3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(1):124-130. doi:10.7150/jca.17437 This issue Cite
Research Paper
Strong Correlation of Indoleamine 2,3-Dioxygenase 1 Expression with Basal-Like Phenotype and Increased Lymphocytic Infiltration in Triple-Negative Breast Cancer
1. Department of Pathology, CHA Bundang Medical Center, CHA University, 59, Yatap-ro, Bundang-gu, Seongnam-si, Gyeonggi-do, 13496, Korea.
2. Department of Surgery, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea.
3. Department of Pathology, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea.
Received 2016-9-2; Accepted 2016-10-30; Published 2017-1-1
Abstract
Indoleamine 2,3-dioxygenase 1 (IDO1) is an immunosuppressive enzyme involved in tumor immune escape. Blockade of the IDO1 pathway is an emerging modality of cancer immunotherapy. Triple-negative breast cancer (TNBC) lacks established therapeutic targets and may be a good candidate for this novel immunotherapeutic agent. The purpose of this study was to evaluate the clinicopathologic characteristics of the IDO1-expressing TNBC subset. A tissue microarray was constructed from 200 patients with TNBC. Immunohistochemistry (IHC) for IDO1 and TNBC molecular subtype-surrogate markers (AR, GCDFP-15, claudin-3, E-cadherin, CK5/6, and EGFR) was performed using this tissue microarray. Real-time polymerase chain reaction was performed to confirm the IDO1 mRNA expression level in 16 fresh-frozen TNBC samples. Two hundred TNBCs were classified into four subtypes based on surrogate IHC results: 22 luminal androgen receptor type (11.0%), 23 claudin-low type (11.4%), 103 basal-like type (51.5%), and 52 mixed type (26.0%). IDO1 positivity (defined as expression of >10% tumor cells) was observed in 37% of all TNBCs. IDO1 IHC expression was well correlated with mRNA expression. IDO1 positivity was significantly associated with smaller tumor size, dense stromal lymphocytic infiltration, and basal-like phenotype; however, it did not affect the patients' prognosis. IDO1 expression in basal-like TNBCs is considered an immune inhibitory signal that counterbalances active immunity and may reflect the high mutational load of these tumors. Our results suggest which patients with TNBC would be more efficaciously treated with IDO1 blockade.
Keywords: triple negative breast carcinoma, molecular subtype, basal-like phenotype, indoleamine 2, 3-dioxygenase 1 (IDO1), immunohistochemistry, tumor infiltrating lymphocyte (TIL).
Introduction
Indoleamine 2,3-dioxygenase 1 (IDO1) is an intracellular enzyme that mediates the first and rate-limiting step of tryptophan catabolism along the kynurenin e pathway. It contributes to immune suppression by tryptophan depletion and accumulation of immunosuppressive catabolites such as kynurenine [1]. Regarding the biologic significance of IDO1, focus was initially placed on its antimicrobial effect in infectious disease [2, 3] and its ability to prevent allogeneic fetal rejection during pregnancy [4].
More recently, IDO1 has been studied in terms of tumor immunity, which is one of several immune checkpoint proteins involved in tumor immune escape. IDO1 pathway activation induces the blocking of differentiation, cell cycle arrest, and functional anergy of effector T cells and promotes the de novo differentiation of regulatory T cells and functional activation of mature regulatory T cells [5]. IDO1 expression by tumor cells themselves has been reported in a range of carcinomas [6-8]. Blockade of the IDO1 immune-inhibitory pathway is an important emerging anticancer modality in the era of cancer immunotherapy, and several IDO1 inhibitors are currently under clinical development [9].
Triple-negative breast cancer (TNBC) lacks the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) and accounts for 10% to 20% of all breast cancers. The clinical management of TNBC is challenging because of its aggressive biologic nature, and no therapeutic molecular targets have been established [10]. Hence, it is highly desirable to recognize patients who are more efficaciously treated with a novel immunotherapeutic agent such as a IDO1 inhibitor. As a heterogeneous disease, TNBC can be classified into the luminal androgen receptor (LAR) type, claudin-low (CL) type, and basal-like (BL) type based on its gene expression profile [11]. Whether the IDO1 expression differs among TNBC subtypes or correlates with the density of tumor infiltrating lymphocytes (TILs) remains unclear. We evaluated the IDO1 expression in a large cohort of patients with TNBC and the clinicopathologic characteristics of the IDO1-expressing TNBC subset.
Materials and methods
Patient selection and collection of clinicopathologic data
This study included 200 patients who underwent surgical treatment and were diagnosed with TNBC at Ewha Womans University Mokdong Hospital from January 2001 to December 2012. Twelve patients who underwent neoadjuvant chemotherapy before surgical treatment were excluded. TNBC was defined as breast cancer that was entirely negative by immunohistochemistry for ER, PR, and HER2 and by fluorescence in situ hybridization for HER2 amplification. Clinical data including patient age at initial diagnosis, local recurrence, systemic recurrence, and patient survival were retrieved from the medical charts. This study was approved by the Institutional Review Board of Ewha Womans University Mokdong Hospital.
Hematoxylin and eosin-stained slides of all cases were reviewed by two pathologists (S. Kim and S.H. Sung). The histologic subtypes were classified based on the World Health Organization classification of breast tumors [12]. The histological grade was assessed using the Nottingham grading system [13]. Tumor staging was based on the 7th American Joint Committee on Cancer criteria [14]. The density of TILs was recorded as the stromal TIL%, which was defined as the area occupied by mononuclear inflammatory cells over the total intratumoral stromal area according to the recommendation by the International TILs Working Group in 2014 [15].
Construction of tissue microarray
A representative area on the hematoxylin and eosin-stained slides was selected for each case, and corresponding spots were marked on the surfaces of the paraffin blocks. A tissue microarray (TMA) was constructed using 3-mm tissue cores obtained from the blocks. A normal breast tissue core was included in each TMA block. Each tissue core was assigned a unique TMA number and was linked to a database containing the clinicopathologic data.
Immunohistochemistry and interpretation
The following antibodies were used for immunohistochemistry (IHC): androgen receptor (AR) (SP107, 1:100; Roche, Rotkreuz, Switzerland), gross cystic disease fluid protein 15 (GCDFP-15) (23 A3, 1:100; Novocastra, Newcastle, UK), claudin-3 (rabbit polyclonal, 1:100; Abcam, Cambridge, UK); E-cadherin (36B5, 1:100; Novocastra), cytokeratin 5/6 (CK5/6) (CK5/6.007, 1:100; Biocare, Concord, CA), epidermal growth factor receptor (EGFR) (31G7, 1:100; Invitrogen, Carlsbad, CA), and IDO1 (clone 10.1, 1:100; Millipore, Darmstadt, Germany). Briefly, 4-μm sections were transferred to adhesive slides and dried at 62°C for 30 min. After incubation with primary antibodies, immunodetection was achieved by the addition of biotinylated anti-mouse immunoglobulin, followed by peroxidase-labeled streptavidin (a component of a streptavidin biotin kit) and 3,3′-diaminobenzidine as the chromogenic substrate. Slides were counterstained with Harris hematoxylin.
IHC was interpreted by reference to the intensity or proportion of tumor cell staining for nuclear antigens (AR and Ki-67), cytoplasmic antigens (GCDFP-15, CK5/6, and IDO1), and membranous antigens (EGFR, claudin-3, and E-cadherin). IHC results for AR, GCDFP-15, CK5/6, and EGFR were considered positive when >1% of tumor cells were stained [16-18]. Claudin-3 and E-cadherin IHC were scored as compared with positive control. The normal breast tissue cores in each TMA block served as a positive control, and the normal ductules and acini entrapped in tumor were internal positive control in some cases. The staining intensity was scored as intense (strong complete membranous staining comparable to normal epithelial cells), moderate (complete membranous staining, clearly visible with the 20× objective), weak (barely perceptible, faint membranous staining), and negative (no staining) [19]. Tumors exhibiting moderate or intense expression in ≥10% of tumor cells were considered positive [20]. IDO1 positivity was defined as when >10% of tumor cells expressed IDO1 [21]. The results of p53 IHC staining were classified as “mutation type” (completely negative or diffuse strong nuclear staining in >50% of tumor cells) or “wild type” (nuclear staining in ≤50% of tumor cells with heterogeneous intensity) [22]. Molecular subtypes were defined by the IHC results as follows: LAR: AR(+) and/or GCDFP-15(+), CL: claudin-3(-) and/or E-cadherin(-), BL: CK5/6(+) and/or EGFR(+), and mixed: tumors with features of two or three types.
Measurement of Ki-67 labeling index
Ki-67-stained slides were captured digitally in hot spots under 200× magnification. Ki-67 labeling indices were measured using digital image analysis software (Tissue Studio 64 Dual, version 3.5; Definiens, Munich, Germany).
Real-time polymerase chain reaction
The mRNA expression level of the IDO1 gene was analyzed in 16 fresh-frozen TNBC tissues (which entirely overlapped with the IHC-tested cases) by real-time polymerase chain reaction (RT-PCR). Total RNA was extracted from fresh frozen tissues using Trizol Reagent (Invitrogen) and reverse-transcribed to cDNA using an iScript™cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). RT-PCR quantification was performed using the TaqMan® Gene Expression Master Mix (Foster City, CA, USA) on a CFX96 Real-Time PCR Detection System (Bio-Rad). The thermal cycling profile was 30 minutes at 50°C, 20.5 minutes at 8°C, and 2 minutes at 95°C followed by 45 cycles of 15 seconds at 95°C and 30 seconds at 60°C. All RT-PCR reactions were run in triplicate. The transcript level of the IDO1 gene was normalized by glyceraldehyde 3-phosphate dehydrogenase gene expression and calculated using the formula 2-ΔCt.
Statistical analysis
Data were analyzed using SPSS software for Windows, version 20.0 (IBM Corp., Armonk, NY). Fisher's exact test or the χ2 test was used to compare categorical variables. The Mann-Whitney test was used to compare continuous variables (stromal TIL% and Ki-67 labeling index). A p-value of <0.05 was considered to reflect statistical significance. Survival analysis was performed using the Kaplan-Meier method with the log-rank test. Multivariate analysis was performed with Cox's regression.
Results
Patient characteristics
The baseline characteristics of the 200 patients with TNBC are summarized in Table 1. Based on the IHC results, 200 TNBCs were classified into LAR type (n = 22, 11.0%), CL type (n = 23, 11.5%), BL type (n = 103, 51.5%), and mixed type (n = 52, 26.0%). The mixed type comprised LAR+CL (n = 8, 4.0%), LAR+BL (n = 21, 10.5%), CL+BL (n = 18, 9.0%), and LAR+CL+BL (n = 5, 2.5%). IDO1 positivity was observed in 37% of all TNBCs. The follow-up duration ranged from 1 to 147 months. After a median follow-up duration of 32 months, tumor recurrence and death from TNBC occurred in 29 (14.5%) and 16 (8.0%) patients, respectively.
Baseline characteristics of patients with TNBC.
| Parameters | Number (%) | |
|---|---|---|
| Age | <50 | 92 (46.0) |
| ≥50 | 108 (54.0) | |
| Tumor stage | pT1 | 85 (42.5) |
| pT2 | 105 (52.5) | |
| pT3 | 10 (5.0) | |
| Nodal stage | pN0 | 137 (68.5) |
| pN1 | 44 (22.0) | |
| pN2 | 9 (4.5) | |
| pN3 | 10 (5.5) | |
| Histologic grade | I | 6 (3.0) |
| II | 41 (20.5) | |
| III | 153 (76.5) | |
| TNBC subtype | Luminal androgen receptor | 22 (11.0) |
| Claudin-low | 23 (11.5) | |
| Basal-like | 103 (51.5) | |
| Mixed | 52 (26.0) | |
| AR | Positive | 62 (31.0) |
| GCDFP-15 | Positive | 37 (18.5) |
| E-cadherin | Negative | 39 (19.5) |
| Claudin-3 | Negative | 29 (14.5) |
| CK5/6 | Positive | 76 (38.0) |
| EGFR | Positive | 103 (51.5) |
| IDO1 | Positive | 74 (37.0) |
| p53 staining pattern | Wild type | 46 (23.0) |
| Mutatnt type | 154 (77.0) | |
| Recurrence | Present | 29 (14.5) |
| Death | Present | 16 (8.0) |
| Follow-up duration | Median months (range) | 32 (1-147) |
Comparison of clinicopathologic characteristics according to IDO1 protein expression
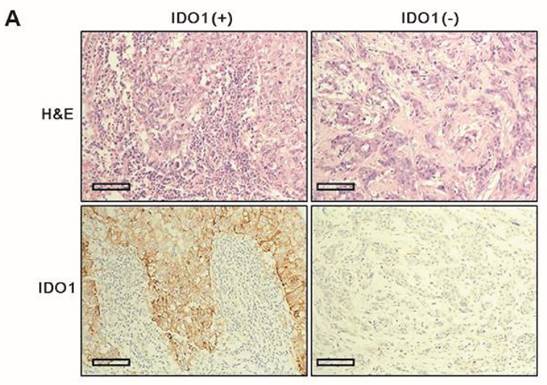
Clinicopathologic parameters were compared between IDO1(+) and IDO1(-) TNBCs (Table 2). IDO1(+) TNBCs were characterized by a smaller tumor size (p = 0.012) and exhibited significantly more dense stromal lymphocytic infiltration than did IDO1(-) TNBCs (p < 0.001) (Fig. 1A). Among the four TNBC subtypes, the BL type most frequently exhibited IDO1 positivity (p = 0.026).
Correlation between IDO1 protein and mRNA expression levels
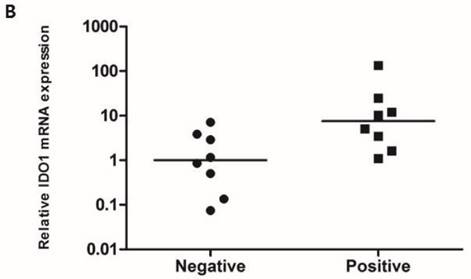
To compare the IDO1 mRNA expression level according to the IDO1 protein expression, RT-PCR was performed with 16 fresh-frozen TNBC samples comprising 8 IDO1(+) and 8 IDO1(-) tumors. The mean expression level of IDO1 mRNA was significantly higher in IDO(+) than in IDO1(-) TNBCs (p = 0.021) (Fig. 1B).
Comparison of clinicopathologic characteristics between patients with and without basal-like phenotype
The BL phenotype was present in 147 (73.5%) patients (103 pure BL type and 44 mixed type). Because many of these patients had the BL phenotype as a component of the mixed subtype, the clinicopathologic differences according to the presence or absence of the BL phenotype were analyzed (Table 3). The BL phenotype was associated with a younger age at the initial diagnosis (p = 0.004), worse histologic grade (p = 0.004), higher stromal TIL density (p = 0.004), higher Ki-67 labeling index (p < 0.001), and a mutant type p53 staining pattern (p < 0.001) than was the lack of a BL phenotype. IDO1 positivity was more frequently observed in TNBCs with a BL phenotype than in those without (p = 0.005).
A. IDO1(+) tumor exhibits conspicuously dense stromal lymphocytic infiltration compared with IDO1(-) tumor (×200 magnification). Bar indicates 100 µm. B. Expression level of IDO1 mRNA is significantly upregulated in IDO1(+) tumors compared with that in IDO1(-) tumors (p = 0.021). The inner bar in the graph indicates the mean value.
Clinicopathologic differences according to IDO1 protein expression.
| Parameters | IDO1 expression | p-value | ||
|---|---|---|---|---|
| Negative (n = 126), n (%) | Positive (n = 74), n (%) | |||
| Age | <50 | 54 (42.9) | 38 (51.4) | 0.304 |
| ≥50 | 72 (57.1) | 36 (48.6) | ||
| Tumor stage | pT1 | 45 (35.7) | 40 (54.1) | 0.012 |
| pT2/T3 | 81 (64.3) | 34 (45.9) | ||
| Nodal stage | pN0 | 81 (64.3) | 56 (75.7) | 0.115 |
| pN1-3 | 45 (35.7) | 18 (24.3) | ||
| Histologic grade | I | 5 (4.0) | 1 (1.4) | 0.600 |
| II | 26 (20.6) | 15 (20.3) | ||
| III | 95 (75.4) | 58 (78.4) | ||
| Stromal TIL % | Q1, Q2, Q3 | 5, 10, 20 | 10, 20, 50 | <0.001 |
| Mean ± SD | 17.2 ± 19.7 | 28.3 ± 24.2 | ||
| Ki-67 labeling index | Q1, Q2, Q3 | 19, 40, 63 | 20, 40, 69 | 0.546 |
| Mean ± SD | 41.3 ± 26.8 | 4.37 ± 2.62 | ||
| TNBC subtype | Luminal androgen receptor | 18 (14.3) | 4 (5.4) | 0.026 |
| Claudin-low | 16 (12.7) | 7 (9.5) | ||
| Basal-like | 55 (43.7) | 48 (64.9) | ||
| Mixed | 37 (29.4) | 15 (20.3) | ||
Q1 the first quartile, Q2 the second quartile, Q3 the third quartile, SD standard deviation.
Clinicopathologic differences according to the presence of basal-like phenotype.
| Parameters | Basal-like phenotype | p-value | ||
|---|---|---|---|---|
| Absent (n=53), n (%) | Present (n=147), n (%) | |||
| Age | <50 | 15 (28.3) | 77 (52.4) | 0.004 |
| ≥50 | 38 (71.7) | 70 (47.6) | ||
| Tumor stage | pT1 | 19 (35.8) | 66 (44.9) | 0.263 |
| pT2/T3 | 34 (64.2) | 81 (55.1) | ||
| Nodal stage | pN0 | 33 (62.3) | 104 (70.7) | 0.301 |
| pN1-3 | 20 (37.7) | 43 (29.3) | ||
| Histologic grade | I | 3 (5.7) | 3 (2.0) | 0.004 |
| II | 18 (34.0) | 23 (15.6) | ||
| III | 32 (60.4) | 121 (82.3) | ||
| Stromal TIL % | Q1, Q2, Q3 | 3, 10, 20 | 5, 15, 40 | 0.007 |
| Mean ± SD | 15.5 ± 19.1 | 23.4 ± 22.8 | ||
| Ki-67 labeling index | Q1, Q2, Q3 | 10, 25, 50 | 25, 45, 70 | <0.001 |
| Mean ± SD | 29.6 ± 26.1 | 46.7 ± 25.3 | ||
| p53 staining pattern | Wild type | 22 (41.5) | 24 (16.3) | <0.001 |
| Mutatnt type | 31 (58.5) | 123 (83.7) | ||
| IDO1 | Negative | 42 (79.2) | 84 (57.1) | 0.005 |
| Positive | 11 (20.8) | 63 (42.9) | ||
Q1 the first quartile, Q2 the second quartile, Q3 the third quartile, SD standard deviation.
Univariate and multivariate analyses for disease-free and overall survival.
| Parameter | Mean DFS months | p-value | HR (95% CI) | p-value | |
| Age | <50 vs. ≥50 | 124 vs. 105 | 0.459 | ||
| Tumor stage | pT1 vs. T2-3 | 135 vs. 101 | 0.016 | 1.800(0.729-4.449) | 0.203 |
| Nodal stage | pN0 vs. N1-3 | 131 vs. 86 | 0.006 | 2.661(1.283-5.519) | 0.009 |
| Histologic grade | I,II vs. III | 122 vs. 122 | 0.617 | ||
| Stromal TIL % | High vs. Low | 117 vs. 110 | 0.002 | 3.549(1.514-8.324) | 0.004 |
| Ki-67 labeling index | Low vs. High | 126 vs. 109 | 0.520 | ||
| Basal-like phenotype | Absent vs. Present | 117 vs. 124 | 0.785 | ||
| IDO1 | Positive vs. Negative | 120 vs. 119 | 0.304 | ||
| Parameter | Mean OS months | p-value | HR (95% CI) | p-value | |
| Age | <50 vs. ≥50 | 132 vs. 112 | 0.713 | ||
| Tumor stage | pT1 vs. T2-3 | 140 vs. 110 | 0.031 | 2.052(0.562-7.490) | 0.277 |
| Nodal stage | pN0 vs. N1-3 | 143 vs. 89 | <0.001 | 10.138(2.886-35.616) | <0.001 |
| Histologic grade | I,II vs. III | 134 vs. 128 | 0.194 | ||
| Stromal TIL % | High vs. Low | 122 vs. 120 | 0.027 | 3.094(0.995-9.619) | 0.051 |
| Ki-67 labeling index | Low vs. High | 133 vs. 115 | 0.509 | ||
| Basal-like phenotype | Absent vs. Present | 131 vs. 129 | 0.529 | ||
| IDO1 | Positive vs. Negative | 128 vs. 126 | 0.459 |
Stromal TIL% and Ki-67 labeling index were split into high or low groups in reference to their respective median values.
Prognostic impact of clinicopathologic parameters
Univariate analysis revealed that an advanced tumor stage (pT2-3), nodal metastasis, and low stromal TIL density significantly predicted shorter disease-free survival (DFS) and overall survival (OS) (Table 4). IDO1 positivity did not affect the DFS and OS. In the Cox proportional hazard model, nodal metastasis was significantly associated with shorter DFS [hazard ratio (HR): 2.661, 95% confidence interval (CI): 1.283-5.519, p = 0.009] and OS (HR: 10.138, 95% CI: 2.886-35.616; p < 0.001). A low stromal TIL density was significantly associated with shorter DFS (HR: 2.661, 95% CI: 1.283-5.519, p = 0.004).
Discussion
The main findings from our results are that IDO1 positivity was unevenly distributed among TNBC subtypes and that IDO1(+) tumors had more active TIL responses than did IDO1(-) tumors. IDO1 positivity was observed in 37.0% of TNBCs and was significantly more prevalent in the BL type than in LAR and CL types. The tight correlation between IDO1 positivity, the BL phenotype, and increased TIL density can be explained in terms of tumor immunogenicity. TIL density is a crucial surrogate reflecting immune responsiveness to tumor peptide. TNBC is more densely infiltrated with lymphocytes than other subtypes [23], which may be partly explained by its high genetic instability and mutational load [24, 25]. Aberrant proteins, termed neoantigens, encoded by mutated genes are more likely recognized by T cells as foreign than are non-mutant proteins and may provoke a more brisk lymphocytic reaction [26]. In this study, we further classified TNBCs into more detailed subtypes (LAR, CL, BL, and mixed types) based on surrogate IHC markers. We found that 73.5% of TNBCs had the BL phenotype, which showed the well-known clinicopathologic characteristics of BL carcinoma, including younger age, high histologic grade, dense lymphocytic infiltration, high Ki-67 proliferation index, and an aberrant p53 staining pattern [27]. BL cancer is strongly associated with BRCA1 dysfunction. Most breast cancers arising in patients with the BRCA1 germline mutation are of the BL type [28]. Additionally, despite the lack of BRCA1 mutation, sporadic BL carcinomas show BRCA1 pathway dysfunction and are phenotypically similar to BRCA1-mutated tumors [29]. Because BRCA1 is involved in DNA mismatch repair through interaction with the mismatch repair proteins MLH1, MSH2, and MSH6 [30], BL-type TNBCs can be considered more mismatch repair-deficient tumors than other types. Dense lymphocytic infiltration in mismatch repair-deficient tumors is a well-known phenomenon shown by colon cancer studies [31, 32]. Thus, the high TIL density of BL-type TNBCs may be partially explained by the mismatch repair deficiency of these tumors.
Burstein et al. [33] proposed four novel TNBC subtypes: the luminal androgen receptor, mesenchymal, BL immunosuppressed, and BL immune-activated types. IDO1 is one of the most highly expressed genes in the BL immune-activated type. In accordance with this result, we found that IDO1 is preferentially expressed in TNBCs with the BL phenotype and a high TIL density. Given that IDO1 is an immunosuppressive enzyme, the positive correlation between IDO1 and active immune reaction seems somewhat paradoxical. However, a recent study showed that microsatellite-instable colon cancers, characterized by defects in mismatch repair and rapid lymphocytic infiltration, showed overexpression of five immune checkpoint proteins: IDO1, CTLA-4, PD-1, PD-L1, and LAG3. This result indicates that the active immune microenvironment is extinguished by immune inhibitory signals as a mechanism to resist tumor elimination [34]. Additionally, a mechanistic study using melanoma proved that IDO1 expression was upregulated by the infiltration of CD8+ T cells [35]. Therefore, IDO1 expression in BL cancer cells can be considered to represent immune-intrinsic negative feedback following the recruitment of activated T cells to the tumor site, rather than being a property of the cancer cells themselves.
A high mutational burden is a proven important predictor of the response to immune checkpoint blockade, including anti-PD-1 [36, 37] and anti-CTLA-4 [38]. The dense immune reaction in BL carcinoma probably reflects the high number of mutation-associated neoantigens. Although the relationship between the mutational burden and responsiveness to IDO1 inhibitor remains unknown, previous results and our findings suggest that TNBC with the BL phenotype is probably the subset in which the IDO1 inhibitor is selectively effective.
In this study, IDO1 positivity was defined as >10% of tumor cells expressing IDO1, and we found that IDO1 mRNA was significantly differentially expressed between IDO1(+) and IDO1(-) TNBCs. This is in agreement with the results of Jacquemier et al. [21], who found that IDO protein and mRNA expression were strongly correlated when using the 10% cut-off value. Although whether IDO1 expression is a biomarker predicting the response to IDO1 inhibitor remains unclear, IDO1 IHC is a reliable method for evaluating IDO1 expression in breast cancer, and 10% is considered a reasonable cut-off value in IHC interpretation.
Our results are similar to those of Jacquemier et al. [21], who reported a strong correlation of IDO1 overexpression with dense lymphocytic infiltration (medullary feature) and the BL subtype. They also found a correlation between IDO1 overexpression and favorable outcomes in the patients with BL breast cancer. In the present study, however, IDO1 expression did not have a prognostic influence on patients with TNBCs of all types, nor when analysis was confined to BL-type TNBC (data not shown).
In conclusion, IDO1 expression was closely associated with TNBC with the BL phenotype and enhanced stromal lymphocytic infiltration, which may reflect the high mutational burden of BL TNBC. Our results suggest which patients with TNBC would be more efficaciously treated with IDO1 immune checkpoint blockade.
Acknowledgements
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea. (HI12C0050).
Conflict of interest
The authors declare that they have no conflicts of interest.
References
1. Takikawa O, Yoshida R, Kido R, Hayaishi O. Tryptophan degradation in mice initiated by indoleamine 2,3-dioxygenase. J Biol Chem. 1986;261:3648-53
2. Yoshida R, Hayaishi O. Induction of pulmonary indoleamine 2,3-dioxygenase by intraperitoneal injection of bacterial lipopolysaccharide. Proc Natl Acad Sci U S A. 1978;75:3998-4000
3. Yoshida R, Urade Y, Tokuda M, Hayaishi O. Induction of indoleamine 2,3-dioxygenase in mouse lung during virus infection. Proc Natl Acad Sci U S A. 1979;76:4084-6
4. Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B. et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191-3
5. Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137-43
6. Nelson BH. IDO and outcomes in ovarian cancer. Gynecol Oncol. 2009;115:179-80
7. Gao YF, Peng RQ, Li J, Ding Y, Zhang X, Wu XJ. et al. The paradoxical patterns of expression of indoleamine 2,3-dioxygenase in colon cancer. J Transl Med. 2009;7:71
8. Pan K, Wang H, Chen MS, Zhang HK, Weng DS, Zhou J. et al. Expression and prognosis role of indoleamine 2,3-dioxygenase in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:1247-53
9. Moon YW, Hajjar J, Hwu P, Naing A. Targeting the indoleamine 2,3-dioxygenase pathway in cancer. J Immunother Cancer. 2015;3:51
10. Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721-8
11. Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61-70
12. Lakhani SR, International Agency for Research on Cancer, World Health Organization. WHO classification of tumours of the breast. 4th ed. Lyon: International Agency for Research on Cancer. 2012
13. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41:154-61
14. Edge SB, American Joint Committee on Cancer. AJCC cancer staging manual. 7th ed. New York; London: Springer. 2010
15. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259-71
16. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z. et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367-74
17. Darb-Esfahani S, von Minckwitz G, Denkert C, Ataseven B, Hogel B, Mehta K. et al. Gross cystic disease fluid protein 15 (GCDFP-15) expression in breast cancer subtypes. BMC Cancer. 2014;14:546
18. Choi J, Jung WH, Koo JS. Clinicopathologic features of molecular subtypes of triple negative breast cancer based on immunohistochemical markers. Histol Histopathol. 2012;27:1481-93
19. Acs G, Lawton TJ, Rebbeck TR, LiVolsi VA, Zhang PJ. Differential expression of E-cadherin in lobular and ductal neoplasms of the breast and its biologic and diagnostic implications. Am J Clin Pathol. 2001;115:85-98
20. Rakha EA, Patel A, Powe DG, Benhasouna A, Green AR, Lambros MB. et al. Clinical and biological significance of E-cadherin protein expression in invasive lobular carcinoma of the breast. Am J Surg Pathol. 2010;34:1472-9
21. Jacquemier J, Bertucci F, Finetti P, Esterni B, Charafe-Jauffret E, Thibult ML. et al. High expression of indoleamine 2,3-dioxygenase in the tumour is associated with medullary features and favourable outcome in basal-like breast carcinoma. Int J Cancer. 2012;130:96-104
22. Alsner J, Jensen V, Kyndi M, Offersen BV, Vu P, Borresen-Dale AL. et al. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta Oncol. 2008;47:600-7
23. Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F. et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol. 2013;31:860-7
24. Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y. et al. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat. 2011;128:23-30
25. Budczies J, Bockmayr M, Denkert C, Klauschen F, Lennerz JK, Gyorffy B. et al. Classical pathology and mutational load of breast cancer - integration of two worlds. J Pathol Clin Res. 2015;1:225-38
26. Disis ML, Stanton SE. Triple-negative breast cancer: immune modulation as the new treatment paradigm. Am Soc Clin Oncol Educ Book. 2015 e25-30
27. Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V. et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157-67
28. Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N. et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482-5
29. Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846-53
30. Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927-39
31. Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527-35
32. Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R. et al. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107-16
33. Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA. et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688-98
34. Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM. et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51
35. Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT. et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116
36. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD. et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-20
37. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124-8
38. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189-99
Author contact
![]() Corresponding authors: Byung-In Moon MD, PhD. Department of Surgery, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea. Phone: +82-2-2650-5584 Fax: +82-2-2644-7984 Email: mbitac.kr. Sun Hee Sung MD, PhD, Department of Pathology, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea. Phone: +82-2-2650-5734 Fax: +82-2-2650-2635 Email: sunhsungac.kr.
Corresponding authors: Byung-In Moon MD, PhD. Department of Surgery, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea. Phone: +82-2-2650-5584 Fax: +82-2-2644-7984 Email: mbitac.kr. Sun Hee Sung MD, PhD, Department of Pathology, Ewha Womans University School of Medicine, 1071, Anyangcheon-ro, Yangcheon-gu, Seoul, 07985, Korea. Phone: +82-2-2650-5734 Fax: +82-2-2650-2635 Email: sunhsungac.kr.

 Global reach, higher impact
Global reach, higher impact