3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(1):1-8. doi:10.7150/jca.16602 This issue Cite
Research Paper
Downregulation of ARID1A, a component of the SWI/SNF chromatin remodeling complex, in breast cancer
1. Department of Surgical Oncology, Gifu University, Gifu University Graduate School of Medicine, Gifu, Japan;
2. Department of Surgery, Kizawa Memorial Hospital, Minokamo, Japan;
3. Department of Pathology and Translational Research, Gifu University Graduate School of Medicine, Gifu, Japan;
4. Department of Breast and Molecular Oncology, Gifu University Graduate School of Medicine, Gifu, Japan.
Received 2016-6-26; Accepted 2016-9-27; Published 2017-1-1
Abstract
Recent studies unraveled that AT-rich interactive domain-containing protein 1A (ARID1A), a subunit of the mammary SWI/SNF chromatin remodeling complex, acts as a tumor suppressor in various cancers. In this study, we first evaluated ARID1A expression by immunohistochemistry in invasive breast cancer tissue specimens and assessed the correlation with the prognosis of patients with breast cancer. Non-tumorous mammary duct epithelial cells exhibited strong nuclear ARID1A staining, whereas different degrees of loss in ARID1A immunoreactivity were observed in many invasive breast cancer cells. We scored ARID1A immunoreactivity based on the sum of the percentage score in invasive cancer cells (on a scale of 0 to 5) and the intensity score (on a scale of 0 to 3), for a possible total score of 0 to 8. Interestingly, partial loss of ARID1A expression, score 2 to 3, was significantly correlated with poor disease free survival of the patients. Subsequently, we performed siRNA-mediated ARID1A knockdown in cultured breast cancer cells followed by comprehensive gene profiling and quantitative RT-PCR. Interestingly, many genes were downregulated by partial loss of ARID1A, whereas RAB11FIP1 gene expression was significantly upregulated by partial loss of ARID1A expression in breast cancer cells. In contrast, a more than 50% reduction in ARID1A mRNA decreased RAB11FIP1gene expression. Immunoblotting also demonstrated that partial downregulation of ARID1A mRNA at approximately 20% reduction significantly increased the expression of RAB11FIP1 protein in MCF-7 cells, whereas, over 50% reduction of ARID1A mRNA resulted in reduction of RAB11FIP1 protein in cultured breast cancer cells. Recent studies reveal that RAB11FIP1 overexpression leads to breast cancer progression. Altogether, the present findings indicated that partial loss of ARID1A expression is linked to unfavorable outcome for patients with breast cancer, possibly due to increased RAB11FIP1 expression.
Keywords: ARID1A, breast cancer, prognosis, chromatin remodeling complex, RAB11FIP1.
Introduction
Condensation of chromosomal DNA by nucleosomes is an essential biological process to store the large amount of DNA in the nucleus, but occludes many regulatory DNA elements. Chromatin remodeling complexes are critical for transcription by facilitating access to packaged DNA [see review in 1]. The SWI/SNF-related, matrix-associated, actin-dependent regulators of chromatin (SMARC), also called BRG1-associated factors (BAFs), have been identified as components of the human SWI/SNF-like chromatin-remodeling protein complexes [2]. AT-rich interactive domain-containing protein 1A (ARID1A; also known as BAF250a, p270, and B120, and SMARCF1) is a subunit of the SWI/SNF chromatin remodeling complex, which possesses DNA binding activity [3-6]. ARID1A contributes to the specific recruitment of its chromatin remodeling activity by binding to transcription factors and transcriptional coactivator/corepressor complexes [7]. As expected by the central role of ARID1A in the chromatin remodeling machinery, the ablation of ARID1A results in early embryonic developmental arrest [8].
In contrast, early pathobiological analysis revealed that mutation in the ARID1A gene results in the development of several malignant tumors [6, 9]. The high prevalence of inactivating ARID1A mutations in endometrial and clear cell carcinomas of the ovary or uterus also suggests that loss of ARID1A function contributes to carcinogenesis in these tumors [10-12]. Furthermore, growing evidence indicates that ARID1A may have a widespread role in the suppression of various tumors [see review in 13]. However, the mechanisms by which mutations in the ARID1A gene drive tumorigenesis are largely unclear. Especially, why insufficient ARID1A expression is linked to tumorigenesis instead of cancer cell death remains a mystery. In addition, the prognosis value of ARID1A expression remains debatable in many cancers.
In this study, we demonstrated that partial loss of ARID1A is related to poor prognosis for patients with invasive breast cancer. Subsequent in vitro comprehensive gene expression analysis unraveled that the expression of more than 80% genes was reduced by partial loss of ARID1A in breast cancer cells. RAB11 family interacting protein 1 (RAB11FIP1, also known as Rab-coupling protein (RCP)), which assists breast cancer progression [14], was significantly increased by the partial loss of ARID1A. The present findings suggest that partial loss of ARID1A might drive mammary gland carcinogenesis, possibly through increasing the RAB11FIP1-mediating pathway.
Materials and methods
Ethical statements
The paraffin-embedded tissues surgically resected from patients with breast cancer were used retrospectively after being used for diagnosis. The need for written informed consent was waived by the Institutional Review Board of the Gifu University Graduate School of Medicine. Instead, the Institutional Review Board requested us to inform the patients that they could refuse the use of their tissue specimens for this study, if they did not want to participate in the present study. The present study was conducted in accordance with the ethical standards of the Helsinki Declaration in 1975, after approval of the Institutional Review Board of the Gifu University Graduate School of Medicine (specific approval number: 25-81).
Antibodies and immunohistochemical staining
A mouse-specific monoclonal (clone PSG3) and conventional rabbit antibodies against ARID1A were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and GeneTex (San Antonio, TX, USA), respectively. We also generated a murine monoclonal antibody against human ARID1A expressed in bacteria. The detailed procedure for preparing recombinant proteins and the establishment of the hybridoma according to the method of Köhler and Milstein were previously described [3]. The characterization of the generated antibody against human ARID1A is described in (Figure S1). A rabbit specific antibody against RAB11FIP1 was purchased from GeneTex.
Archived pathological tissue specimens from 127 invasive ductal carcinomas were used in this study. All tissue specimens were obtained surgically, fixed in 10% buffered formalin, and embedded in paraffin. Tissues were immunostained with antibodies using the ImmPRESS™ polymerized reporter enzyme staining system (Vector laboratories, Inc. Burlingame, CA, USA) as previously reported [15].
Evaluation of immunohistochemical staining and statistical analysis
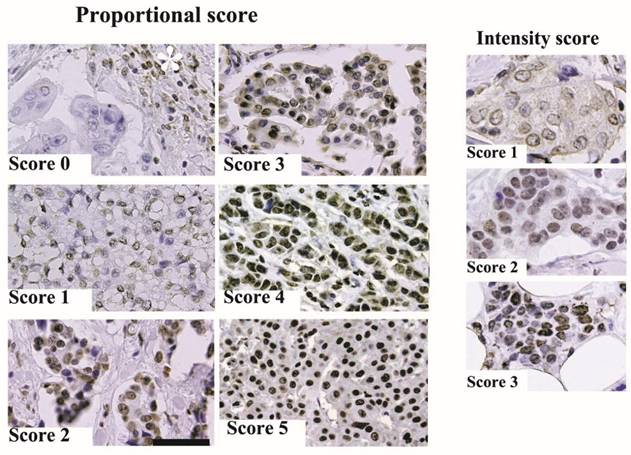
We scored the immunohistochemical staining results as a percentage of ARID1A immunoreactivity in breast cancer cells. The fraction of nuclear ARID1A-positive stained cancer cells was scored after examining six high-power fields (40×) in one tissue section for each case. The proportional intensity was scored on a scale of 0 to 5 (Score 0: less than 50%, score 1: 50-75%, score 2: 75-90%, score 3: 90-95%, score 4: 95-98%, and score 5: over 98% invasive cancer cells exhibited ARID1A immunoreactivity). The intensity score was determined by the ARID1A immunoreactivity of invasive cancer cells, Score 0: negative; score 1: weak compared to ARID1A immunoreactivity in adipose nuclear cells; score 2: equal intensity to adipose nuclear cells; and score 3: stronger than ARID1A immunoreactivity in adipose nuclear cells. The total score was the sum of the intensity + proportional scores.
Curves for disease free survival were drawn using the Kaplan-Meier method and the differences in survival rates were compared using the log-rank test for univariate survival analysis. Multivariate analysis was performed using the Cox proportional hazards model and the R statistics system. A p value of <0.05 was considered statistically significant.
Cell culture and siRNA-mediated RNA interference
A breast cancer cell line, MDA-MB-157 (ATCC HTB-24) was obtained from the ATCC (Manassas, VA, USA). MCF-7 and MDA-MB-231 breast cancer cells were obtained from Japan Health Science Research Resources Bank (JCRB) (Osaka, Japan). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco Life Technologies, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (FBS). Cells were passaged in our laboratory for no more than 6 months after resuscitation.
The detailed procedure for siRNA silencing of a target gene has been previously described [16]. In this study, we employed the Origene (Origene Technologies, Inc. Rockville, MD, USA) siRNAs. 5′-rArGrArUrCrArCrCrArArGrUrUrGrUrArUrGrArGrCrUrGGG-3′ (SR305421A), 5′-rArCrArGrArArGrArArUrGrArUrCrCrArUrUrUrGrUrGrGTG-3′ (SR305421B), and 5′-rArGrArCrUrArCrArArUrGrUrArUrCrArArCrArGrCrArACA-3′ (SR305421C). A Trilencer-27 Universal scrambled negative control siRNA (Origene) was used as a non-silencing control. siRNAs were transfected into cells using lipofectamine™ RNAiMAX following the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). At 48 h after transfection, the cells were used for subsequent studies.
cDNA microarray assay, reverse transcription polymerase chain reaction (RT-PCR), and quantitative real-time RT-PCR
We utilized the Human Whole Genome DNA Microarray system (SurePrint G3 Human 8x60K ver. 2.0, Agilent Technologies, Santa Clara, CA, USA) to obtain the altered gene expression profile for MDA-MB-157 cells in which the ARID1A gene was silenced by siRNA. cDNA synthesis from the total RNA and subsequent PCR were performed using a RT-PCR kit (Takara, Ohtsu, Japan). The procedure was performed according to the manufacturer's instructions, as previously described [15]. Real-time PCR reactions were performed using the SYBR Green reaction kit according to the manufacturer's instructions (Roche Diagnostics, GmbH, Mannheim, Germany) in a LightCycler (Roche Diagnostics). cDNA (2 μL each) was diluted to a volume of 20 μL with the PCR mix containing a final primer concentration of 0.2 pmol.
The following primers were used for real-time RT-PCR: RAB11FIP1-forward 5′-AGAGCTGTCAAGCCCCGACTTCATCCTG-3′; RAB11FIP1-reverse 5′-AGCTGCGCATATGCAAATGCAGGGTCCG-3′; GAPDH-forward 5′-GAAATCCCATCACCATCTTCCAGG-3′; GAPDH-reverse 5′-GAGCCCCAGCCTTCTCCATG-3′; ARID1A-forward 5′-CATGTCCTATGAGCCAAATAAGGATCC-3′; ARID1A-reverse 5′-GAATAACATCCCCGAGCTGGGTTGGAA-3′.
To ensure that the SYBR green was not incorporated into primer dimers or non-specific amplicons during the real-time PCR runs, the PCR products were analyzed by polyacrylamide gel electrophoresis in preliminary experiments. Single bands at the expected sizes were obtained in all instances. The samples were cultured in triplicate and the expression of each target gene was analyzed by the 2-ΔΔCT method described by Livak and Schmittgen [17] using the LightCycler system. The ΔCT values were normalized to GAPDH for each triplicate set in both the Trilencer-27 Universal scrambled negative control siRNA-treated (control) and si-ARID1A-treated groups. The values for the si-ARID1A -treated group were then calculated for each target gene as the fold change relative to the mean values for the control group (control; set to 1.0). The standard deviations were then computed for the triplicate sets, namely, the 3 target genes and the fold changes are presented. In addition, Student's t-tests were performed to determine significant differences. P < 0.05 was considered statistically significant.
Plasmid and transfection
Expression vector containing human ARID1A cDNA under the CMV promoter [18] was kindly provided by Dr. Kato H (Rockefeller University, NY, NY). Cells were transfected using the N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) transfection reagent (Boehringer Mannheim, Indianapolis, IN, USA) according to the manufacturer protocol, as previously described [16]. After G418 selection, three independent ARID1A-overexpressing MCF-7 and MDA-MB-157 cell lines were obtained.
Western blotting
Western blotting was performed as previously described [19], according to Towbin et al. [20]. The separated proteins were transferred onto polyvinylidene difluoride membranes (Millipore Co., Bedford, MA, USA) and probed with an anti-ARID1A antibody, anti-RAB11FIP1 antibody, or anti-GAPDH antibody (Sigma-Aldrich, St. Louis, MO, USA). Immunoreactivity was assessed using a western blotting detection kit (Promega, Madison, WI, USA). Immunoblot bands were quantified by densitometry using LI-COR C-DiGit Blot Scanner imaging software (LI-Cor Biosciences, Lincoln, NE) and were normalized to the GAPDH band.
α5β1Integrin expression and Matrigel invasion assay
Cell surface integrin α5β1 expression was evaluated by immunocytochemical staining using specific antibody (Biorbyt LLC. San Francisco, CA) as previously reported [21].
The invasiveness of the cultured cells was determined using 24-well BD BioCoat Matrigel Invasion Chamber Plates (BD Falcon) according to the manufacturer protocol as previously described [16]. Briefly, 1 × 105 cells were placed in the upper compartment of the invasion chamber. After 48 h of incubation with DMEM containing 10% (lower chamber) or 0% (upper chamber) FBS, the non-invading cells were gently removed from the filter by scrubbing with a cotton-tipped swab. The cells on the lower surface of the filter were counted under a microscope. Experiments were performed in triplicate, and mean and standard deviation values were calculated.
Results
ARID1A expression in invasive breast carcinoma tissue specimens
In this study, we mainly used a newly generated murine monoclonal antibody to determine ARID1A expression by immunohistochemical staining. In addition, we used two commercially available antibodies, a conventional rabbit antibody and murine monoclonal antibody against ARID1A, on several tissue specimens to confirm the results. We obtained similar results with the three antibodies.
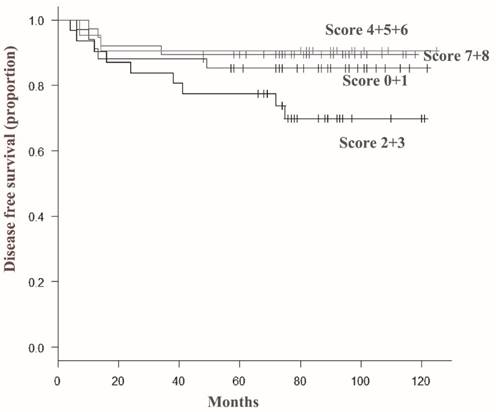
Representative results from the immunohistochemical staining are shown in Figure 1. In this study, we examined ARID1A expression in archived pathological tissue specimens from patients with invasive ductal carcinoma. ARID1A immunoreactivity was scored as the sum of proportional intensity (Score 0 to 5) and intensity score (Score 0 to 3). Subsequently, ARID1A immunoreactivity was classified into four groups (total score 0 or 1, 2 or 3, 4 to 6, and 7 or 8) and subsequently correlated with the prognosis of the patients. The results are summarized in Figure 2. Surprisingly, the disease free survival rate of patients with score 2 or 3 was significantly worse than that of the other groups. Unexpectedly, severe loss of ARID1A immunoreactivity (score 0 or 1) or no loss of ARID1A immunoreactivity (score 7 or 8) had no significant effect on the prognosis of the patients. In the multivariate Cox regression model, nodal metastasis (p=0.0017), triple negative (estrogen receptor-negative, progesterone receptor-negative and HER2-negative) (p=0.043), and partial loss of ARID1A (p=0.0385), were independent prognostic factors.
Representative immunohistochemical staining. (A) ARID1A immunoreactivity was scored according to proportional intensity (Score 0 to 5) and intensity score (Score 0 to 3) as described in Materials and methods. The scale bar represents 100 µm. The asterisk indicates ARID1A immunoreactivity of lymphocytes.
Disease-free survival curves according to total score of ARID1A immunoreactivity in invasive ductal carcinoma cells. The disease free survival rate of patients with a score of 2 or 3 (indicated as 2+3) was significantly lower than that of patients in other groups. In contrast, severe loss of ARID1A immunoreactivity (score 0 or 1) or minimal loss of ARID1A immunoreactivity (score 7 or 8) had no significant effect on the prognosis of the patients.
Partial ARID1A downregulation upregulates RAB11FIP1 expression
In order to examine the molecular mechanism linked to poor prognosis of patients with partial loss of ARID1A in breast cancer cells, we further performed in vitro experiments. By using Human Whole Genome DNA Microarray system (Agilent Technologies), we determined that an approximately 20% reduction of ARID1A mRNA level resulted in the reduction, less than 50%, of the expression of about 772 genes, while it upregulated by 2 folds the expression of 468 genes in MDA-MB-157 breast cancer cells (Figure S2 and Figure S3). Microarray data are deposited in the GEO public database (Array data accession number: GSE72669).
Interestingly, RAB11FIP1 mRNA expression was significantly upregulated by the 20% reduction of ARID1A mRNA level. Since RAB11FIP1 is a human breast cancer-promoting gene with Ras-activating function, we further examined the relation between ARID1A and RAB11FIP1 expression.
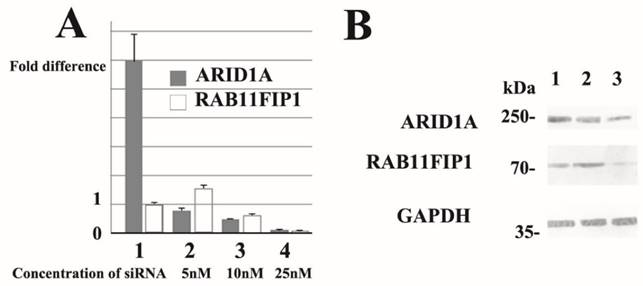
Subsequently, we performed dose-dependent siRNA downregulation of ARID1A mRNA in several breast cancer cell lines and evaluated RAB11FIP1 mRNA expression. Notably, partial loss of ARID1A significantly increased RAB11FIP1 mRNA expression in MCF-7 and MDA-MB-231 cells. Over 50% reduction of ARID1A gene rather reduced RAB11FIP1 mRNA expression in MCF-7 and MDA-MB-231 cells. Subsequent western-blotting analysis confirmed that RAB11FIP1 was overexpressed by partial loss of ARID1A, while loss of ARID1A at a detectable level reduced RAB11FIP1 expression in cultured breast cancer cells. Representative results are presented in Figure 3.
Over 90% reduction of ARID1A gene resulted in significant loss of RAB11FIP1 mRNA in MCF-7 and MDA-MB-231 cells (Figure 3A). Forced overexpression of ARID1A did not significantly alter RAB11FIP1 mRNA expression in MCF-7 and MDA-MB-157 cells (Figure 3A).
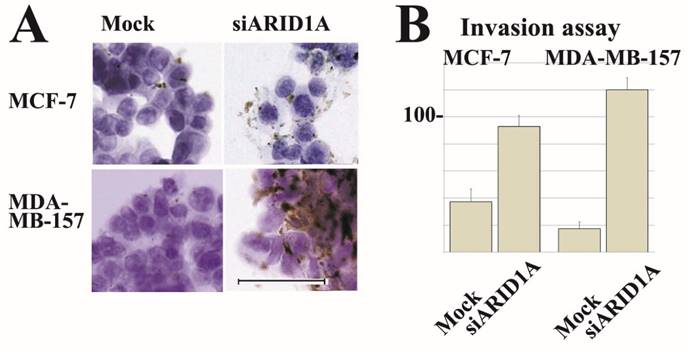
Partial loss of ARID1A expression increased cell surface localization of integrin α5β1 in both, MCF-7 and MDA-MB-157 cells. Furthermore, partial loss of ARID1A expression significantly increased Matrigel-invasion activity of MCF-7 and MDA-MB-157 cells. Representative images are shown in Figure 4.
Discussion
ARID1A gene is somatically mutated in many endometrial carcinomas [11, 12]. However, a recent meta-scale analysis clearly demonstrates that ARID1A gene mutations correlate with better survival in uterine endometrial carcinoma [22]. Meta-scale analysis did not indicate any prognosis values of ARID1A gene mutation in breast cancer [ 22]. In contrast, Zhao et al. reported that ARID1A deletion was detected as an independent prognostic factor in breast cancer [23]. However, mutation of the ARID1A gene is not frequent in breast cancer, approximately 5% [24]. Recently, Zhang et al. reported that promoter hypermethylation of the ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers [25]. These findings encouraged us to score the ARID1A immunoreactivity of invasive breast cancer to evaluate the prognosis value of ARID1A in invasive breast cancers.
Unexpectedly, we determined that partial loss of ARID1A immunoreactivity was significantly related to poor disease free survival rate for patients with invasive breast cancer. In most cases in this group (total score 2 or 3), a weak ARID1A immunoreactivity intensity was detected in over 50% cancer cells.
Representative quantitative reverse transcription polymerase chain reaction (RT-PCR) and immunoblotting results. siRNA treatment reduced ARID1A mRNA (representative data of quantitative RT-PCR ) and ARID1A protein (western-immunoblotting) in breast cancer MCF-7 cells. Results using SR305421A siRNA is shown. Similar results were obtained using other siRNAs (SR305421B and SR305421C). None of the three ARID1A-overexpressing MCF-7 exhibited loss or up-regulation of ARID1A mRNA (lane 1). Partial downregulation (approximately 20%) of ARID1A mRNA resulted in a significant increased (lane 2), whereas 50% ARID1A mRNA reduction caused a decrease in RAB11FIP1 gene expression in MCF-7 cells (lane 3). Moreover, >90% reduction of ARID1A gene resulted in significant loss of RAB11FIP1 mRNA in MCF-7 cells (lane 4). Values are means ± S.D. (n = 3) after 48 h. Statistical analysis was performed by using Student's t test for unpaired observations (P<0.01). Additionally, we performed quantitative RT-PCR experiments using MDA-MB-231 cells and obtained similar results. Partial downregulation (approximately 20%) of ARID1A mRNA significantly increased the expression of RAB11FIP1 protein in MCF-7 cells (Lane 2). In contrast, >50% reduction of ARID1A mRNA resulted in reduction of RAB11FIP1 protein (Lane 3). Lane 1 shows the protein band representing control siRNA treated cells. Concentration of siRNA used in the samples run in Lanes 1 and 2 was 5nM, while 25nM in Lane 3 at final concentration, respectively. In this immunoblot, the results using a monoclonal antibody (clone PSG3) to probe ARID1A is shown. We also obtained similar results using a monoclonal antibody generated in our laboratory. The densitometry values of bands in Lane 2 are 0.726 (ARID1A) and 1.40 (RAB11FIP1), while values in Lane 3 are 0.468 (ARID1A) and 0.678 (RAB11FIP1). The densitometry data are 'fold change' as compared with control (Lane 1) after normalization with respective loading control (GAPDH).
Representative immunocytochemical staining and invasion assay results. siRNA treatment reduced ARID1A mRNA in breast cancer MCF-7 and MDA-MB-157 cells. Results using SR305421A siRNA is shown. Similar results were obtained using other siRNAs (SR305421B and SR305421C). (A) Partial downregulation (approximately 20%) of ARID1A mRNA resulted in a significant increase of integrin α5β1 expression on surface membrane in both MCF-7 and MDA-MB-157 cells. The scale bar represents 50 µm. (B) Partial downregulation of ARID1A mRNA resulted in a significant increase of invasion activity of both MCF-7 and MDA-MB-157 cells. Values are means ± S.D. (n = 3) after 48 h. Statistical analysis was performed by using Student's t test for unpaired observations. (P<0.01).
Subsequent comprehensive gene expression analysis unraveled that most protein-encoding genes were repressed by a 20% reduction of the ARID1A mRNA level in breast cancer cells. However, no tumor suppressor encoding gene was identified among the genes repressed by the partial ARID1A-depletion in breast cancer cells. In contrast, we determined that RAB11FIP1 gene expression was significantly upregulated by the 20% reduction of the ARID1A mRNA level. In the present microarray analysis, we found that 468 genes were up-regulated over 2-fold by partial down-regulation of ARID1A. However, we also noted that signal intensity of Immunoglobulin λ gene expression was 0.06 (normalized value) in 20%-ARID1A-reduced MDA-MB-157 cells. Except for this noise, approximately 165 gene sequences were up-regulated by partial down-regulation of ARID1A. However, most of these sequences included miscRNA, lincRNAs, or unknown gene sequences, which are out of the scope of this study. Finally, we identified RAB11FIP1 as a candidate molecule in partial down-regulation of ARID1A-mediated breast carcinogenesis.
Quantitative RT-PCR confirmed that approximately 20% reduction in ARID1A mRNA significantly increased RAB11FIP1 gene expression in MCF-7 breast cancer cells. We also employed two other siRNA sequences to exclude off-target effects and obtained similar results. Representative data are shown in Fig 3A. Notably, a higher reduction in ARID1A mRNA decreased RAB11FIP1 gene expression. Western-blotting analysis also indicated that partial ARID1A reduction affected RAB11FIP1 expression (Fig 3B).
We also performed immunohistochemical staining using anti-RAB11FIP1 antibody on representative tissue specimens from patients with poor DFS (ARID1A score 2 or 3) and from patients with no disease recurrence after 120 months (AIRD1A score 7 or 8). As demonstrated in (Figure S4), RAB11FIP1 immunoreactivity was stronger in specimens with ARID1A score of 2 or 3 than in those with AIRD1A score 7 or 8.
RAB11FIP1 (also known as Rab-coupling protein, RCP) is often overexpressed in breast cancer as a prime oncogene candidate [14]. RAB11FIP1 plays a role in the Rab-11 mediated recycling of vesicles, i.e., endocytic sorting, trafficking of proteins, and transport between the recycling endosome and the trans-Golgi network [26-28]. RAB11FIP1 plays roles in endosomal trafficking and receptor sorting, including trafficking of integrin α5β1. Integrin α5β1 is a receptor for fibronectin and contributes to cancer cell invasion, metastasis, and resistance to anticancer drugs and is believed to decrease survival in patients [29, 30]. In the present study, we also found that partial down-regulation of ARID1A increased RAB11FIP1 expression, resulted in accumulation of integrin α5β1 on breast cancer cell surface membrane, and significantly up-regulated invasion activity.
Notably, 50% or >90% reduction of ARID1A gene significantly reduced RAB11FIP1 mRNA in breast cancer cells in vitro. The exact molecular mechanism, which was responsible for up-regulation of RAB11FIP1 expression via partial loss of ARID1A expression, remains unclear.
We speculate that insufficient chromatin remodeling, probably caused by partial ARID1A reduction, may alter the three-dimensional structure of the promoter region of the RAB11FIP1 gene. As a result, partial loss of ARID1A increases RAB11FIP1 expression and may result in poor prognosis in patients with invasive breast cancer.
In turn, RAB11FIP1 could be a novel target for the development of therapeutics for patients with breast cancer harboring partial ARID1A loss.
In summary, the present study indicates the important pathobiological role of partial loss of ARID1A in invasive breast cancer.
Supplementary Material
Additional File 1Figure S1, S3-S4.
Figure S2.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436-447
2. Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117-2130 PMID: 8804307
3. Takeuchi T, Chen BK, Qiu Y, Sonobe H, Ohtsuki Y. Molecular cloning and expression of a novel human cDNA containing CAG repeats. Gene. 1997;204:71-77
4. Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879-8888
5. Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137-3146
6. Takeuchi T, Nicole S, Misaki A, Furihata M, Iwata J, Sonobe H, Ohtsuki Y. Expression of SMARCF1, a truncated form of SWI1, in neuroblastoma. Am J Pathol. 2001;158:663-672
7. Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396-420
8. Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656-6661
9. Wang X, Nagl NG Jr, Flowers S, Zweitzig D, Dallas PB, Moran E. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636-642
10. Jones S, Wang TL, Shih IeM, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228-231
11. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih IeM, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532-1543
12. Wiegand KC, Lee AF, Al-Agha OM, Chow C, Kalloger SE, Scott DW, Steidl C, Wiseman SM, Gascoyne RD, Gilks B, Huntsman DG. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328-333
13. Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35-43
14. Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY, Leong WY, Chan YS, Palanisamy N, Liu ET, Karuturi KM, Lim B, Miller LD. RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest. 2009;119:2171-2183
15. Morikawa A, Takeuchi T, Kito Y, Saigo C, Sakuratani T, Futamura M, Yoshida K (2015) Expression of beclin-1 in the microenvironment of invasive ductal carcinoma of the breast. correlation with prognosis and the cancer-stromal interaction. PLoS One. 2015;10:e0125762
16. Takeuchi T, Adachi Y, Nagayama T. WWOX-binding molecule, transmembrane protein 207, is related to the invasiveness of gastric signet-ring cell carcinoma. Carcinogenesis. 2012;33:548-554
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402-428
18. Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeu17chi T, Ohtsuki Y, Sugano S, de Bruijn DR, Chait BT, Roeder RG. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem. 2002;277:5498-505
19. Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, Ohtsuki Y. Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489-1497
20. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350-4354
21. Taniuchi K, Nishimori I, Takeuchi T, Fujikawa-Adachi K, Ohtsuki Y, Onishi S. Developmental expression of carbonic anhydrase-related proteins VIII, X, and XI in the human brain. Neuroscience. 2002;112:93-99
22. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12major cancer types. Nature. 2013;502:333-339
23. Zhao J, Liu C, Zhao Z. ARID1A: a potential prognostic factor for breast cancer. Tumour Biol. 2014;35:4813-4819
24. Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, Hruban RH, Eshleman JR, Iacobuzio-Donahue CA, Goggins M, Maitra A, Malek SN, Powell S, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100-103
25. Zhang X, Sun Q, Shan M, Niu M, Liu T, Xia B, Liang X, Wei W, Sun S, Zhang Y, Liu XS, Song Q, Yang Y, Ma Y, Liu Y, Yang L, Ren Y, Zhang G, Pang D. Promoter hypermethylation of ARID1A gene is responsible for its low mRNA expression in many invasive breast cancers. PLoS One. 2013;8:e53931
26. Jing J, Junutula JR, Wu C, Burden J, Matern H, Peden AA, Prekeris R. FIP1/RCP binding to Golgin-97 regulates retrograde transport from recycling endosomes to the trans-Golgi network. Mol Biol Cell. 2010;21:3041-3053
27. Rainero E, Caswell PT, Muller PA, Grindlay J, McCaffrey MW, Zhang Q, Wakelam MJ, Vousden KH, Graziani A, Norman JC. Diacylglycerol kinase α controls RCP-dependent integrin trafficking to promote invasive migration. J Cell Biol. 2012;196:277-295
28. Baetz NW, Goldenring JR. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol Biol Cell. 2013;24:643-658
29. Morello V, Cabodi S, Sigismund S, Camacho-Leal MP, Repetto D, Volante M, Papotti M, Turco E, Defilippi P. β1 integrin controls EGFR signaling and tumorigenic properties of lung cancer cells. Oncogene. 2011;30:4087-4096
30. Paul NR, Allen JL, Chapman A, Morlan-Mairal M, Zindy E, Jacquemet G, Fernandez del Ama L, Ferizovic N, Green DM, Howe JD, Ehler E, Hurlstone A, Caswell PT. α5β1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J Cell Biol. 2015;210:1013-1031
Author contact
![]() Corresponding author: Fax: +81-058-230-6361; E-mail: takeutitac.jp.
Corresponding author: Fax: +81-058-230-6361; E-mail: takeutitac.jp.

 Global reach, higher impact
Global reach, higher impact