3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(15):2341-2345. doi:10.7150/jca.16947 This issue Cite
Short Research Communication
Antimicrobial peptide LL-37 participates in the transcriptional regulation of melanoma cells
1. Departamento de Análises Clínicas e Toxicológicas, Instituto de Ciências Farmacêuticas, Universidade de São Paulo, São Paulo, Brazil.
2. Active Motif Incorporation, Carlsbad, California, USA.
3. Instituto Israelita de Ensino e Pesquisa, Hospital Israelita Albert Einstein, São Paulo, Brazil.
4. Departamento de Emergências Clínicas, Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil.
5. Departamento de Oncologia, Faculdade de Medicina da Universidade de São Paulo, São Paulo, Brazil.
Received 2016-7-22; Accepted 2016-9-30; Published 2016-11-26
Abstract
Antimicrobial peptides are an ancient family of molecules that emerged millions of years ago and have been strongly conserved during the evolutionary process of living organisms. Recently, our group described that the human antimicrobial peptide LL-37 migrates to the nucleus, raising the possibility that LL-37 could directly modulate transcription under certain conditions. Here, we showed evidence that LL-37 binds to gene promoter regions, and LL-37 gene silencing changed the transcriptional program of melanoma A375 cells genes associated with histone, metabolism, cellular stress, ubiquitination and mitochondria.
Keywords: LL-37, transcription factors, antimicrobial peptides, cancer.
Introduction
LL-37 is one of the most studied antimicrobial peptides (AMPs) and plays a significant role in several biological processes. Despite its bactericidal capacity, LL-37 can bind to lipopolysaccharide [1, 2] and other pathogen-associated patterns [3], activate membrane receptors [4-6], induce chemotaxis [7, 8], stimulate angiogenesis [9], promote wound healing [10-12], trigger the inflammasome [13], and induce apoptosis and other mechanisms of cell death [14, 15], among other immunoregulatory and non-immune functions [16]. Recently, our group demonstrated that LL-37 migrates to the nucleus and colocalizes with genomic DNA [17]. We hypothesized that this peptide could also directly modulate transcription under certain conditions.
Material and Methods
Chromatin Immunoprecipitation (ChIP)
Cells were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated and DNA sheared to an average length of 300-500 bp. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, proteinase K and heat for de-crosslinking, followed by ethanol precipitation. Resulting DNA was quantified on a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
An aliquot of chromatin (30 ug) was precleared with protein A agarose beads (Invitrogen, USA). Genomic regions of interest were isolated using 10 ug of antibody against cathelicidin (ab69192; Abcam, USA). Complexes were washed, eluted from beads with SDS buffer and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
To validate selected LL-37 binding sites identified by ChIP-Seq assays, quantitative PCR (qPCR) was performed in triplicate on specific genomic regions using SYBR Green Supermix (Bio-Rad, USA). Resulting signals were normalized for primer efficiency by performing qPCR for each primer pair using Input DNA.
DNA Sequencing
Illumina sequencing libraries were prepared from ChIP and Input DNA by standard consecutive enzymatic steps of end-polishing, dA-addition and adaptor ligation. After a final amplification step, resulting DNA libraries were quantified and sequenced on NextSeq 500 (75 nt reads, single end). Reads were aligned to the human genome (hg19) using the BWA algorithm (default settings). Duplicate reads were removed and only uniquely mapped reads (mapping quality ≥ 25) were used for further analysis. Alignments were extended in silico at 3′-ends to a length of 200 bp, which is the average genomic fragment length in the size-selected library, and assigned to 32-nt bins along the genome. The number of fragments in each bin was determined to identify the density of fragments and extended tags. Resulting histograms (genomic “signal maps”) were stored in bigWig files. Peak locations were determined using the MACS algorithm v1.4.2 with a cutoff P-value of 1×10-7 [18, 19]. Signal maps and peak locations were used as input data to the Active Motifs proprietary analysis program, which creates Excel tables containing detailed information on sample comparison, peak metrics, peak locations and gene annotations. Peaks were annotated with genes if located ≤ 10 kb from gene regions.
LL-37 gene silencing
Cells were plated at 2.5×105 cells/well in a 6-well plate overnight. LL-37 Silencer Selected Pre-designed short interfering RNA (siRNA) or negative scramble siRNA (Ambion®) (10 nM each) was combined with 5 µL of Lipofectamine™ RNAiMAX reagent for 20 min. Opti-MEM® I Reduced Serum Medium (Invitrogen) was added at a final volume of 2.5 mL/well after cells were rinsed with PBS. After 48 h, experimental assays were performed.
RNA extraction and microarray experiments
Six independent samples of A375 cells were subjected to gene silencing or a scrambled sequence of the siRNA target sequence. Cells were then lysed in TRIzol reagent and total RNA was isolated following the manufacturer's protocol (Life Technologies, Carlsbad, CA, USA). RNA integrity and concentration were assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA), and total RNA was stored at -80°C until use.
Expression levels of protein-coding genes and long intergenic non-coding RNAs were evaluated by DNA microarray using the SurePrint G3 Human GE V2 Microarray (design ID #G4851B) and Low Input Quick Amp Labeling kit, following a two-color labeling protocol (Agilent Technologies). Cyanine-3 labeled RNA from samples and cyanine-5 labeled reference RNA (Universal Human Reference RNA, Agilent Technologies) were combined and hybridized to microarrays following manufacturer's protocols.
Data preprocessing and filtering
Microarrays were scanned using the SureScan Microarray Scanner (Agilent Technologies) and images were processed using Feature Extraction Software v12 (Agilent Technologies) to determine feature intensities and ratios. Each dataset was preprocessed using the R/Bioconductor arrayQualityMetrics package [20]. Probes that represent the same gene were collapsed by taking their mean divided by the square root of all probes matching the gene. Preprocessing involved background correction and log2 transformation. Data normalization and transformation including quantile normalization and Z-scores are detailed below.
Normalization
After data filtering and preprocessing raw data, quantile normalization was performed using perl scripts and pseudo code. Z-score transformation was calculated using Math::Complex and Statistics::Descriptive perl modules.
Microarray analyses
Differential gene expression analyses were performed using limma R package [21, 22] using a cutoff P-value ≤ 0.01 and mean fold-change > 1.25. Network enrichment analyses were performed using Ingenuity Pathway Analysis suite (IPA; www.qiagen.com/ingenuity) and NetworkAnalyst program (http://www.networkanalyst.ca/) [23]. IPA Upstream Analysis was used to identify upstream regulators and predict whether they were activated or inhibited, given observed gene expression changes in the expression dataset.
Microarray Quality controls
For unintended experimental factors (batch effects), we computed mean absolute distances between microarray data using data from all probes without filtering. Furthermore, we generated a PCA that reflects similarity between microarray arrangements of overall data.
LL-37 gene expression in A375 melanoma cell lines
Differential expression (DE) analysis in the microarray of A375 cell lines (control and LL-37 silenced) showed 379 significantly downregulated and 505 upregulated genes. These results were intersected and 64 downregulated and 80 upregulated genes were obtained. LL-37 demonstrated the lowest decreased fold change value, confirming the experimental design.
Results and Discussion
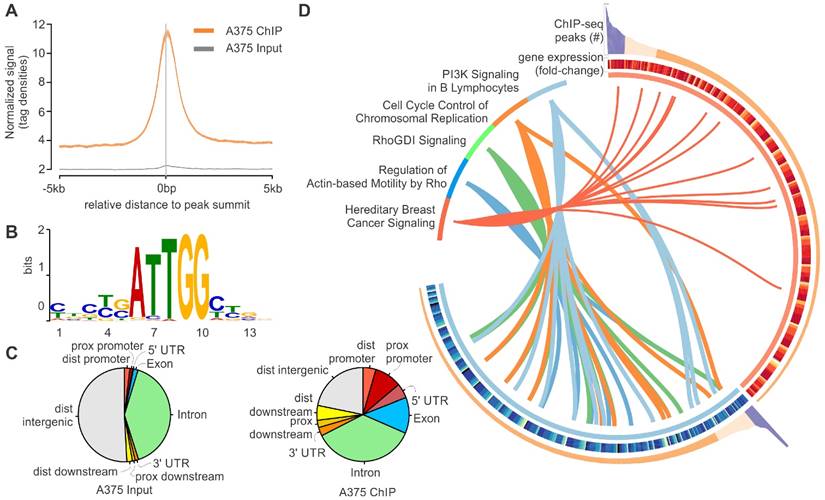
To test this hypothesis, we performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-Seq) of LL-37 in immortalized skin malignant melanoma A375 cells. Peak regions for aligned reads distributed across all target regions, transcriptional start sites, are presented in Fig. 1A. De novo motif analysis was performed using MEME software. The top 1,000 peak sequences (60-bp surrounding peak summits) were used as input (Fig. 1B). Analysis showed peak regions with values > 100 were associated with 460 genes. Peak locations relative to genomic features for A375 cell lines, and a similar number of random locations as control, are shown in Fig. 1C.
LL-37 gene silencing using siRNAs identified differentially expressed genes (DEGs); 379 and 505 genes were down- and upregulated, respectively. As expected, LL-37 was the greatest downregulated gene, confirming successful LL-37 gene silencing. Network enrichment analyses using the Ingenuity Pathway Analysis suite (www.ingenuity.com) revealed that these DEGs are associated with PI3K signaling, cell cycle control and Rho signaling (Fig. 1D).
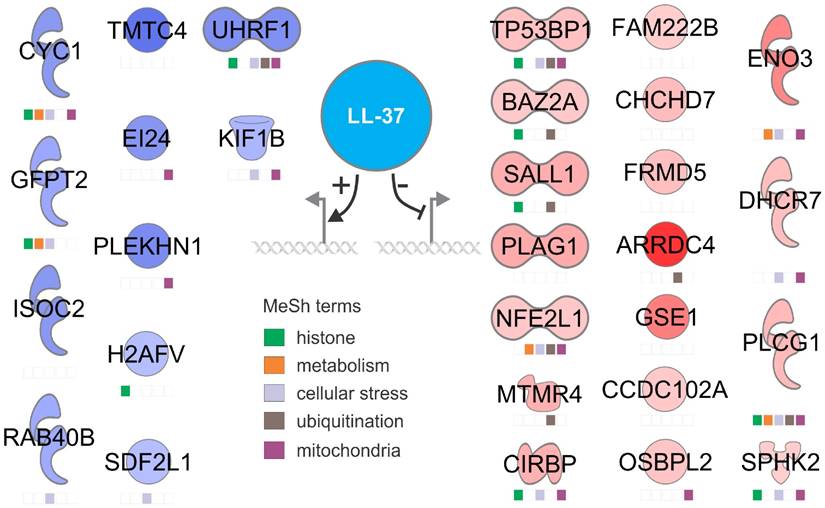
Eleven upregulated and 18 downregulated genes were confirmed by ChIP-Seq experiments. Manual functional annotation revealed that these genes are associated with histone, metabolism, cellular stress, ubiquitination and mitochondria (Fig. 2). We have not confirmed the results with synthetic LL-37, because the effects of exogenous and endogenous LL-37 are different. In fact, exogenous LL-37 is cytotoxic, it is hard to mimmick the effects of endogenous cathelicidin using exogenous LL-37 [24]. RNA silencing is the gold standard in this case.
(A) Average density plot of tag distributions across peak regions. Note the strong enrichment for the A375 ChIP-Seq sample and essentially flat distribution for the Input. (B) De novo motif analysis using MEME software. The top 1,000 peak sequences (60-bp surrounding peak summits) were used as input. (C) Genomic locations for A375 cells and random control peaks were intersected with locations of known genomic features. Note that peaks were preferentially located in promoter and 5'-UTR regions (enrichment over random distribution 10- to 20-fold for A375 cells, in red). (D) Schematic model integrating ChIP-Seq and DNA microarray results with network enrichment analyses.
A working model showing the most relevant up- and downregulated genes by LL-37 at the nuclear level. Functions were assigned to each gene through active PubMed database search using MeSH terms.
The role of LL-37 in cancer has been under intense investigation and both promotion and inhibition of tumor growth have been described, emphasizing its contradictory mechanisms of action. Indeed, LL-37 overexpression has been demonstrated to promote tumor development in ovarian, lung and breast cancers and suppress tumorigenesis in colon and gastric cancers [25, 26]. Interestingly, Kim et al. reported that LL-37 promotes proliferation, migration and invasion in vitro in melanoma cells [27].
By showing that LL-37 may bind to nuclear DNA in a specific manner, we provided evidence, for the first time, that this antimicrobial peptide may also act as a transcription factor, controlling the expression of several genes in malignant melanoma. Further experiments are necessary to confirm our findings and reveal the specific transcriptional program regulated by LL-37. We have preliminary data in SKMEL5 melanoma cells and the results are different, putting in evidence that LL-37 exerts different functions at the nucleus depending on the cell type and tumor cell line.
Acknowledgements
FPS and HIN are supported by the National Council for Scientific and Technological Development and the Sao Paulo Research Foundation.
Conflict of Interest
The authors have no financial or ethical conflicts of interest to declare.
References
1. Ciornei CD, Sigurdardottir T, Schmidtchen A, Bodelsson M. Antimicrobial and chemoattractant activity, lipopolysaccharide neutralization, cytotoxicity, and inhibition by serum of analogs of human cathelicidin LL-37. Antimicrobial agents and chemotherapy. 2005;49:2845-50
2. Larrick JW, Hirata M, Balint RF, Lee J, Zhong J, Wright SC. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infection and immunity. 1995;63:1291-7
3. Ries M, Schuster P, Thomann S, Donhauser N, Vollmer J, Schmidt B. Identification of novel oligonucleotides from mitochondrial DNA that spontaneously induce plasmacytoid dendritic cell activation. Journal of leukocyte biology. 2013;94:123-35
4. Wewers MD, Sarkar A. P2X(7) receptor and macrophage function. Purinergic signalling. 2009;5:189-95
5. Coffelt SB, Tomchuck SL, Zwezdaryk KJ, Danka ES, Scandurro AB. Leucine leucine-37 uses formyl peptide receptor-like 1 to activate signal transduction pathways, stimulate oncogenic gene expression, and enhance the invasiveness of ovarian cancer cells. Molecular cancer research: MCR. 2009;7:907-15
6. Yin J, Yu FS. LL-37 via EGFR transactivation to promote high glucose-attenuated epithelial wound healing in organ-cultured corneas. Investigative ophthalmology & visual science. 2010;51:1891-7
7. Tjabringa GS, Ninaber DK, Drijfhout JW, Rabe KF, Hiemstra PS. Human cathelicidin LL-37 is a chemoattractant for eosinophils and neutrophils that acts via formyl-peptide receptors. International archives of allergy and immunology. 2006;140:103-12
8. Niyonsaba F, Iwabuchi K, Someya A, Hirata M, Matsuda H, Ogawa H. et al. A cathelicidin family of human antibacterial peptide LL-37 induces mast cell chemotaxis. Immunology. 2002;106:20-6
9. Salvado MD, Di Gennaro A, Lindbom L, Agerberth B, Haeggstrom JZ. Cathelicidin LL-37 induces angiogenesis via PGE2-EP3 signaling in endothelial cells, in vivo inhibition by aspirin. Arteriosclerosis, thrombosis, and vascular biology. 2013;33:1965-72
10. Gonzalez-Curiel I, Trujillo V, Montoya-Rosales A, Rincon K, Rivas-Calderon B, deHaro-Acosta J. et al. 1,25-dihydroxyvitamin D3 induces LL-37 and HBD-2 production in keratinocytes from diabetic foot ulcers promoting wound healing: an in vitro model. PloS one. 2014;9:e111355
11. Steinstraesser L, Lam MC, Jacobsen F, Porporato PE, Chereddy KK, Becerikli M. et al. Skin electroporation of a plasmid encoding hCAP-18/LL-37 host defense peptide promotes wound healing. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22:734-42
12. Girnita A, Zheng H, Gronberg A, Girnita L, Stahle M. Identification of the cathelicidin peptide LL-37 as agonist for the type I insulin-like growth factor receptor. Oncogene. 2012;31:352-65
13. Salzer S, Kresse S, Hirai Y, Koglin S, Reinholz M, Ruzicka T. et al. Cathelicidin peptide LL-37 increases UVB-triggered inflammasome activation: possible implications for rosacea. Journal of dermatological science. 2014;76:173-9
14. Nagaoka I, Suzuki K, Niyonsaba F, Tamura H, Hirata M. Modulation of neutrophil apoptosis by antimicrobial peptides. ISRN microbiology. 2012;2012:345791
15. von Kockritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M. et al. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111:3070-80
16. Pinheiro da Silva F, Machado MC. Antimicrobial peptides: clinical relevance and therapeutic implications. Peptides. 2012;36:308-14
17. Pinheiro da Silva F, Medeiros MC, Dos Santos AB, Ferreira MA, Garippo AL, Chammas R. et al. Neutrophils LL-37 migrate to the nucleus during overwhelming infection. Tissue & cell. 2013;45:318-20
18. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE. et al. Model-based analysis of ChIP-Seq (MACS). Genome biology. 2008;9:R137
19. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-60
20. Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics-a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415-6
21. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article3
22. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47
23. Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nature protocols. 2015;10:823-44
24. Pinheiro da Silva F, Gallo RL, Nizet V. Differing effects of exogenous or endogenous cathelicidin on macrophage toll-like receptor signaling. Immunology and cell biology. 2009;87:496-500
25. Piktel E, Niemirowicz K, Wnorowska U, Watek M, Wollny T, Gluszek K. et al. The Role of Cathelicidin LL-37 in Cancer Development. Archivum immunologiae et therapiae experimentalis. 2015
26. Kuroda K, Okumura K, Isogai H, Isogai E. The Human Cathelicidin Antimicrobial Peptide LL-37 and Mimics are Potential Anticancer Drugs. Frontiers in oncology. 2015;5:144
27. Kim JE, Kim HJ, Choi JM, Lee KH, Kim TY, Cho BK. et al. The antimicrobial peptide human cationic antimicrobial protein-18/cathelicidin LL-37 as a putative growth factor for malignant melanoma. The British journal of dermatology. 2010;163:959-67
Author contact
![]() Corresponding author: Fabiano Pinheiro da Silva, MD, PhD, Faculdade de Medicina da Universidade de São Paulo, Laboratório de Emergências Clínicas (LIM-51), Av. Dr. Arnaldo, 455 sala 3189, CEP 01246-000, São Paulo - SP, Brazil. Tel.: +55 11 3061 8480; fax: +55 11 3061 8480. E-mail address: pinheirofabianocom (F. Pinheiro da Silva).
Corresponding author: Fabiano Pinheiro da Silva, MD, PhD, Faculdade de Medicina da Universidade de São Paulo, Laboratório de Emergências Clínicas (LIM-51), Av. Dr. Arnaldo, 455 sala 3189, CEP 01246-000, São Paulo - SP, Brazil. Tel.: +55 11 3061 8480; fax: +55 11 3061 8480. E-mail address: pinheirofabianocom (F. Pinheiro da Silva).

 Global reach, higher impact
Global reach, higher impact