | 4,4 | 4,0 | Yes | Death |
| neoadjuvant | CIS+PEM | 69 | m | 3 | 0 | 0 | 3 | 42,6 | 40,8 | No | Death |
| neoadjuvant | CIS+PEM | 69 | f | 2 | 0 | 0 | 2 | 27,3 | 9,7 | Yes | Death |
| neoadjuvant | CIS+PEM | 73 | m | 4 | 2 | 0 | 4 | 24,0 | 5,9 | No | Death |
CARBO … Carboplatin; CIS … Cisplatin; PEM…Pemetrexed.
For dichotomous variables (e.g. gender) the Wilcoxon Mann-Whitney rank sum test was used. Correlations between mRNA-expression and TNM-criteria were tested by using the Spearman's rank correlation test. Kaplan-Meier analysis was done for the assessment of associations between gene expression and progression-free survival (PFS) or overall survival (OS). Significant differences in PFS or OS between groups were verified by COXPH-model using Wald-test, likelihood-ratio test and Score (logrank) test.
The level of statistical significance was defined as p≤0.05.
Results
Overall Expression of Targets
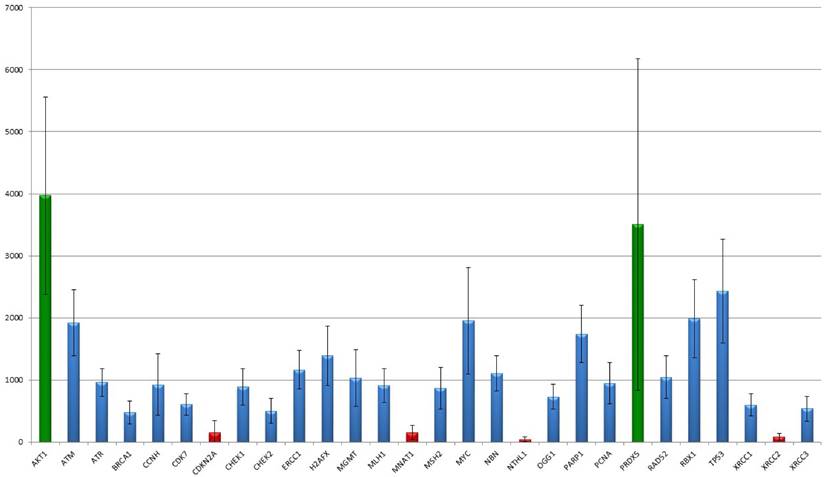
The 366 analysed targets were narrowed down by their profile resulting in 30 members important for DNA-damage recognition and repair. In their overall expression, two markers stick out as they show a high expression of more than 2,500 median detected counts (AKT1: median counts 3513, range 1,804-7,493; PRDX5: median counts 2,873, range 417-10,850). Seven markers showed low expression less than 500 counts in median (BRCA1, CDC25A, CDKN2A, MNAT1, NTHL1, TP73, XRCC2), in particular NTHL1 (median: 37 counts; range 0-113) and XRCC2 (median: 68 counts; range 0-194) show a basal to absent gene expression but still differences between the tumours were detected. The remaining 21 targets revealed medial count numbers, ranging from 500 to 2,500 median counts. Median, mean and range of all 30 markers are summarized in table 2 and results are graphically illustrated in figure 1.
Correlation to Clinicopathological Data
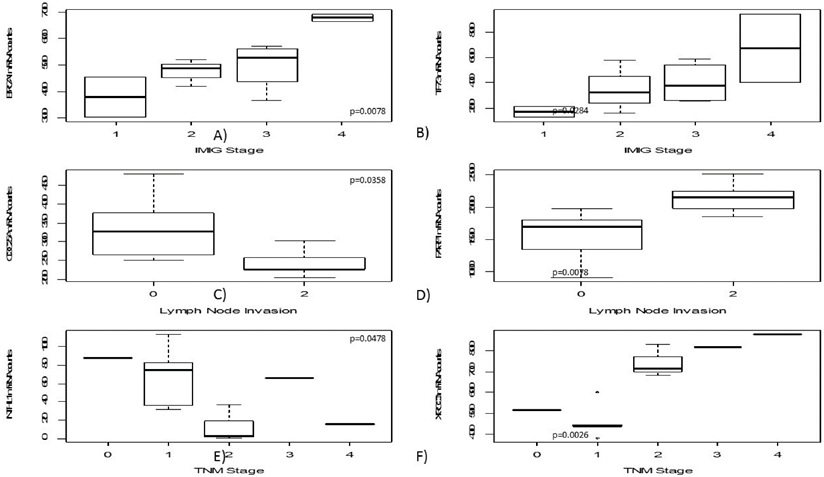
High mRNA levels of BRCA1 and TP73 were significantly correlated to higher IMIG-Stage (p=0.0067; p= 0.0284). Furthermore, a low CDC25A gene expression was correlated to lymph node invasion (p=0.0358). In contrast, high PARP1 expression levels were significantly associated with lymph node spread (p=0.0078).
Similarly, lower NTHL1 gene expression (p=0.0478) and higher XRCC3 gene expression levels were significantly associated with higher T-stage (p=0.0026). An increase of gene expression during platin-based chemotherapy was determined for H2AFX (p=0.0100), PCNA (p=0.0242) and XRCC1 (p=0.0387).
With increasing patients age, MSH2 (p=0.0267), RBX1 (p=0.0390) as well as XRCC1 (p=0.0030) expression decreased. No correlation between one of the investigated markers and the appearance of distant metastasis was found. No significant association to patient's gender or storage duration of the analysed paraffin tissues was detected.
Results are summarized and illustrated in figure 2.
Summary of median count number, mean count number and count range of each measured target. Of note, AKT1, PRDX5 and TP53 show outstanding high, and BRCA1, CDC25A, CDKN2A, MNAT1, NTHL1, TP73 and XRCC2 a low to basal gene expression.
| Median | Mean | Minimum | Maximum | |
|---|---|---|---|---|
| AKT1 | 3,513 | 3,969 | 1,804 | 7,493 |
| ATM | 1,920 | 1,921 | 758 | 3,128 |
| ATR | 1,006 | 960 | 456 | 1,291 |
| BRCA1 | 480 | 479 | 0 | 900 |
| CCNH | 739 | 925 | 206 | 2,066 |
| CDC25A | 278 | 275 | 31 | 633 |
| CDK7 | 607 | 580 | 213 | 939 |
| CDKN2A | 108 | 153 | 0 | 890 |
| CHEK1 | 830 | 887 | 332 | 1,654 |
| CHEK2 | 446 | 501 | 195 | 1,120 |
| ERCC1 | 1,191 | 1,164 | 639 | 1,965 |
| H2AFX | 1,284 | 1,389 | 762 | 2,497 |
| MGMT | 934 | 1,026 | 223 | 2,012 |
| MLH1 | 924 | 911 | 213 | 1,541 |
| MNAT1 | 127 | 155 | 0 | 388 |
| MSH2 | 834 | 871 | 427 | 2,021 |
| MYC | 1,724 | 1,952 | 1,001 | 4,821 |
| NBN | 1,091 | 1,110 | 470 | 1,696 |
| NTHL1 | 37 | 46 | 0 | 113 |
| OGG1 | 767 | 730 | 313 | 1,234 |
| PARP1 | 1,769 | 1,740 | 912 | 2,899 |
| PCNA | 809 | 948 | 377 | 1,540 |
| PRDX5 | 2,694 | 2,996 | 1,043 | 5,678 |
| RAD52 | 1,013 | 1,045 | 320 | 1,915 |
| RBX1 | 1,817 | 1,985 | 938 | 3,551 |
| TP53 | 2,446 | 2,435 | 897 | 4,128 |
| TP73 | 327 | 347 | 0 | 943 |
| XRCC1 | 589 | 599 | 230 | 969 |
| XRCC2 | 68 | 84 | 0 | 194 |
| XRCC3 | 503 | 536 | 0 | 900 |
Associations to Clinical Outcome
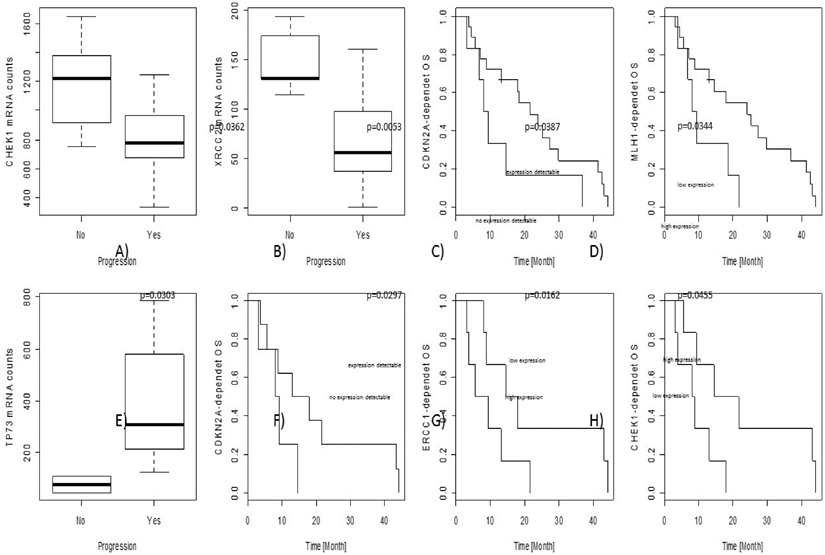
In the overall cohort, associations between gene expression level and objective tumour progression were found for CHEK1 (p=0.0362) and XRCC2 (p=0.0053). No associations with PFS were observed; nevertheless, CDKN2A (p=0.0387) as well as MLH1 (p=0.0344) gene expression levels influenced OS independently.
In the cohort of adjuvant treated patients, some predictive and prognostic markers were identified. Increased TP73 gene expression was significantly associated with progression of the tumour during therapy (p=0.0303). CDKN2A (p=0.0297) as well as CHEK1 (p=0.0162) and ERCC1 (p=0.0455) gene expression levels were significantly correlated to OS.
Results are graphically illustrated in figure 3 and a list of all significant p-values is given in table 3.
The figure shows the distribution of NanoString mRNA counts over for each target. AKT1 as well as PRDX5 showed the strongest gene expression level. By the way, the disparities of PRDX5 are notable ranging between 1,000 and 5,800 counts. CDKN2A, MNAT1, NTHL1 and XRCC2 show the lowest mean expression levels. Interestingly, for both CDKN2A and XRCC2 the standard deviation clearly exceeds the median count number, indicating differences between the samples.
In this figure boxplots regarding clinicopathological parameters are shown, including IMIG-stage (A+B), N-status (C+D) as well as TNM-stage (E+F). Both BRCA1 and TP73 show increasing gene expression with higher IMIG-stage. CDC25A gene expression decreases in lymph-metastatic tumours whereas PARP1 gene expression increases in those. Interestingly, strong XRCC3 gene expression was just found in pT2 or higher tumours.
Prognostic and predictive results are illustrated in the figure. The upper line (A-D) show results from the overall collective, the lower line (E-H) results from the adjuvant cohort only. In particular, CDKN2A, ERCC1 and CHEK1 seem to be predictive markers for platin-response in the adjuvant-treated patients, gaining a real survival benefit.
List of all significant p-values after Bonferroni-correction for multiple testing. For the adjuvant treated subgroup only, just predictive parameters as survival or progression were tested.
| Gene | Variable | Test-Method | p-Value | Likelihood ratio test | Wald test | Score (logrank) test | Z | rho | |
|---|---|---|---|---|---|---|---|---|---|
| Overall Cohort | CDC25A | N-Status | Spearman's rank correlation rho | 0,0358 | - | - | - | - | -0,6351 |
| PARP1 | N-Status | Spearman's rank correlation rho | 0,0078 | - | - | - | - | 0,7506 | |
| BRCA1 | IMIG-Stage | Spearman's rank correlation rho | 0,0078 | - | - | - | - | 0,7505 | |
| TP73 | IMIG-Stage | Spearman's rank correlation rho | 0,0284 | - | - | - | - | 0,6561 | |
| NTHL1 | TNM-Stage | Spearman's rank correlation rho | 0,0478 | - | - | - | - | -0,6068 | |
| XRCC3 | TNM-Stage | Spearman's rank correlation rho | 0,0026 | - | - | - | - | 0,8090 | |
| CDK7 | Age | Spearman's rank correlation rho | 0,0450 | - | - | - | - | -0,4148 | |
| MSH2 | Age | Spearman's rank correlation rho | 0,0267 | - | - | - | - | -0,4548 | |
| OGG1 | Age | Spearman's rank correlation rho | 0,0148 | - | - | - | - | -0,4957 | |
| PRDX5 | Age | Spearman's rank correlation rho | 0,0301 | - | - | - | - | -0,4461 | |
| RBX1 | Age | Spearman's rank correlation rho | 0,0390 | - | - | - | - | -0,4261 | |
| XRCC1 | Age | Spearman's rank correlation rho | 0,0030 | - | - | - | - | -0,5887 | |
| H2AFX | Therapy | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0100 | - | - | - | 2,5403 | - | |
| MYC | Therapy | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0205 | - | - | - | -2,3094 | - | |
| PCNA | Therapy | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0242 | - | - | - | 2,2517 | - | |
| XRCC1 | Therapy | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0387 | - | - | - | 2,0785 | - | |
| CHEK1 | Progression | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0362 | - | - | - | -2,0969 | - | |
| XRCC2 | Progression | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0053 | - | - | - | -2,6679 | - | |
| - | PFS | Cox Proportional Hazard Model | - | - | - | - | - | - | |
| CDKN2A | OS | Cox Proportional Hazard Model | - | 0,0387 | 0,0263 | 0,0214 | - | - | |
| MLH1 | OS | Cox Proportional Hazard Model | - | 0,0344 | 0,0382 | 0,0401 | - | - | |
| Adjuvant Cohort | TP73 | Progression | Exact Wilcoxon Mann-Whitney Rank Sum Test | 0,0303 | - | - | - | 2,1483 | - |
| - | PFS | Cox Proportional Hazard Model | - | - | - | - | - | - | |
| CDKN2A | OS | Cox Proportional Hazard Model | - | 0,0297 | 0,0433 | 0,0460 | - | - | |
| CHEK1 | OS | Cox Proportional Hazard Model | - | 0,0162 | 0,0246 | 0,0283 | - | - | |
| ERCC1 | OS | Cox Proportional Hazard Model | - | 0,0455 | 0,0479 | 0,0458 |
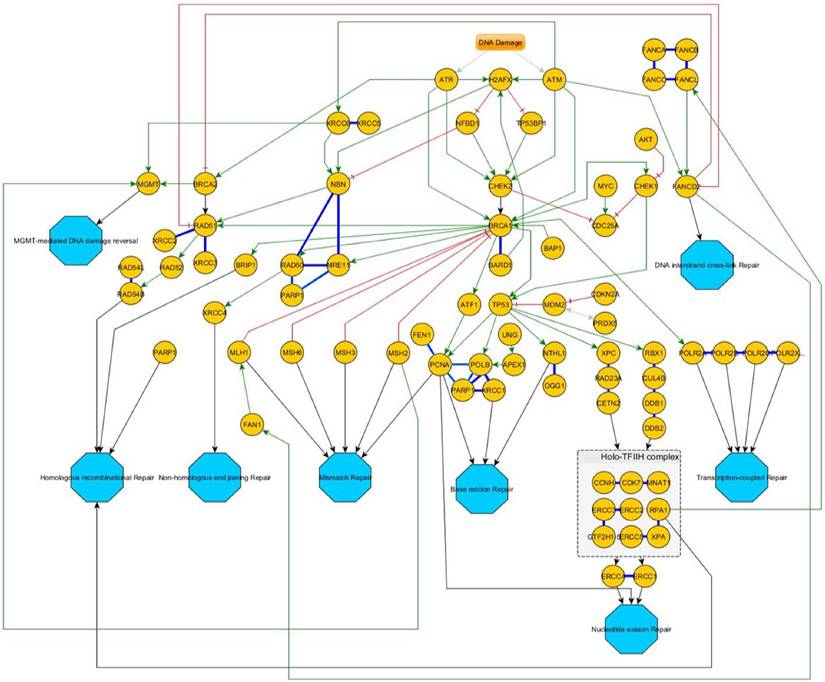
Overview of the DNA-damage response pathway-network. After an initial stress signal (DNA-damage), ATR and ATM transmit these signals downstream over CHEK1/2 to BRCA1/2 and then distribute to the different pathways. Green Arrows indicate activating signals; red cross-marks indicate inhibitory signals. Blue bold lines indicate complexes. If a protein is involved in different independent complexes, different shades of bold-blue lines indicate the different complexes. Turquoise octagons indicate the different pathway end-points.
Discussion
We present a study focussing on a mRNA screening for DNA-damage response associated targets aiming to understand the mechanisms of platinum-based therapy-induced apoptosis and to identify possible predictive markers.
An overview of the corresponding DNA-repair pathways is given in figure 4.
Besides pemetrexed, platinum-compounds are standard chemotherapeutic agents for the treatment of MPM [24]. Besides, Platinum-containing regimens have a greater activity than non-platinum containing combinations in MPM [56].
Several studies have investigated carboplatin and cisplatin as single-agent chemotherapy. Cisplatin resulted in a response rate of merely 14% and a median survival of below 8 months [13, 14]. Carboplatin resulted in similar response rates ranging from 6 to 16% [13, 14]. The reasons for the rather poor efficacy of platinum-compounds remain largely unknown.
DNA-Damage Recognition and TP53 Inhibitory Network
DNA double-strand breaks (DSBs) and DNA replication stress activate the ATM signalling pathways, which transduce the signal to downstream ATR/CHEK1 pathway [57, 58]. CHEK1 directly activates TP53 and thereby triggers TP53-induced apoptosis and senescence in response to DNA damage. Previous studies showed that cells lacking CHEK1 were more sensitive to gemcitabine in a TP53-independent manner [58, 59]. To date, the role of the ATM/CHEK2 pathway in cells treated with platin-containing compounds is unknown [58]. Our results confirm this relationship. CHEK1 was associated with both progression during therapy and OS in the adjuvant treated sub-cohort. Therefore, CHEK1 gene expression seems to be a strong predictive marker for patients' response and outcome under platin-based chemotherapy. We recommend validation of this result in a larger prospective study to bring CHEK1 to a presumable clinical application.
Of note, a direct inhibition of CHEK1 by small molecules already showed to promising results in vitro in breast cancer [60], in vivo in hepatic carcinoma [61] as well as in mesothelioma [62].
In contrast to other solid tumours, mutations of the TP53 gene are extremely rare in MPM, so other mechanisms such as deletion of the locus or methylation contribute to inactivation of TP53 [63-66]. For instance, overexpression of MDM2 can lead to a loss of TP53 regulatory function in cancer cells by increased proteasomal degradation of TP53 [67-72]. Gene amplification can lead to increased MDM2 protein expression. However, many tumours present with high MDM2 protein levels without an increased gene copy number [73-77].
In previous studies, we have demonstrated that MDM2 is overexpressed in approximately 20-30% of MPM and this finding was restricted to epitheloid MPM or the epithelioid components of biphasic MPM [78, 79]. Moreover, patients with MDM2-positive MPM presented with a significantly decreased OS compared to MDM2-negative MPM [78, 79]. The physiological inhibitor of MDM2 is P14/ARF and loss of P14/ARF activity may have a similar effect as loss of TP53 [80-83]. P14/ARF may control TP53 transcription, represses TP53 degradation that is not MDM2-mediated and stimulates TP53 activity [84]. Additionally, loss of P14/ARF activity seems to occur in a reciprocal manner to TP53 loss and seems to be typical for tumours that are TP53 wild type [83]. In a previous study, we found CDKN2A (gene locus coding for P14/ARF) gene expression associated with OS and PFS in platinum-treated MPM patients [79]. In concordance, CDKN2A gene expression is significantly associated with OS in this cohort, and additionally in the adjuvant cohort only. This indicates that CDKN2A gene expression could be used as marker for platin-response prediction. Additionally, TP73 shows an increasing expression with increasing IMIG-stages and associates significantly with progression in pleural mesothelioma.
Nucleotide Excision Repair (NER), Mismatch Repair (MMR) and Base Excision Repair (BER)
The impact of ERCC1 expression on treatment response in MPM is discussed controversially. Recent studies have suggested that the expression levels of DNA-damage repair enzyme-related genes, including ERCC1 mRNA, are associated with an overall survival benefit during cisplatin-based chemotherapy in MPM [12, 37, 51]. Yet, investigation of ERCC1 protein levels in a phase II clinical trial revealed a significant correlation between negative ERCC1 status and long PFS in a cohort of 54 MPM patients undergoing treatment with cisplatin and vinorelbine [21]. Our findings are in line with these earlier studies, as ERCC1 gene expression level clearly impact the OS rate, but influence neither PFS nor progression.
During the recombinational repair processing of interstrand cross-links, MSH2 cooperates with several components of DNA damage repair pathways, including ERCC1 [24]. MutL homologue 1 (MLH1) protein is also a key component in the MMR pathway being involved in mismatch strand excision and subsequent repair. In a previous study, we showed that MLH1 protein levels were associated with PFS [37]. Additionally, in the recent study MHL1 gene expression was found as a prominent marker for OS.
As DNA lesions occurring in mesothelioma are in majority single strand breaks (SSBs), the BER pathway plays a crucial role in survival of these cells [43, 44]. Several studies have investigated BER family members in tumorigenesis and pathophysiology of MPM [41, 62, 85-90]. In particular, PARP1 was upregulated and activated in human mesothelial cells exposed to asbestos [87, 88]. PARP1 inhibition resulted in an accumulation of SSB lesions induced by direct asbestos exposure in human mesothelial cells [90]. Interestingly, beside its role in DNA repair, PARP1 has been found to induce apoptosis and cell death following asbestos exposure [87-89, 91]. In this study, the analysed tumour samples formed two distinct expression patterns with respect to PARP1 gene expression levels. Interestingly, high PARP1 expression associated with lymph node invasion.
Recombination Repair System
Homologue recombination (HR) and non-homologous end-joining (NHEJ) are of great importance, since DNA DSBs induce chromosomal deletions and translocations which are common in MPM [45, 92, 93]. Upregulation of various genes encoding HR protein members, including RAD50, RAD54L, RAD21, BRCA1 and BRCA2 could be detected by gene expression analysis in MPM [62, 94]. Our results indicate that a higher gene expression of BRCA1 is highly associated with increasing clinical malignancy (indicated by higher IMIG-stage). Furthermore, a reduced XRCC2 gene expression, acting downstream of the MRN-complex [95], correlated with with progressive disease more often. Similarly, increased XRCC3 gene expression, forming a complex together with XRCC2, was observed in tumours showing higher TNM-stage.
Cellular Response to Platin-Induced DNA-Damage
DNA damage, e.g. induced by platin-compounds, is first recognized by ATM and ATR. This leads to a subsequent phosphorylation of histone H2AFX, activating the different downstream cascades of cellular DNA damage response [96, 97]. Deregulation of H2AFX expression and modification has been reported to be associated with a variety of human malignancies [98-103]. Surprisingly, studies exploring H2AFX expression in MPM are still lacking. In this study, we observed an increase of expression induced by platinum-based chemotherapy, leading to the assumption that activation of H2AFX expression plays a crucial role for early DNA damage response. Furthermore, PCNA and XRCC1, forming a central complex together with the DNA polymerase beta (POLB) in BER [41], show the same increase during therapy, indicating the activation of H2AFX downstream targets during the accumulation of DNA SSBs and DSBs.
Conclusion
DNA damage response plays a crucial role in response to platin-based chemotherapeutic regimes. In particular, CHEK1, XRCC2 and TP73 are strongly associated with tumour progression. Therefore, we suggest that these markers are used along with staging and restaging. MLH1, CHEK1, ERCC1 and most promising CDKN2A are prognostic markers for OS in MPM. Therefore, we suggest that these markers are at staging and upon diagnosis of MPM. TP73, CDKN2A, CHEK1 and ERCC1 seem to be also predictive markers in adjuvant treated MPMs. Those should be validated prospectively, hopefully finding their way into clinical and pathological practice, leading to an enhanced clinical management and improved clinical outcome.
Conflicts of interest
All authors state that they have no conflicts of interest to declare.
References
1. Goudar RK. Review of pemetrexed in combination with cisplatin for the treatment of malignant pleural mesothelioma. Therapeutics and clinical risk management. 2008;4:205-11
2. Liu Z, Klominek J. Regulation of matrix metalloprotease activity in malignant mesothelioma cell lines by growth factors. Thorax. 2003;58:198-203
3. Weill H, Hughes JM, Churg AM. Changing trends in US mesothelioma incidence. Occupational and environmental medicine. 2004;61:438-41
4. Hazarika M, White RM Jr, Booth BP, Wang YC, Ham DY, Liang CY. et al. Pemetrexed in malignant pleural mesothelioma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:982-92
5. van Ruth S, Baas P, Zoetmulder FA. Surgical treatment of malignant pleural mesothelioma: a review. Chest. 2003;123:551-61
6. Vorobiof DA, Mafafo K. Malignant pleural mesothelioma: medical treatment update. Clinical lung cancer. 2009;10:112-7 doi:10.3816/CLC.2009.n.014
7. Lee AY, Raz DJ, He B, Jablons DM. Update on the molecular biology of malignant mesothelioma. Cancer. 2007;109:1454-61 doi:10.1002/cncr.22552
8. Herndon JE, Green MR, Chahinian AP, Corson JM, Suzuki Y, Vogelzang NJ. Factors predictive of survival among 337 patients with mesothelioma treated between 1984 and 1994 by the Cancer and Leukemia Group B. Chest. 1998;113:723-31
9. Ceresoli GL, Zucali PA, Favaretto AG, Grossi F, Bidoli P, Del Conte G. et al. Phase II study of pemetrexed plus carboplatin in malignant pleural mesothelioma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:1443-8 doi:10.1200/JCO.2005.04.3190
10. Ceresoli GL, Zucali PA, Mencoboni M, Botta M, Grossi F, Cortinovis D. et al. Phase II study of pemetrexed and carboplatin plus bevacizumab as first-line therapy in malignant pleural mesothelioma. British journal of cancer. 2013;109:552-8 doi:10.1038/bjc.2013.368
11. Ceresoli GL, Castagneto B, Zucali PA, Favaretto A, Mencoboni M, Grossi F. et al. Pemetrexed plus carboplatin in elderly patients with malignant pleural mesothelioma: combined analysis of two phase II trials. British journal of cancer. 2008;99:51-6 doi:10.1038/sj.bjc.6604442
12. Zucali PA, Giovannetti E, Destro A, Mencoboni M, Ceresoli GL, Gianoncelli L. et al. Thymidylate synthase and excision repair cross-complementing group-1 as predictors of responsiveness in mesothelioma patients treated with pemetrexed/carboplatin. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:2581-90 doi:10.1158/1078-0432.CCR-10-2873
13. Tomek S, Manegold C. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. Lung Cancer. 2004;45(Suppl 1):S103-19 doi:10.1016/j.lungcan.2004.04.020
14. Tomek S, Emri S, Krejcy K, Manegold C. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. British journal of cancer. 2003;88:167-74 doi:10.1038/sj.bjc.6600673
15. Rosell R, Taron M, Barnadas A, Scagliotti G, Sarries C, Roig B. Nucleotide excision repair pathways involved in Cisplatin resistance in non-small-cell lung cancer. Cancer control: journal of the Moffitt Cancer Center. 2003;10:297-305
16. Olaussen KA, Mountzios G, Soria JC. ERCC1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Current opinion in pulmonary medicine. 2007;13:284-9 doi:10.1097/MCP.0b013e32816b5c63
17. Booton R, Ward T, Ashcroft L, Morris J, Heighway J, Thatcher N. ERCC1 mRNA expression is not associated with response and survival after platinum-based chemotherapy regimens in advanced non-small cell lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2007;2:902-6 doi:10.1097/JTO.0b013e318155a637
18. Friboulet L, Barrios-Gonzales D, Commo F, Olaussen KA, Vagner S, Adam J. et al. Molecular Characteristics of ERCC1-Negative versus ERCC1-Positive Tumors in Resected NSCLC. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:5562-72 doi:10.1158/1078-0432.CCR-11-0790
19. Fujii T, Toyooka S, Ichimura K, Fujiwara Y, Hotta K, Soh J. et al. ERCC1 protein expression predicts the response of cisplatin-based neoadjuvant chemotherapy in non-small-cell lung cancer. Lung Cancer. 2008;59:377-84 doi:10.1016/j.lungcan.2007.08.025
20. Hubner RA, Riley RD, Billingham LJ, Popat S. Excision repair cross-complementation group 1 (ERCC1) status and lung cancer outcomes: a meta-analysis of published studies and recommendations. PloS one. 2011;6:e25164. doi:10.1371/journal.pone.0025164
21. Zimling ZG, Sorensen JB, Gerds TA, Bech C, Andersen CB, Santoni-Rugiu E. Low ERCC1 expression in malignant pleural mesotheliomas treated with cisplatin and vinorelbine predicts prolonged progression-free survival. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2012;7:249-56 doi:10.1097/JTO.0b013e318233d6a9
22. Bhagwat NR, Roginskaya VY, Acquafondata MB, Dhir R, Wood RD, Niedernhofer LJ. Immunodetection of DNA repair endonuclease ERCC1-XPF in human tissue. Cancer research. 2009;69:6831-8 doi:10.1158/0008-5472.CAN-09-1237
23. Kerr KM. Personalized medicine for lung cancer: new challenges for pathology. Histopathology. 2012;60:531-46 doi:10.1111/j.1365-2559.2011.03854.x
24. Kamal NS, Soria JC, Mendiboure J, Planchard D, Olaussen KA, Rousseau V. et al. MutS homologue 2 and the long-term benefit of adjuvant chemotherapy in lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:1206-15 doi:10.1158/1078-0432.CCR-09-2204
25. Tada Y, Hiroshima K, Shimada H, Shingyoji M, Suzuki T, Umezawa H. et al. An intrapleural administration of zoledronic acid for inoperable malignant mesothelioma patients: a phase I clinical study protocol. SpringerPlus. 2016;5:195. doi:10.1186/s40064-016-1893-2
26. Kidane B, Hirpara D, Yasufuku K. Photodynamic Therapy in Non-Gastrointestinal Thoracic Malignancies. International journal of molecular sciences. 2016:17 doi:10.3390/ijms17010135
27. Porpodis K, Zarogoulidis P, Boutsikou E, Papaioannou A, Machairiotis N, Tsakiridis K. et al. Malignant pleural mesothelioma: current and future perspectives. Journal of thoracic disease. 2013;5(Suppl 4):S397-406 doi:10.3978/j.issn.2072-1439.2013.08.08
28. Zarogoulidis K, Zarogoulidis P, Darwiche K, Tsakiridis K, Machairiotis N, Kougioumtzi I. et al. Malignant pleural effusion and algorithm management. Journal of thoracic disease. 2013;5(Suppl 4):S413-9 doi:10.3978/j.issn.2072-1439.2013.09.04
29. Zarogoulidis P, Domvri K, Huang H, Zarogoulidis K. Gene therapy for lung cancer malignant pleural effusion: current and future nano-biotechnology. Translational lung cancer research. 2012;1:234-7 doi:10.3978/j.issn.2218-6751.2012.08.01
30. Zarogoulidis P, Chatzaki E, Hohenforst-Schmidt W, Goldberg EP, Galaktidou G, Kontakiotis T. et al. Management of malignant pleural effusion by suicide gene therapy in advanced stage lung cancer: a case series and literature review. Cancer gene therapy. 2012;19:593-600 doi:10.1038/cgt.2012.36
31. Cobo M, Isla D, Massuti B, Montes A, Sanchez JM, Provencio M. et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:2747-54 doi:10.1200/JCO.2006.09.7915
32. Martin SA, McCarthy A, Barber LJ, Burgess DJ, Parry S, Lord CJ. et al. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO molecular medicine. 2009;1:323-37 doi:10.1002/emmm.200900040
33. Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y. et al. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer cell. 2010;17:235-48 doi:10.1016/j.ccr.2009.12.046
34. Lo YL, Hsiao CF, Jou YS, Chang GC, Tsai YH, Su WC. et al. Polymorphisms of MLH1 and MSH2 genes and the risk of lung cancer among never smokers. Lung Cancer. 2011;72:280-6 doi:10.1016/j.lungcan.2010.10.009
35. Vageli D, Daniil Z, Dahabreh J, Karagianni E, Vamvakopoulou DN, Ioannou MG. et al. Phenotypic mismatch repair hMSH2 and hMLH1 gene expression profiles in primary non-small cell lung carcinomas. Lung Cancer. 2009;64:282-8 doi:10.1016/j.lungcan.2008.09.018
36. Kim M, Kang HG, Lee SY, Lee HC, Lee EB, Choi YY. et al. Comprehensive analysis of DNA repair gene polymorphisms and survival in patients with early stage non-small-cell lung cancer. Cancer science. 2010;101:2436-42 doi:10.1111/j.1349-7006.2010.01699.x
37. Ting S, Mairinger FD, Hager T, Welter S, Eberhardt WE, Wohlschlaeger J. et al. ERCC1, MLH1, MSH2, MSH6, and betaIII-tubulin: resistance proteins associated with response and outcome to platinum-based chemotherapy in malignant pleural mesothelioma. Clinical lung cancer. 2013;14:558-67 e3. doi:10.1016/j.cllc.2013.04.013
38. Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366-74 doi:10.1038/35077232
39. Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutation research. 2000;462:129-35
40. Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS letters. 2000;476:73-7
41. Toumpanakis D, Theocharis SE. DNA repair systems in malignant mesothelioma. Cancer letters. 2011;312:143-9 doi:10.1016/j.canlet.2011.08.021
42. Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008;18:27-47 doi:10.1038/cr.2008.8
43. Jaurand MC. Mechanisms of fiber-induced genotoxicity. Environmental health perspectives. 1997;105(Suppl 5):1073-84
44. Ollikainen T, Linnainmaa K, Kinnula VL. DNA single strand breaks induced by asbestos fibers in human pleural mesothelial cells in vitro. Environmental and molecular mutagenesis. 1999;33:153-60
45. Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell research. 2008;18:134-47 doi:10.1038/cr.2007.111
46. San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annual review of biochemistry. 2008;77:229-57 doi:10.1146/annurev.biochem.77.061306.125255
47. Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nature reviews Cancer. 2008;8:193-204 doi:10.1038/nrc2342
48. Travis WD, World Health Organization, International Agency for Research on Cancer, International Association for the Study of Lung Cancer, International Academy of Pathology. Pathology and genetics of tumours of the lung, pleura, thymus and heart. Lyon, Oxford: IARC Press, Oxford University Press (distributor). 2004
49. Sobin LH, Gospodarowicz MK, Wittekind C, International Union against Cancer. TNM classification of malignant tumours. 7th ed. Chichester, West Sussex, UK; Hoboken, NJ: Wiley-Blackwell. 2010
50. Rusch VW. A proposed new international TNM staging system for malignant pleural mesothelioma. From the International Mesothelioma Interest Group. Chest. 1995;108:1122-8
51. Byrne MJ, Nowak AK. Modified RECIST criteria for assessment of response in malignant pleural mesothelioma. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2004;15:257-60
52. Ceresoli GL, Chiti A, Zucali PA, Cappuzzo F, De Vincenzo F, Cavina R. et al. Assessment of tumor response in malignant pleural mesothelioma. Cancer treatment reviews. 2007;33:533-41 doi:10.1016/j.ctrv.2007.07.012
53. Mairinger FD, Walter RF, Werner R, Christoph DC, Ting S, Vollbrecht C. et al. Activation of angiogenesis differs strongly between pulmonary carcinoids and neuroendocrine carinomas and is crucial for carcinoid tumourgenesis. Journal of Cancer. 2014;5:465-71 doi:10.7150/jca.9235
54. Walter RF, Mairinger FD, Werner R, Ting S, Vollbrecht C, Theegarten D. et al. SOX4, SOX11 and PAX6 mRNA expression was identified as a (prognostic) marker for the aggressiveness of neuroendocrine tumors of the lung by using next-generation expression analysis (NanoString). Future oncology. 2015;11:1027-36 doi:10.2217/fon.15.18
55. Walter RF, Werner R, Ting S, Vollbrecht C, Theegarten D, Christoph DC. et al. Identification of deregulation of apoptosis and cell cycle in neuroendocrine tumors of the lung via NanoString nCounter expression analysis. Oncotarget. 2015
56. Kelly RJ, Sharon E, Hassan R. Chemotherapy and targeted therapies for unresectable malignant mesothelioma. Lung Cancer. 2011;73:256-63 doi:10.1016/j.lungcan.2011.04.014
57. Okazaki T, Jiao L, Chang P, Evans DB, Abbruzzese JL, Li D. Single-nucleotide polymorphisms of DNA damage response genes are associated with overall survival in patients with pancreatic cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:2042-8 doi:10.1158/1078-0432.CCR-07-1520
58. Karnitz LM, Flatten KS, Wagner JM, Loegering D, Hackbarth JS, Arlander SJ. et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Molecular pharmacology. 2005;68:1636-44 doi:10.1124/mol.105.012716
59. Morgan MA, Parsels LA, Parsels JD, Mesiwala AK, Maybaum J, Lawrence TS. Role of checkpoint kinase 1 in preventing premature mitosis in response to gemcitabine. Cancer research. 2005;65:6835-42 doi:10.1158/0008-5472.CAN-04-2246
60. Abdel-Fatah TM, Middleton FK, Arora A, Agarwal D, Chen T, Moseley PM. et al. Untangling the ATR-CHEK1 network for prognostication, prediction and therapeutic target validation in breast cancer. Molecular oncology. 2015;9:569-85 doi:10.1016/j.molonc.2014.10.013
61. Gao Q, Zhou J, Huang X, Chen G, Ye F, Lu Y. et al. Selective targeting of checkpoint kinase 1 in tumor cells with a novel potent oncolytic adenovirus. Molecular therapy: the journal of the American Society of Gene Therapy. 2006;13:928-37 doi:10.1016/j.ymthe.2005.12.009
62. Roe OD, Anderssen E, Sandeck H, Christensen T, Larsson E, Lundgren S. Malignant pleural mesothelioma: genome-wide expression patterns reflecting general resistance mechanisms and a proposal of novel targets. Lung Cancer. 2010;67:57-68 doi:10.1016/j.lungcan.2009.03.016
63. Papp T, Schipper H, Pemsel H, Bastrop R, Muller KM, Wiethege T. et al. Mutational analysis of N-ras, p53, p16INK4a, p14ARF and CDK4 genes in primary human malignant mesotheliomas. International journal of oncology. 2001;18:425-33
64. Papp T, Schipper H, Pemsel H, Unverricht M, Muller KM, Wiethege T. et al. Mutational analysis of the PTEN/MMAC1 tumour suppressor gene in primary human malignant mesotheliomas. Oncology reports. 2001;8:1375-9
65. Toyooka S, Kishimoto T, Date H. Advances in the molecular biology of malignant mesothelioma. Acta medica Okayama. 2008;62:1-7
66. Kumar K, Rahman Q, Schipper H, Matschegewski C, Schiffmann D, Papp T. Mutational analysis of 9 different tumour-associated genes in human malignant mesothelioma cell lines. Oncology reports. 2005;14:743-50
67. Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206-8 doi:10.1038/378206a0
68. Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell death and differentiation. 2006;13:927-34 doi:10.1038/sj.cdd.4401912
69. Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203-6 doi:10.1038/378203a0
70. Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG. et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature genetics. 2001;29:92-5 doi:10.1038/ng714
71. Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer cell. 2006;10:501-14 doi:10.1016/j.ccr.2006.10.010
72. Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature reviews Cancer. 2006;6:909-23 doi:10.1038/nrc2012
73. Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S. et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nature medicine. 2012;18:1239-47 doi:10.1038/nm.2863
74. Pishas KI, Al-Ejeh F, Zinonos I, Kumar R, Evdokiou A, Brown MP. et al. Nutlin-3a is a potential therapeutic for ewing sarcoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:494-504 doi:10.1158/1078-0432.CCR-10-1587
75. Gilkes DM, Pan Y, Coppola D, Yeatman T, Reuther GW, Chen J. Regulation of MDMX expression by mitogenic signaling. Molecular and cellular biology. 2008;28:1999-2010 doi:10.1128/MCB.01633-07
76. Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C. et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61-6 doi:10.1038/nature05194
77. McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J. et al. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PloS one. 2012;7:e42739. doi:10.1371/journal.pone.0042739
78. Mairinger FD, Walter RF, Ting S, Vollbrecht C, Kollmeier J, Griff S. et al. Mdm2 protein expression is strongly associated with survival in malignant pleural mesothelioma. Future oncology. 2014;10:995-1005 doi:10.2217/fon.13.261
79. Walter RF, Mairinger FD, Ting S, Vollbrecht C, Mairinger T, Theegarten D. et al. MDM2 is an important prognostic and predictive factor for platin-pemetrexed therapy in malignant pleural mesotheliomas and deregulation of P14/ARF (encoded by CDKN2A) seems to contribute to an MDM2-driven inactivation of P53. British journal of cancer. 2015;112:883-90 doi:10.1038/bjc.2015.27
80. Kanellou P, Zaravinos A, Zioga M, Spandidos DA. Deregulation of the tumour suppressor genes p14(ARF), p15(INK4b), p16(INK4a) and p53 in basal cell carcinoma. The British journal of dermatology. 2009;160:1215-21 doi:10.1111/j.1365-2133.2009.09079.x
81. Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071-83 doi:10.1016/j.cell.2005.03.037
82. Miao L, Song Z, Jin L, Zhu YM, Wen LP, Wu M. ARF antagonizes the ability of Miz-1 to inhibit p53-mediated transactivation. Oncogene. 2010;29:711-22 doi:10.1038/onc.2009.372
83. Huang Y, Tyler T, Saadatmandi N, Lee C, Borgstrom P, Gjerset RA. Enhanced tumor suppression by a p14ARF/p53 bicistronic adenovirus through increased p53 protein translation and stability. Cancer research. 2003;63:3646-53
84. Van Maerken T, Rihani A, Dreidax D, De Clercq S, Yigit N, Marine JC. et al. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Molecular cancer therapeutics. 2011;10:983-93 doi:10.1158/1535-7163.MCT-10-1090
85. Singhal S, Wiewrodt R, Malden LD, Amin KM, Matzie K, Friedberg J. et al. Gene expression profiling of malignant mesothelioma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9:3080-97
86. Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. The Biochemical journal. 1997;325( Pt 1):1-16
87. Yang H, Rivera Z, Jube S, Nasu M, Bertino P, Goparaju C. et al. Programmed necrosis induced by asbestos in human mesothelial cells causes high-mobility group box 1 protein release and resultant inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12611-6 doi:10.1073/pnas.1006542107
88. Broaddus VC, Yang L, Scavo LM, Ernst JD, Boylan AM. Asbestos induces apoptosis of human and rabbit pleural mesothelial cells via reactive oxygen species. The Journal of clinical investigation. 1996;98:2050-9 doi:10.1172/JCI119010
89. Upadhyay D, Kamp DW. Asbestos-induced pulmonary toxicity: role of DNA damage and apoptosis. Exp Biol Med (Maywood). 2003;228:650-9
90. Ollikainen T, Puhakka A, Kahlos K, Linnainmaa K, Kinnula VL. Modulation of cell and DNA damage by poly(ADP)ribose polymerase in lung cells exposed to H(2)O(2) or asbestos fibres. Mutation research. 2000;470:77-84
91. Dong HY, Buard A, Levy F, Renier A, Laval F, Jaurand MC. Synthesis of poly(ADP-ribose) in asbestos treated rat pleural mesothelial cells in culture. Mutation research. 1995;331:197-204
92. Taguchi T, Jhanwar SC, Siegfried JM, Keller SM, Testa JR. Recurrent deletions of specific chromosomal sites in 1p, 3p, 6q, and 9p in human malignant mesothelioma. Cancer research. 1993;53:4349-55
93. Neragi-Miandoab S, Weiner S, Sugarbaker DJ. Incidence of atrial fibrillation after extrapleural pneumonectomy vs. pleurectomy in patients with malignant pleural mesothelioma. Interactive cardiovascular and thoracic surgery. 2008;7:1039-42 doi:10.1510/icvts.2008.181099
94. Balsara BR, Bell DW, Sonoda G, De Rienzo A, du Manoir S, Jhanwar SC. et al. Comparative genomic hybridization and loss of heterozygosity analyses identify a common region of deletion at 15q11.1-15 in human malignant mesothelioma. Cancer research. 1999;59:450-4
95. Kostyrko K, Bosshard S, Urban Z, Mermod N. A role for homologous recombination proteins in cell cycle regulation. Cell Cycle. 2015;14:2853-61 doi:10.1080/15384101.2015.1049784
96. Chen WT, Alpert A, Leiter C, Gong F, Jackson SP, Miller KM. Systematic identification of functional residues in mammalian histone H2AX. Molecular and cellular biology. 2013;33:111-26 doi:10.1128/MCB.01024-12
97. Katsube T, Mori M, Tsuji H, Shiomi T, Wang B, Liu Q. et al. Most hydrogen peroxide-induced histone H2AX phosphorylation is mediated by ATR and is not dependent on DNA double-strand breaks. Journal of biochemistry. 2014;156:85-95 doi:10.1093/jb/mvu021
98. Sone K, Piao L, Nakakido M, Ueda K, Jenuwein T, Nakamura Y. et al. Critical role of lysine 134 methylation on histone H2AX for gamma-H2AX production and DNA repair. Nature communications. 2014;5:5691. doi:10.1038/ncomms6691
99. Hu S, Zhang G, Xu J, Zhu X, Lu X, Jiang L. et al. Disparity expression of gammaH2AX in papillary thyroid cancer and nodular goiter. Clinical laboratory. 2014;60:2031-7
100. Yang H, Luo J, Liu Z, Zhou R, Luo H. MicroRNA-138 Regulates DNA Damage Response in Small Cell Lung Cancer Cells by Directly Targeting H2AX. Cancer investigation. 2015;33:126-36 doi:10.3109/07357907.2015.1006329
101. Van Oorschot B, Oei AL, Nuijens AC, Rodermond H, Hoeben R, Stap J. et al. Decay of gamma-H2AX foci correlates with potentially lethal damage repair and P53 status in human colorectal carcinoma cells. Cellular & molecular biology letters. 2014;19:37-51 doi:10.2478/s11658-013-0113-0
102. Fernandez MI, Gong Y, Ye Y, Lin J, Chang DW, Kamat AM. et al. gamma-H2AX level in peripheral blood lymphocytes as a risk predictor for bladder cancer. Carcinogenesis. 2013;34:2543-7 doi:10.1093/carcin/bgt270
103. Bhatia A, Dey P, Goel S, Singh G. Expression of gammaH2AX may help in defining a genetically more stable subtype of infiltrating ductal carcinoma of breast. The Indian journal of medical research. 2013;137:759-66
Author contact
![]() Corresponding author: Paul Zarogoulidis, M.D, Ph. D. Pulmonary Department-Oncology Unit, “G. Papanikolaou” General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece. Fax: 00302310992424 Mobile: 00306977271974 E-mail: pzarogcom.
Corresponding author: Paul Zarogoulidis, M.D, Ph. D. Pulmonary Department-Oncology Unit, “G. Papanikolaou” General Hospital, Aristotle University of Thessaloniki, Thessaloniki, Greece. Fax: 00302310992424 Mobile: 00306977271974 E-mail: pzarogcom.

 Global reach, higher impact
Global reach, higher impact