3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(15):2179-2186. doi:10.7150/jca.16609 This issue Cite
Research Paper
Association of common variants in MTAP with susceptibility and overall survival of osteosarcoma: a two-stage population-based study in Han Chinese
1. Department of Orthopedic, the First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, China;
2. Department of Rheumatology and Immunology, the Fifth Hospital of Xi'an, Xi'an, China.
3. Department of Energy, Environment and Chemical Engineering, Washington University, Saint Louis, MO, USA.
4. Department of Orthopedic, Xijing Orthopedic Hospital, the Fourth Military Medical University, Xi'an, China;
5. Department of Orthopedic, Hong-hui Hospital, Xi'an Jiaotong University College of Medicine, Xi'an, China;
6. Department of Orthopedic, the Second Affiliated Hospital of Xi'an Jiaotong University, Xi'an, China.
* These authors contributed equally to this work.
Received 2016-6-26; Accepted 2016-9-4; Published 2016-10-25
Abstract
Osteosarcoma (OS) is a common malignant tumor, which exists widely in the bone of children and adolescents, and genetic factors may influence its susceptibility. Recently, the gene MTAP has been reported to be associated with OS in a Caucasian population. To investigate the association of common variants in MTAP with OS risk in Han Chinese individuals, we designed a two-stage case-control study with 392 OS patients and 1,578 unrelated healthy controls of Han Chinese individuals. A total of 17 tagging single nucleotide polymorphisms (SNPs) were firstly genotyped in the discovery stage, and single-SNP association and haplotypic association analyses have been performed. The SNP rs7023329 was found to be strongly associated with the OS risk (adjusted P = 0.002908), and the results of odds ratios (ORs) and 95% confidence intervals (CI) revealed increased risks from A allele of the SNP on OS (OR=1.33, 95% CI=1.13-1.62). The results were confirmed with a similar pattern in the validation stage (adjusted P = 0.006737, OR=1.49, 95% CI=1.11-2.00). Moreover, haplotypic analyses indicated that one haplotype block containing rs7023329 was significantly associated with OS risk in both stages (both global P<0.0001). The statistically significant association between the rs7023329 genotype and poor survival in OS patients was also observed. To sum up, our results prove that MTAP plays an important role in the etiology of OS, suggesting this gene as a potential genetic modifier for OS development.
Keywords: Osteosarcoma susceptibility, Common variants, MTAP, Case-control studies, Han Chinese
Introduction
Osteosarcoma (OS) is a rare bone cancer, which is derived from mesenchymal tissues and usually occurs in the long bones of the body such as the distal femur, proximal tibia, and humeral metaphysis. OS is one of the most common bone malignancies in children and adolescents, with an annual incidence of approximately 5/1,000,000, and it always has detectable metastases at diagnosis frequently leading to a poor prognosis generally 1. Although the fundamental mechanism underlying the development of OS is still not well understood, previous studies have suggested that the development of OS is a complex, multistep and multifactorial process and many environmental and genetic factors are involved throughout the development of this cancer 2,3. However, not all individuals exposed to similar risk factors would develop OS, which suggests that genetic variations may influence OS susceptibility. Recently, many genome-wide association studies (GWASs) have reported some evidence related to genetic susceptibility of OS, and some results were consistently replicated in different studies 4,5. Although genetic association analysis has provided a promising approach to the genetics of complex diseases, such as psychiatry disorders 6-10, current results account for only a small percentage of the estimated heritability with the lack of their systematic biological interpretation 11. Give of genetic heterogeneity existing in different populations, identifying individual candidate genes/variants with small effects on disease risks and follow-up studies are essential to confirm those findings and extend them 12.
During the past several years, molecular genetic studies have brought a wealth of new information of some susceptibility loci in chromosome 9 for multiple tumors, such as CDKN2A/B (cyclin-dependent kinase inhibitors 2A/B), ANRIL (antisense non coding RNA) and MTAP (methylthioadenosine phosphorylase), which are all involved in cell cycle, protein synthesis and tumors suppression 13-15. The MTAP gene located at 9p21 is flanked by CDKN2A and miR-31, and the gene is frequently co-deleted with the CDKN2A and CDKN2B in many different tumors 16-19. Moreover, Pasmant et al20 found that the regulatory sequences, such as enhancers, were enriched in the region of 9p21.3, which can regulate the expression of genes (MTAP-CDKN2A/2B/CDKN2BAS/-IFNα21). MTAP is a ubiquitous enzyme that catalyzes the phosphorolysis of the nucleoside MTA (methylthioadenosine), which is generated during the synthesis of polyamines spermidine and spermine. In addition, it is also an essential enzyme in the salvage pathway of adenine and in methionine synthesis 21-23. It has further been reported that the genetic polymorphisms of MTAP associated with some cancers including OS in Caucasians and ischemic stroke and myocardial infarction in Han Chinese population 24-27. Loss of MTAP expression would exert a tumor-promoting effect, suggesting that MTAP may function as a tumor suppressor gene 28-30. MTAP-deficient cells are reported to be more sensitive than MTAP-positive cells to inhibitors of de novo purine synthesis and to methionine deprivation 31,32. In MTAP-negative tumor cells, it has also been evidenced that conversion proceeds and the tumor cells can be selectively killed 31,32.
Since MTAP has been linked closely with many tumors, MTAP has gained much attention among many candidate genes predisposing to OS. Because the underlying biological mechanisms of OS remain largely unknown, the contribution of MTAP to OS has not been elucidated, despite evidence of a strongly significant association within the Caucasian population. Thus, to improve the current understanding about the role of MTAP in the OS predisposition, it is necessary to investigate the consequences of genetic diversity in other ethnic populations. Currently, the role of MTAP in OS susceptibility in the Han Chinese population has not been previously evaluated. Therefore, we carried out the two-stage case-control study to further examine the associations between MTAP and the OS risk in Han Chinese individuals. The aim of the present study was to assess the relationship of common variants in MTAP with the increased risk of OS and provide clues as to the nature of the mechanisms involved in the etiology of OS.
Material and methods
Study Subjects
A two-stage population-based case-control study was conducted. All of 284 OS cases and 981 healthy controls were recruited from 3 different hospitals (The First Affiliated Hospital of Xi'an Jiaotong University, The Second Affiliated Hospital of Xi'an Jiaotong University and Xi'an Honghui Hospital) and designed as the discovery stage (the first stage) in this study. Subsequently, independent 108 patients with OS and 597 healthy controls additionally collected from another hospital (Xijing Hospital) were considered as the subjects of the validation stage (the second stage). All patients were histologically/pathologically confirmed by two experienced pathologists. All the healthy controls were recruited from the general health check-up centers in these hospitals, and they had been under the health screening. Each subject was interviewed using a standard questionnaire by a trained nurse to collect medical histories and demographic characteristics. The clinical characteristics of controls were matched to the gender and age distribution with the OS cases, as presented in Table 1. All subjects were of Chinese Han ethnicity and recruited from the city of Xi'an in Shaanxi Province (China) May 2010 to July 2015. Based on self-report and medical records regarding their own and their paternal grandparents' place of birth, we excluded individuals or their three-generation families not born in Xi'an. This study was performed in accordance with the ethical guidelines of the Declaration of Helsinki (version 2002) and was approved by the Xi'an Jiaotong University Ethics Committee. All participants have written (signed) informed consent forms.
Distributions of selected variables in osteosarcoma patients and healthy controls.
| Variables | The Discovery Stage | P-value | The Validation Stage | P-value | ||
|---|---|---|---|---|---|---|
| OS(N=284) | Control(N=981) | OS (N=108) | Control(N=597) | |||
| Mean age (years) | 21.00±4.7 | 21.02±4.3 | 0.956 | 21.25 (±5.4) | 21.26 (±5.0) | 0.990 |
| ≤20 | 165 | 563 | 0.832 | 61 | 340 | 0.928 |
| >20 | 119 | 418 | 47 | 257 | ||
| Gender | ||||||
| Male | 146 (51.4%) | 498 (50.8%) | 0.848 | 56 (51.9%) | 308 (51.6%) | 0.960 |
| Female | 138 (48.6%) | 483 (49.2%) | 52 (48.1%) | 289 (48.4%) | ||
| Cancer family history | ||||||
| Yes | 203 (71.5%) | 685 (69.8%) | 0.592 | 70 (64.8%) | 381 (63.8%) | 0.843 |
| No | 81 (28.5%) | 296 (30.2%) | 38 (35.2%) | 216 (36.2%) | ||
| Tumor location | ||||||
| Long tubular bones | 231 (81.3%) | 83 (76.9%) | ||||
| Axial skeleton | 53 (18.7%) | 25 (23.1%) | ||||
| Pathological fracture | ||||||
| Yes | 55 (19.4%) | 23 (21.3%) | ||||
| No | 229 (80.6%) | 85 (78.7%) | ||||
| Metastasis | ||||||
| Yes | 89 (31.3%) | 31 (28.7%) | ||||
| No | 195 (68.7%) | 77 (71.3%) | ||||
| Enneking stages | ||||||
| I | 30 (10.6%) | 10 (9.3%) | ||||
| II | 213 (75%) | 79 (73.1%) | ||||
| III | 41 (14.4%) | 19 (17.6) | ||||
The genomic location of selected SNP markers across the MTAP gene and their relationship with gene exons.
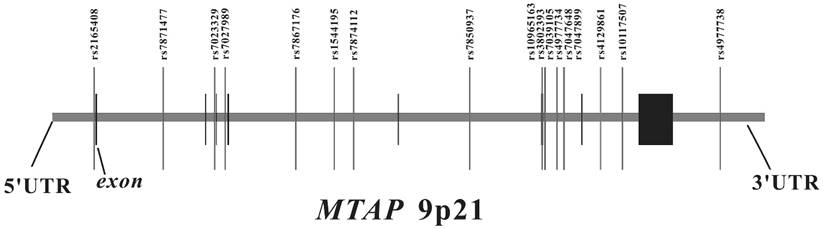
SNP selection and genotyping
We searched for SNPs with minor allele frequencies (MAF) over 0.01 between 10 kb upstream and 10 kb downstream of MTAP in the HapMap CHB database and found 45 SNPs. Afterwards, MAF ≥ 0.01 with pair-wise tagging and r2 ≥ 0.9 were used as the cutoff criteria during tag SNPs selection and resulted in 17 tag SNPs covering the region of MTAP for our study (Figure 1). Peripheral blood was drawn from a vein into a sterile tube containing ethylenediamine tetraacetic acid (EDTA), and the genomic DNA was isolated from peripheral blood leukocytes according to the manufacturer's protocol (Genomic DNA kit, Axygen Scientific Inc., California, USA). SNPs genotyping was performed at the Sequenom MassARRAY platform with the iPLEX GOLD chemistry (Sequenom, San Diego, CA, USA) based on the manufacturer's protocols. Briefly, SNPs were genotyped using high-throughput, matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry. Next, the resulting spectra were processed using Typer Analyzer software (Sequenom), and genotype data were generated from the samples 33. As the final genotype call rate of each SNP exceeded 99.7% and the overall genotyping call rate reached 99.9%, further statistical analyses were considered to be reliable. For quality control, the status of both case and control samples was blind during all genotyping processes. Meanwhile, 5% of random samples were repeated and the results were 100% concordant.
Statistical analyses
Student's t test and chi-square (χ2) test were conducted to analyze the differences in the distribution of characteristics between the OS patients and the healthy controls. Hardy-Weinberg equilibrium (HWE) for each SNP was estimated by using Haploview v4.2. Allelic and genotypic association tests were performed with the genetic analysis software Plink v1.9 and a logistic model was implemented for each SNP to investigate the association between genetic polymorphisms and OS risk. The Bonferroni correction was applied to address the multiple comparison problems and the strength of association between MTAP and OS risk was assessed by odds ratios (ORs) and 95% confidence intervals (CIs) from the logistic model with adjustments for age, gender and family history. For the single SNP association analyses, P-value was corrected using Bonferroni correction (corrected P-value = P × n, the number of SNPs). In addition to the single SNP association analyses, we also conducted haplotype-based analyses. The inter-marker relationship was determined by performing pair-wise linkage disequilibrium (LD) with Haploview v4.2. The haplotype frequencies were estimated by using GENECOUNTING v2.2. Besides, the haplotypic association analyses included a likelihood ratio test followed by permutation testing was performed for all haplotypes 34. The Kaplan-Meier method was used to plot the OS curves. All tests were two-tailed, and a P-value of 0.05 was selected as a threshold for significance in all analyses.
Results
Baseline characteristics of subjects
A two-stage approach was utilized in our study, which included 392 OS patients and 1,578 healthy controls in both stages. Age, gender, cancer family history, tumor location, pathological fracture, metastasis and Enneking stage were summarized in Table 1. In both stages, OS cases and healthy controls were matched by age with the mean age, and there were no significant differences regarding gender and family history between cases and controls (Table 1). All OS cases were divided into some subgroups according to their tumor location, pathological fracture, metastasis and Enneking stage (Table 1). In each stage, there were more patients with long tubular bones, metastasis and no pathological fracture, and the highest numbers of OS cases were in stage II, compared to stage I and III.
Allelic and genotypic association of SNPs with OS risk
Seventeen SNPs in MTAP were genotyped in the discovery dataset. The results of the single SNP-based association analyses of these SNPs in the discovery stage, including the Hardy-Weinberg equilibrium (HWE) test, were summarized in Tables 2 and Table S1. All genotype distributions among the subjects were in agreement with HWE (Table 2 and Table S1). We observed significant association signals for two SNPs (rs7023329 and rs7027989; adjusted P = 0.002908 and 0.04427, respectively), but it was only rs7023329 that still remained the association with OS risk after Bonferroni correction (Table 2). Genotypic association analyses also confirmed the result of rs702339 with a similar pattern (adjusted P = 0.002885). The other 15 SNPs did not significantly differ in either allele or genotype distribution (Table S1).
Allele and genotype frequency of single SNP association analysis in both stages.
| SNP Affection | H-WE P-value | Allelic Frequency (%) | Allelic P-value | Adjusted P-value * | Genotype Frequency (%) | Genotypic P-value | Adjusted P-value * | OR* 95%CI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| The Discovery Stage | |||||||||||
| rs7023329 | A | G | AA | AG | GG | ||||||
| OS | 0.759 | 56.69 | 43.31 | 0.002033 | 0.002908 | 31.69 | 50.00 | 18.31 | 0.008446 | 0.002885 | 1.33 |
| CTR | 0.919 | 49.34 | 50.66 | 0.049436 | 24.26 | 50.15 | 25.59 | 0.049045 | 1.13-1.62 | ||
| rs7027989 | G | A | GG | GA | AA | ||||||
| OS | 0.348 | 78.35 | 21.65 | 0.023912 | 0.04427 | 62.32 | 32.04 | 5.63 | 0.053070 | 0.07106 | 1.50 |
| CTR | 0.997 | 82.52 | 17.48 | 68.09 | 28.85 | 3.06 | 0.99-2.11 | ||||
| The Validation Stage | |||||||||||
| rs7023329 | A | G | AA | AG | GG | ||||||
| OS | 0.743 | 57.87 | 42.13 | 0.006589 | 0.006737 | 34.26 | 47.22 | 18.52 | 0.020154 | 0.007089 | 1.49 |
| CTR | 0.801 | 47.82 | 52.18 | 0.047159 | 22.61 | 50.42 | 26.97 | 0.049623 | 1.11-2.00 | ||
| rs7027989 | G | A | GG | GA | AA | ||||||
| OS | 0.853 | 77.78 | 22.22 | 0.105079 | 0.1054 | 60.19 | 35.19 | 4.63 | 0.120548 | 0.2914 | 1.32 |
| CTR | 0.895 | 82.41 | 17.59 | 67.84 | 29.15 | 3.02 | 0.79-2.21 | ||||
OS: osteosarcoma; CTR: control; CI: confidence interval; OR: odds ratio.
Risk allele and significant P values were in italic bold, and corrected P values were underlined after Bonferroni correction. OR referred to the risk allele odds ratio in cases and controls.
* Obtained in logistic regression models with adjustment for age, gender and family history.
Relationship between genotypes of rs7023329 within MTAP gene and clinicopathological features of patients with osteosarcoma.
| rs7023329 | The Discovery Stage | The Validation Stage | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n=284 | AA | GG | AG | P-value | n=108 | AA | GG | AG | P-value | |
| Mean age (years) | 90 | 52 | 142 | 0.695 | 37 | 20 | 51 | 0.796 | ||
| ≤20 | 165 | 55 | 28 | 82 | 61 | 21 | 10 | 30 | ||
| >20 | 119 | 35 | 24 | 60 | 47 | 16 | 10 | 21 | ||
| Gender | 0.968 | 0.854 | ||||||||
| Male | 146 | 47 | 26 | 73 | 56 | 20 | 11 | 25 | ||
| Female | 138 | 43 | 26 | 69 | 52 | 17 | 9 | 26 | ||
| Cancer family history | 0.104 | 0.849 | ||||||||
| Yes | 81 | 23 | 10 | 48 | 70 | 25 | 12 | 33 | ||
| No | 203 | 67 | 42 | 94 | 38 | 12 | 8 | 18 | ||
| Tumor location | 0.459 | 0.713 | ||||||||
| Long tubular bones | 231 | 77 | 41 | 113 | 83 | 32 | 11 | 40 | ||
| Axial skeleton | 53 | 13 | 11 | 29 | 25 | 5 | 9 | 11 | ||
| Pathological fracture | 0.983 | 0.314 | ||||||||
| Yes | 55 | 18 | 10 | 27 | 23 | 6 | 4 | 13 | ||
| No | 229 | 72 | 42 | 115 | 85 | 31 | 16 | 38 | ||
| Metastasis | 0.157 | 0.483 | ||||||||
| Yes | 89 | 33 | 11 | 45 | 31 | 8 | 6 | 17 | ||
| No | 195 | 57 | 41 | 97 | 77 | 29 | 14 | 34 | ||
| Enneking stages | 0.118 | 0.358 | ||||||||
| I | 30 | 12 | 8 | 10 | 10 | 5 | 2 | 3 | ||
| II | 213 | 65 | 41 | 107 | 79 | 24 | 17 | 38 | ||
| III | 41 | 13 | 3 | 25 | 19 | 8 | 1 | 10 | ||
Next, we performed single SNP association analyses for the two SNPs (rs7023329 and rs7027989) with 5 other surrounding SNPs (rs2165408, rs7871477, rs7867176, rs1544195 and rs7874112) in the validation dataset. The significant association of the SNP rs7023329 with OS risk was replicated (adjusted P = 0.006737) (Table 2), indicating that the A allele was an OS risk allele, and the association with OS risk was still survival after Bonferroni correction (Table 2). Moreover, genotypic association analyses also validated the conclusion (adjusted P = 0.007089). The allelic or genotype distributions of the other five SNPs did not significantly differ (Table S1). A similar positive effect of SNP on OS risk was further revealed by logistic regression regarding odds ratios (OR) and 95% confidence interval (CI) in both stages (Table 2), after adjustments for age, gender and cancer family history.
Relationship between rs7023329 genotypes and clinicopathological characteristics
The relationships between the associated SNP rs7023329 genotypes and clinicopathological parameters were evaluated, and the results were presented in Table 3. However, there are not any obvious differences in the relations between age, gender, cancer family history, tumor location, pathological fracture, metastasis and Enneking stage, respectively, and rs7023329 genotypes in both stages (Table 3).
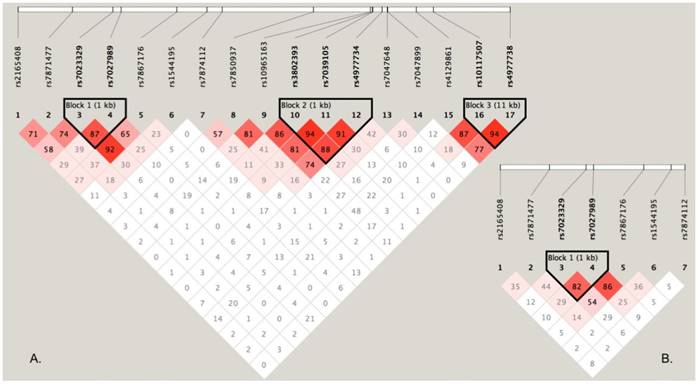
Haplotypic association analyses
To perform haplotype-based association analyses, we examined LD structure of 17 SNPs using the data from the discovery stage and identified three LD blocks, of which one block consisted of two SNPs (rs7023329 and rs7027989, Figure 2A). The LD block was also constructed in the validation stage (Figure 2B). Subsequent haplotypic association analyses were used to test the LD block mentioned above. As presented in Table 4, significant P values (global P < 0.0001, respectively) were obtained in both stages. Some haplotypes in the LD block were positively associated with OS risk. For example, the haplotype AA was significantly associated with OS as a risk haplotype. Because of the higher frequencies in healthy controls, the haplotype GG might protect against OS (Table 4).
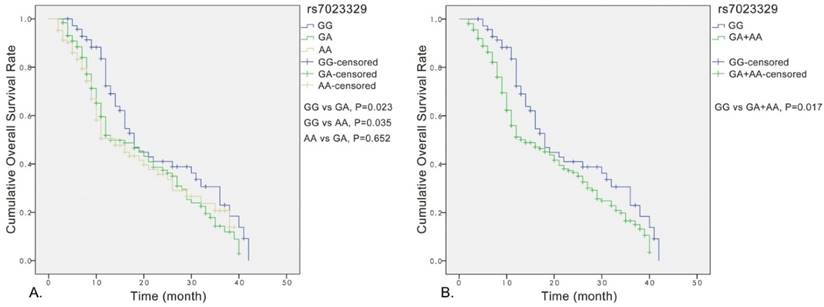
Analyses for the effect of rs7023329 on prognosis of OS patients
Significant association was observed between the rs7023329 and the overall survival of OS patients, and the rs7023329 may be therefore an independent prognostic factor for OS (Figure 3A). The survival analyses of rs7023329 indicated that OS patients carrying the AG+AA genotypes exhibited a worse survival rate compared with patients with the homozygous GG genotype (P = 0.017; Figure 3B). Patients with the AG+AA genotypes had a median survival time of 14.0 months, while patients with homozygote GG genotype had the best outcome (median survival time: 18.0 months).
LD structure based on two-stage genotype datasets. The LD blocks are indicated as shaded matrices, and LD blocks 1 in the discovery stage (A) were confirmed in the validation stage (B).
Kaplan-Meier analyses for the overall survival of OS patients with the rs7023329.
Haplotypes frequency and association analyses.
| Haplotype | Frequency (%) in the discovery stage | Frequency (%) in the validation stage | ||||||
|---|---|---|---|---|---|---|---|---|
| rs7023329-rs7027989 | OS | CTR | P-value 1 | Global P 2 | OS | CTR | P-value 1 | Global P 2 |
| AG | 53.85 | 48.59 | 0.02729* | <0.0001 | 52.51 | 46.90 | 0.12843 | <0.0001 |
| GG | 24.50 | 33.93 | <0.0001 | 25.27 | 35.52 | 0.00343 | ||
| GA | 18.81 | 16.73 | 0.24745 | 16.86 | 16.66 | 0.94181 | ||
| AA | 2.85 | 0.75 | <0.0001 | 5.36 | 0.93 | <0.0001 | ||
OS: osteosarcoma; CTR: control. Haplotypes in italics were the significant ones in the study, and significant P values were in italic bold.
1. Based on 10000 permutations. 2. Based on comparison of frequency distribution of all haplotypes for the combination of SNPs.
* The P-value could not be significant after Bonferroni correction (corrected P = 0.02729×4 = 0.10916).
Discussion
The predominant hypothesis in recent years has been that the genetic architecture of OS involves several common variants of small effects and possibly also rare variants with much larger effects 35. In our hospital-based case-control study, the role of common variants within MTAP gene in OS risk as well as the interaction of the associated SNP rs7023329 with clinical and demographic factors in the development of OS was assessed. The allele and genotype distributions of the SNP rs7023329 were significantly different between the case and control groups in two stages and the ORs and 95% CI also suggested the positive effects of the associated SNP on OS risk. Indeed, analyses of only some SNPs are not sufficient to draw a conclusion 36. Our haplotype analyses also evidenced that the rs7023329 was significantly associated with OS. These results indicated that rs7023329 may be involved in susceptibility to the disease with an increased risk of OS. To the best of our knowledge, it is the first study to investigate the possible role of the SNP rs7023329 as a risk factor for OS and find the relationship between MTAP variations and susceptibility to OS development in the Han Chinese population.
To date, inaugural mechanism of OS was considered as a complex process and was not clear, but it was universally acknowledged that environment carcinogens could induce genomic polymorphism. It was reported that MTAP had a broad expression profile in the mammalian and was a key enzyme in the methionine salvage pathway 24,37. Loss of MTAP expression can exert a tumor promoting effect, suggesting that MTAP may function as a tumor suppressor gene 21,30,31,38. Moreover, the genetic polymorphism of MTAP was also considered as one of the potential genetic factors in various tumors 29,39-41. However, the exact mechanism by which the MTAP genetic polymorphisms affect the susceptibility of OS remains elusive. In our study, given the potential effect of the LD block on regulating gene transcription, we hypothesized that certain SNPs might influence the expression of MTAP in an unpredicted manner. Alternatively, the SNP rs7023329 might coexist in LD with certain undiscovered variants and participate with their regulation machinery to confer a risk for OS. No experimental evidence validates this hypothesis, and future functional studies are warranted to clarify this point. Multiple lines of evidence support that our results are unlikely to be artificial. First, significant signals were captured by both single SNP and haplotype-based analysis in two independent datasets. Second, the problem of population stratification has been avoided, as all samples of Han descent are from the same geographical region and there is no migration history for all subjects within their three previous generations. Moreover, the SNP rs7023329 in MTAP has been recently found to strongly associate with OS risk in Caucasians 26. Similar conclusions were reached in two different ethnic populations (Han Chinese and Caucasian), reaffirming the association of MTAP with OS susceptibility.
Although our results raised the possibility that common variants in the MTAP are involved in the development of OS, several limitations should be noted in the present study. First, the hospital-based case-control study design often produced selection biases and information biases. The subjects may not be representative of the general population. However, geographic region is a good proxy for genetic matching in the Han Chinese population. Therefore, population stratification may be effectively avoided in our study because of samples from the same geographical region 42,43. Second, the relative small sample size might miss some weak gene-disease association and gene-environment interactions. Last, functional experiments are necessary to clarify the underlying molecular mechanisms of the association signal between the SNP rs7023329 within MTAP and OS in the future, and many unmeasured environmental and genetic factors also need to be taken into account in future studies. Therefore, all of our findings should be considered preliminary, and additional follow-up studies are required to confirm these results in other ethnic populations.
In summary, our two-stage case-control study indicates MTAP is a susceptible gene for OS risk in the Han Chinese population. A strong association between SNP rs7023329 and OS risk was revealed through several sets of analyses in two stages. Further functional studies would be desired to uncover fundamental characteristics of pathogenic mutations and to clarify the exact effect of MTAP on OS development, which could facilitate potential applications in the clinic.
Supplementary Material
Table S1.
Acknowledgements
This work was totally supported by the Clinical Education Funds of the First Affiliated Hospital of Xi'an Jiaotong University (Grant No.15ZD09). The funding sources had no role in the design of this study, the collection, analysis and interpretation of data, the writing of the report, or the decision to submit the paper for publication.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mirabello L, Yeager M, Mai PL. et al. Germline tp53 variants and susceptibility to osteosarcoma[J]. J Natl Cancer Inst. 2015;107:163-170 doi:10.1093/jnci/djv101
2. Powers M, Zhang W, Lopez-Terrada D. et al. The molecular pathology of sarcomas[J]. Cancer Biomark. 2010;9:475-491 doi:10.3233/CBM-2011-0170
3. Bovee JV, Hogendoorn PC. Molecular pathology of sarcomas: concepts and clinical implications[J]. Virchows Archiv. 2010;456:193-199 doi:10.1007/s00428-009-0828-5
4. Hansen MF, Seton M, Merchant A. Osteosarcoma in paget's disease of bone[J]. Journal of Bone and Mineral Research. 2006;21:P58-P63 doi:10.1359/jbmr.06s211
5. Ferris ITJ, Berbel TO, Ortega GJ. et al. [risk factors for pediatric malignant bone tumors][J]. An Pediatr (Barc). 2005;63:537-547
6. Guan F, Zhang C, Wei S. et al. Association of PDE4B polymorphisms and schizophrenia in Northwestern Han Chinese[J]. Hum Genet. 2012;131(7):1047-1056 doi: 10.1007/s00439-011-1120-8
7. Guan F, Zhang B, Yan T. et al. Mir137 gene and target gene cacna1c of mir-137 contribute to schizophrenia susceptibility in han chinese[J]. Schizophrenia Research. 2014;152:97-104 doi:10.1016/j.schres.2013.11.004
8. Chen G, Guan F, Lin H. et al. Genetic analysis of common variants in the HDAC2 gene with schizophrenia susceptibility in Han Chinese[J]. J Hum Genet. 2015;60(9):479-484 doi: 10.1038/jhg.2015.66
9. Guan F, Li L, Qiao C. et al. Evaluation of genetic susceptibility of common variants in CACNA1D with schizophrenia in Han Chinese[J]. Sci Rep. 2015:10 5:12935. doi: 10.1038/srep12935
10. Zhang B, Guan F, Chen G. et al. Common variants in SLC1A2 and schizophrenia: Association and cognitive function in patients with schizophrenia and healthy individuals[J]. Schizophrenia Research. 2015;169(1-3):128-134 doi: 10.1016/j.schres.2015.10.012
11. Guan F, Zhang T, Liu X. et al. Evaluation of voltage-dependent calcium channel gamma gene families identified several novel potential susceptible genes to schizophrenia[J]. Sci Rep. 2016;6:24914. doi:10.1038/srep24914
12. Jia X, Zhang T, Li L. et al. Two-stage additional evidence support association of common variants in the HDAC3 with the increasing risk of schizophrenia susceptibility[J]. Am J Med Genet B Neuropsychiatr Genet. 2016 doi:10.1002/ajmg.b.32491
13. Karlsson EK, Sigurdsson S, Ivansson E. et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near cdkn2a/b[J]. Genome Biol. 2013;14:R132. doi:10.1186/gb-2013-14-12-r132
14. Zhi LQ, Ma W, Zhang H. et al. Association of recql5 gene polymorphisms and osteosarcoma in a chinese han population[J]. Tumour Biol. 2014;35:3255-3259 doi:10.1007/s13277-013-1425-4
15. Wang W, Luo H, Wang A. Expression of survivin and correlation with pcna in osteosarcoma[J]. Journal of Surgical Oncology. 2006;93:578-584 doi:10.1002/jso.20507
16. Illei PB, Rusch VW, Zakowski MF. et al. Homozygous deletion of cdkn2a and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas[J]. Clinical Cancer Research. 2003;9:2108-2113
17. Watanabe F, Takao M, Inoue K. et al. Immunohistochemical diagnosis of methylthioadenosine phosphorylase (mtap) deficiency in non-small cell lung carcinoma[J]. Lung Cancer. 2009;63:39-44 doi:10.1016/j.lungcan.2008.04.019
18. Hustinx SR, Hruban RH, Leoni LM. et al. Homozygous deletion of the mtap gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy[J]. Cancer Biology & Therapy. 2005;4:83-86
19. Huang HY, Li SH, Yu SC. et al. Homozygous deletion of mtap gene as a poor prognosticator in gastrointestinal stromal tumors[J]. Clinical Cancer Research. 2009;15:6963-6972 doi:10.1158/1078-0432.CCR-09-1511
20. Pasmant E, Sabbagh A, Vidaud M. et al. Anril, a long, noncoding rna, is an unexpected major hotspot in gwas[J]. Faseb Journal. 2011;25:444-448 doi:10.1096/fj.10-172452
21. Toone EJ. Advances in enzymology and related areas of molecular biology. Preface[J]. Adv Enzymol Relat Areas Mol Biol. 2011;78:ix-xi
22. Backlund PJ, Smith RA. 5'-methylthioadenosine metabolism and methionine synthesis in mammalian cells grown in culture[J]. Biochem Biophys Res Commun. 1982;108:687-695
23. Kamatani N, Nelson-Rees WA, Carson DA. Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme[J]. Proc Natl Acad Sci U S A. 1981;78:1219-1223
24. Gu F, Pfeiffer RM, Bhattacharjee S. et al. Common genetic variants in the 9p21 region and their associations with multiple tumours[J]. Br J Cancer. 2013;108:1378-1386 doi:10.1038/bjc.2013.7
25. Stefanaki I, Panagiotou OA, Kodela E. et al. Replication and predictive value of snps associated with melanoma and pigmentation traits in a southern european case-control study[J]. PLoS One. 2013;8:e55712. doi:10.1371/journal.pone.0055712
26. Kvaskoff M, Whiteman DC, Zhao ZZ. et al. Polymorphisms in nevus-associated genes mtap, pla2g6, and irf4 and the risk of invasive cutaneous melanoma[J]. Twin Res Hum Genet. 2011;14:422-432 doi:10.1375/twin.14.5.422
27. Falchi M, Bataille V, Hayward NK. et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi[J]. Nature Genetics. 2009;41:915-919 doi:10.1038/ng.410
28. Becker AP, Scapulatempo-Neto C, Menezes WP. et al. Expression of methylthioadenosine phosphorylase (mtap) in pilocytic astrocytomas[J]. Pathobiology. 2015;82:84-89 doi:10.1159/000430956
29. Li CF, Fang FM, Kung HJ. et al. Downregulated mtap expression in myxofibrosarcoma: a characterization of inactivating mechanisms, tumor suppressive function, and therapeutic relevance[J]. Oncotarget. 2014;5:11428-11441
30. Miyazaki S, Nishioka J, Shiraishi T. et al. Methylthioadenosine phosphorylase deficiency in japanese osteosarcoma patients[J]. International Journal of Oncology. 2007;31:1069-1076
31. Bertino JR, Waud WR, Parker WB. et al. Targeting tumors that lack methylthioadenosine phosphorylase (mtap) activity: current strategies[J]. Cancer Biology & Therapy. 2011;11:627-632 doi:10.4161/cbt.11.7.14948
32. Lubin M, Lubin A. Selective killing of tumors deficient in methylthioadenosine phosphorylase: a novel strategy[J]. PLoS One. 2009;4:e5735
33. Guan F, Wei S, Feng J. et al. Association study of a new schizophrenia susceptibility locus of 10q24.32-33 in a Han Chinese population[J]. Schizophrenia Research. 2012;138(1):63-68 doi: 10.1016/j.schres.2012.03.030
34. Guan F, Wei S, Zhang C. et al. A population-based association study of 2q32.3 and 8q21.3 loci with schizophrenia in Han Chinese[J]. J Psychiatr Res. 2013;47(6):712-717 doi: 10.1016/j.jpsychires.2013.01.025
35. Wang WY, Barratt BJ, Clayton DG. et al. Genome-wide association studies: theoretical and practical concerns[J]. Nature Reviews Genetics. 2005;6:109-118
36. Yang H, Zhang B, Zhu J. et al. 4q22.1 Contributes to Bone Mineral Density and Osteoporosis Susceptibility in Postmenopausal Women of Chinese Han Population[J]. PLoS One. 2013;8(11):e80165. doi: 10.1371/journal.pone.0080165
37. Appleby TC, Erion MD, Ealick SE. The structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase at 1.7 a resolution provides insights into substrate binding and catalysis[J]. Structure. 1999;7:629-641
38. Su CY, Chang YC, Chan YC. et al. Mtap is an independent prognosis marker and the concordant loss of mtap and p16 expression predicts short survival in non-small cell lung cancer patients[J]. European Journal of Surgical Oncology. 2014;40:1143-1150 doi:10.1016/j.ejso.2014.04.017
39. Maccioni L, Rachakonda PS, Bermejo JL. et al. Variants at the 9p21 locus and melanoma risk[J]. Bmc Cancer. 2013;13:325. doi:10.1186/1471-2407-13-325
40. Yang XC, Zhang Q, Chen ML. et al. Mtap and cdkn2b genes are associated with myocardial infarction in chinese hans[J]. Clinical Biochemistry. 2009;42:1071-1075 doi:10.1016/j.clinbiochem.2009.02.021
41. Bishop DT, Demenais F, Iles MM. et al. Genome-wide association study identifies three loci associated with melanoma risk[J]. Nature Genetics. 2009;41:920-925 doi:10.1038/ng.411
42. Guan F, Niu Y, Zhang T. et al. Two-stage association study to identify the genetic susceptibility of a novel common variant of rs2075290 in ZPR1 to type 2 diabetes[J]. Sci Rep. 2016;6:29586. doi: 10.1038/srep29586
43. Guan F, Zhang T, Li L. et al. Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population[J]. Schizophrenia Research. 2016 doi: 10.1016/j.schres.2016.07.004
Author contact
![]() Corresponding author: Meng Li, Ph.D, Department of Orthopedic, the First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, 710061, China. Tel.: +86-029-85323935; Fax: +86-029-85323935, E-mail: drleemoncom.
Corresponding author: Meng Li, Ph.D, Department of Orthopedic, the First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, 710061, China. Tel.: +86-029-85323935; Fax: +86-029-85323935, E-mail: drleemoncom.

 Global reach, higher impact
Global reach, higher impact