3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(1):131-139. doi:10.7150/jca.16239 This issue Cite
Research Paper
Proteins Involved in HER2 Signalling Pathway, Their Relations and Influence on Metastasis-Free Survival in HER2-Positive Breast Cancer Patients Treated with Trastuzumab in Adjuvant Setting
1. Department of Applied Radiobiology, Maria Sklodowska-Curie Memorial Cancer Centre and Institute of Oncology, Cracow Branch, Cracow, Poland.
2. Department of Systemic and Generalized Malignancies, Maria Sklodowska-Curie Memorial Cancer Centre and Institute of Oncology, Cracow Branch, Cracow, Poland.
3. Department of Tumour Pathology, Maria Sklodowska-Curie Memorial Cancer Centre and Institute of Oncology, Cracow Branch, Cracow, Poland.
Received 2016-5-20; Accepted 2016-9-19; Published 2017-1-1
Abstract
Aim: Resistance to trastuzumab (which is a standard therapy for breast cancer patients with HER2 overexpression) is associated with higher risk of progression or cancer death, and might be related to activation of signalling cascades (PI3K/AKT/mTOR, Ras/Raf/MAPK) and decreased level of their inhibitors.
Material and methods: Formalin-fixed paraffin-embedded tumour specimens from 118 HER2-overexpressing breast cancer patients treated with radical local therapy and trastuzumab in adjuvant setting were used for the assessment of: (1) PIK3CA gene mutations (p.H1047R and p.E545K) by qPCR, and (2) expression of Ki-67, EGFR, MUC4, HER3 and PTEN by immunohistochemistry.
Results: Lower Ki-67LI was observed in EGFR-immunonegative and in PTEN-immunopositive tumours. MUC4-immunonegative tumours more frequently were PTEN- and HER3-immunonegative. Favourable metastasis-free survival was observed in patients with tumours characterized by Ki-67LI≤50% (p=0.027), HER3 immunonegativity or PTEN immunopositivity (vs. tumours with HER3 expression and lack of PTEN expression, p=0.043), additionally, the trend was observed for patients with pN0+pN1 pathological tumour stage (vs. pN2+pN3) (p=0.086). Cox model revealed that independent negative prognostic factors were: (i) Ki-67LI>50% (p=0.014, RR=4.6, 95% CI 1.4-15.4), (ii) HER3 immunopositivity together with PTEN immunonegativity (p=0.034, RR=3.7, 95% CI 1.1-12.5).
Conclusion: The results of our study suggest that combined analysis of HER3 and PTEN expression might bring information on trastuzumab sensitivity in the group of HER2-positive breast cancer patients treated with trastuzumab in adjuvant setting.
Keywords: HER2-overexpressing breast cancer, PTEN, HER3, MUC4, PIK3CA mutations.
Introduction
Breast cancer is one of the most common malignancies in women. Overexpression of human epidermal growth factor receptor type 2 (HER2), associated with poorer prognosis, is detected in 20-25% of invasive breast cancer cases [1].
Trastuzumab, which is an antibody against the IV domain of extracellular fragment of HER2, is currently applied in adjuvant setting for radically treated HER2-overexpressing breast cancer patients [2]. However, previous studies demonstrated that only 10-34% of patients with advanced breast cancer responded to trastuzumab in monotherapy [3]. This finding suggests that blocking only HER2 receptor might be not enough for silencing all HER2-related signalling pathways and preventing tumour progression. There are apparently some other targets regulating HER2 signaling pathways, such as PI3K/AKT/mTOR and Ras/Raf/MAPK.
There are several most often discussed potential mechanisms involved in gaining resistance to trastuzumab treatment. One of them is overexpression of MUC4 protein, which binds with HER2 and masks epitopes recognized by trastuzumab [4 - 7]. Other potential causes for ineffectiveness of trastuzumab therapy might be activation of HER2 signalling cascades caused by: (i) interaction of HER2 with HER3, HER4, EGFR [5 - 8], (ii) PIK3CA (phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit) activating mutations [5, 8], (iii) downregulation of PTEN protein expression [4 - 8]. These molecular mechanisms are only a few of all possible pathways generating trastuzumab resistance [4 - 8].
The aforementioned molecularly-determined resistance might be potentially responsible for higher risk of progression or cancer death in trastuzumab-treated patients with HER2-overexpressing breast cancer. It is hardly unlikely that patients' survival rate could be influenced only by a single factor. The most probable is a joint effect of two or more events. Therefore, the aim of our study was to analyse the prognostic value of potential factors involved in trastuzumab resistance separately and in combination. We analysed metastasis-free survival of HER2-positive breast cancer patients treated with trastuzumab in adjuvant setting according to PIK3CA mutation status, Ki-67, PTEN, MUC4, HER3 and EGFR expression.
Material and Methods
Patients
One hundred eighteen patients with invasive ductal breast cancer (T≥1, N≥0, M0) with overexpression of HER2 were included into the study. Patients underwent radical surgery between 2007 and 2011 at the Department of Surgical Oncology, Centre of Oncology, Cracow Branch and did not receive neoadjuvant chemotherapy or radiotherapy. The mean age of patients was 53 ± 0.9 (mean ± SE) (range 31 - 79) years.
Patients received chemotherapy and trastuzumab in adjuvant setting. Hormonotherapy was applied in 59 patients with tumours presenting estrogen/progesterone receptor (ER/PR) expression. Clinical and histological characteristics of cases is presented in Table 1. The study was approved by Ethical Committee at the Regional Medical Chamber in Cracow (decision from 4th December 2013). No specific consent was needed for this study, as this was a retrospective study performed on archived tissues with no direct patient contact, no modification of diagnostic or treatment procedures and no personal patients' data revealed.
Clinicopathological characteristics of invasive ductal breast cancer patients (n=118).
| Parameter | Category | N (%) |
|---|---|---|
| pT1 | 1 | 43 (38.7) |
| 2 | 67 (60.4) | |
| 3 | 1 (0.9) | |
| pN | 0 | 53 (44.9) |
| 1 | 37 (31.4) | |
| 2 | 19 (16.1) | |
| 3 | 9 (7.6) | |
| Grade2 | 2 | 34 (29.6) |
| 3 | 81 (70.4) | |
| Surgery | BCS | 41 (34.7) |
| Madden | 76 (64.4) | |
| Mastectomy + sentinel node | 1 (0.9) | |
| Hormonal therapy | Not administered | 59 (50.0) |
| Administered | 59 (50.0) | |
| Radiotherapy | Not administered | 22 (18.6) |
| Administered | 96 (81.4) | |
| Chemotherapy3 | TAC | 1 (0.9) |
| ACT | 42 (35.9) | |
| AC | 69 (60.0) | |
| FAC | 5 (4.3) |
1 pT was not assessed in 7 cases, 2 grade was not assessed in 3 cases, 3 chemotherapy was not applied in one case, BCS - breast-conserving surgery, TAC: docetaxel, doxorubicin, and cyclophosphamide, ACT: doxorubicin, cyclophosphamide and paclitaxel, AC: doxorubicin and cyclophosphamide, FAC: 5-fluorouracil, doxorubicin and cyclophosphamide.
Material
Archival tumour specimens were re-examined independently by two pathologists (A.H.-L., A.A.) to confirm histological diagnosis and tumour grade.
Immunohistochemistry (IHC) staining
Status of ER, PR, HER2 expression was evaluated during diagnostic procedures and retrieved from patients' files. Tumours with >1% of tumour cells expressing ER or PR were considered as immunopositive and in these cases hormonal therapy was applied. [9]. Overexpression of HER2 was assessed using HercepTest (Dako Denmark A/S, Glostrup Denmark). In case of expression assessed as +2 (unclear result), amplification of HER2 gene was tested using fluorescence in situ hybridization (FISH) - PathVysion HER2 DNA Probe (Abbot Molecular). Overexpression of HER2 protein/amplification of HER2 gene was evaluated according to recommended standards [10].
Expression of PTEN, MUC4, EGFR, HER3 and Ki67 was assessed immunohistochemically. The sections from formalin-fixed paraffin-embedded tissues were cut at 4 μm, mounted on SuperFrost® Plus (Menzel-Gläser, Germany) slides, and then deparaffinized and hydrated through a series of xylenes and alcohols. Details on IHC procedures are presented in Table 2. Shortly after antigen unmasking and exogenous peroxidases quenching (30 min. in 0.3% H2O2 diluted in methanol), non-specific binding of antibodies was blocked with UltraVision Protein Block (Thermo Scientific, Fremont, USA). Following incubation with primary antibody, for protein visualization, BrightVision detection system (Immunologic, Duiven, The Netherlands) and DAB (Vector Laboratories, Inc., Burlingame, USA) were applied. Hematoxylin was used for nuclear counterstaining.
We were not able to obtain IHC results for all proteins in all cases (Table 2). In some cases insufficient amount of tissue in paraffin blocks or small fragments of tumour tissue hindered obtaining reliable results.
Detailed information on immunohistochemical procedures.
| Antigen | Clone | Manufacturer | Antigen retrieval | Dilution | Number of positive/all stained cases | ||
|---|---|---|---|---|---|---|---|
| PTEN | 138G6 | Cell Signaling1 | TRS, pH=6.1 DAKO, 50 min., 96oC | 1:75 a | 21/111 | ||
| Ki-67 | MIB-1 | DAKO2 | 1:100 b | 114/114 | |||
| MUC4 | 1G8 | BioGenex3 | 1:60 b | 36/108 | |||
| HER3 | SP71 | ThermoFisher Scientific4 | 1:100 b | 32/113 | |||
| EGFR | H11 | DAKO2 | Proteinase K, 10 min., RT | 1:200 b | 20/111 | ||
1 Cell Signaling, Danvers, USA.
2 DakoCytomation Denmark A/S, Glostrup, Denmark.
3 BioGenex, Fremont, USA.
4 Thermo Fisher Scientific, Fremont, USA.
Incubation with primary antibody: a 1 h at 37oC; b overnight, 4oC.
IHC evaluation
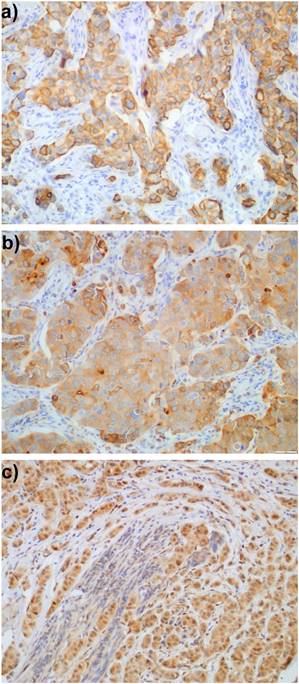
Expression of proteins was evaluated exclusively in the invasive component of the tumours; ductal carcinoma in situ (DCIS) was not included in the evaluation. EGFR expression in more than 1% of tumour cells was determined as immunopositivity, similarly to Gori et al. [11]. To assess cytoplasmic staining (HER3, PTEN, MUC4) a quaternary scale was employed, taking into account the intensity of staining: 0 - lack of staining, 1 - weak, 2 - moderate, and 3 - strong staining. HER3 staining was mostly observed in cytoplasm, although sometimes the outline of cell membranes was visible. Usually, membranous staining was accompanied by cytoplasmic pattern. HER3 was considered immunopositive (Fig. 1a) when in more than 10% of tumour cells a weak to moderate staining of the entire membrane was detected, equivalently to Park et al. [12] or strong cytoplasmic staining (clearly distinguished from stroma, score 3) was present. Because of a very limited number of papers concerning MUC4 expression in breast cancer treated with trastuzumab, we decided to define immunopositivity of MUC4 (Fig. 1b) as score >1 (moderate and strong staining). Tumours with strong cytoplasmic expression of PTEN (score 3) (Fig. 1c) were classified as PTEN-immunopositive as defined by Beelen et al. [13]. Ki-67 labelling index (Ki-67LI) was assessed in about 5 high power fields in >1000 tumour cells and was calculated as the percentage of cells with nuclear Ki-67 immunopositivity.
Expression of: a) HER3, b) MUC4, and c) PTEN in breast cancer cells.
qPCR analysis of PIK3CA mutation status
Detailed procedure of DNA isolation and qPCR procedure (with PCR reaction mixture components and thermocycling conditions) was described elsewhere [14]. DNA was isolated from formalin-fixed, paraffin-embedded tumour tissue blocks using ReliaPrep FFPE gDNA Miniprep System from Promega Corp. qPCR reactions were performed using ViiA 7 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) and commercially available kits (Applied Biosystems, Foster City, CA, USA): (i) PIK3CA 775 TaqMan® Mutation Detection Assay (Assay ID Hs00000831_mu) to establish the H1047R PIK3CA mutation status, (ii) PIK3CA 763 TaqMan® Mutation Detection Assay (Assay ID Hs00000824_mu) to establish the E545K PIK3CA mutation status, (iii) PIK3CA TaqMan Mutation Detection Reference Assay (Assay ID Hs00001025_rf) as reference, assessing conservative fragment of PIK3CA gene. Data from qPCR were analysed using Mutation Detector Software.
Statistical analysis
For continuous variables descriptive statistics were used to determine mean values and standard errors of means (SE) and Mann-Whitney test for testing differences between two groups. Relationships between categorical variables were analysed using Yates test for 2x2 tables. Metastasis-free survival (MFS) was calculated from the time of surgery to clinically- or radiographically-confirmed presence of metastases. The probability of survival was estimated using the Kaplan-Meier method. The log-rank test was applied to: (i) investigate the statistical significance of the differences observed between two groups and (ii) establish the cut-off point for Ki-67LI. P<0.05 was considered significant in all statistical procedures. STATISTICA 10 software, (StatSoft, Inc., Tulsa, OK, USA) was used for analysis of data.
Results
Relations between PTEN, Ki-67, MUC4, HER3, EGFR expression, PIK3CA mutations and clinical parameters
Among the investigated group of HER2-expressing breast cancers, there were 59 (50.0%) cases without expression of both estrogen and progesterone receptors. EGFR imunopositivity was detected in 20 (18.0%) cases, HER3 in 32 (28.3%), MUC4 in 36 (33.3%) and PTEN in 21 (18.9%) cases. In the studied group there were 14 (14.0%) tumours with H1047R mutation and 2 (2.0%) with E545K mutation in PIK3CA gene. The presence of H1047R mutation and E545K mutation analysed separately was not related to pT, pN, tumour grade, expression of EGFR, Ki-67, MUC4, PTEN, and HER3. For further analysis we divided the studied group into two subgroups: with at least one mutation and without any, however, no relations between PIK3CA mutational status and other parameters were found. Ki-67LI was assessed in 114 cases. The mean Ki-67LI was 36.3% ± 1.3 (mean ± SE) (range 0.0 - 74.0).
Lower value of Ki-67LI was observed in EGFR immunonegative tumours (34.53% vs. 42.11%) as well as in PTEN immunopositive tumours (30.32% vs. 37.63%) (Table 3). Higher Ki-67LI was observed in G3 and ER/PR immunonegative carcinomas (Table 3), however those relationships did not reach statistical significance.
Relations between studied parameters.
| Parameter | Category | All | Ki-67LI | PIK3CA mutations (N) | EGFR (N) | PTEN (N) | HER3 (N) | MUC4 (N) | ER/PR (N) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | (N) mean±SD | - | + | - | + | - | + | - | + | - | + | - | + | ||
| pT | 1 | 43 | (42) 34.80±13.77 | 34 | 4 | 31 | 7 | 29 | 10 | 31 | 10 | 26 | 11 | 22 | 21 |
| 2+3 | 68 | (67) 37.97±13.89 | 44 | 11 | 55 | 12 | 56 | 10 | 48 | 19 | 43 | 22 | 34 | 34 | |
| pN | 0+1 | 90 | (87) 35.90±13.61 | 65 | 13 | 67 | 16 | 69 | 16 | 65 | 22 | 57 | 27 | 48 | 42 |
| 2+3 | 28 | (27) 37.67±15.41 | 19 | 3 | 24 | 4 | 21 | 5 | 16 | 10 | 15 | 9 | 11 | 17 | |
| G | 2 | 34 | (31) 32.53±1.55 | 24 | 6 | 30 | 3 | 26 | 5 | 22 | 10 | 22 | 9 | 12 | 22 |
| 3 | 81 | (80) 37.79±14.85 | 58 | 10 | 58 | 17 | 61 | 16 | 57 | 21 | 48 | 26 | 46 | 35 f | |
| ER/PR | - and - | 59 | (58) 37.66±15.22 | 41 | 8 | 37 | 18 | 47 | 11 | 40 | 17 | 34 | 21 | ||
| + or + | 59 | (56) 34.93±12.62 | 43 | 8 | 54 | 2 c | 43 | 10 | 41 | 15 | 38 | 15 | |||
| MUC4* | - | 72 | (71) 35.60±14.22 | 51 | 9 | 59 | 9 | 61 | 9 | 59 | 13 | ||||
| + | 36 | (35) 37.63±14.21 | 26 | 5 | 25 | 10 | 24 | 12 d | 17 | 17 e | |||||
| HER3 | - | 81 | (81) 37.74±15.19 | 59 | 11 | 62 | 14 | 64 | 14 | ||||||
| + | 32 | (31) 33.22±9.93 | 21 | 5 | 25 | 6 | 24 | 7 | |||||||
| PTEN | - | 90 | (88) 37.63±13.86 | 62 | 14 | 69 | 18 | ||||||||
| + | 21 | (21) 30.32±13.51 a | 16 | 2 | 18 | 2 | |||||||||
| EGFR | - | 91 | (88) 34.53±12.66 | 66 | 12 | ||||||||||
| + | 20 | (20) 42.11±18.07 b | 12 | 3 | |||||||||||
| PIK3CA mutations ** | - | 84 | (81) 34.78±14.41 | ||||||||||||
| + | 16 | (15) 35.61±14.43 | |||||||||||||
PIK3CA: phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit; ER: estrogen receptor; PR: progesterone receptor; PTEN: phosphatase and tensin homolog deleted on chromosome ten; EGFR: epidermal growth factor receptor; MUC4: mucin 4; HER3: human epidermal growth factor receptor 3.
*: “+”immunopositive cases, “-“ immunonegative cases; ** “-“ tumours without H1047R or E545K PIK3CA mutations “+” tumours with H1047R or/and E545K PIK3CA mutations.
p from Mann-Whitney test: a: p=0.035; b: p=0.022; p from Yates test c: p<0.001; d: p=0.025; e: p=0.001; f: p=0.057.
MUC4 immunonegative tumours more frequently were PTEN and HER3 immunonegative than MUC4-positive ones (Table 3).
Survival analysis
Survival analysis was conducted in a group of 116 patients. From this analysis we excluded one patient without follow up data and one patient who refused chemotherapy.
Mean time of follow-up was 58.4 months (range 13.6 - 98.6). For Ki-67LI, minimal p-value method from log-rank test was used to establish the cut-off point. We selected 50%, as the cut-off point, indicating maximal difference in survival rate between groups stratified according to Ki-67LI. Studied group consist of HER2-overexpressing carcinomas, which are typically characterized by high proliferation, therefore Ki-67LI cut-off point (50%) in this study is relatively high. In our previous study analysing all subtypes, lower cut-off point was established [15].
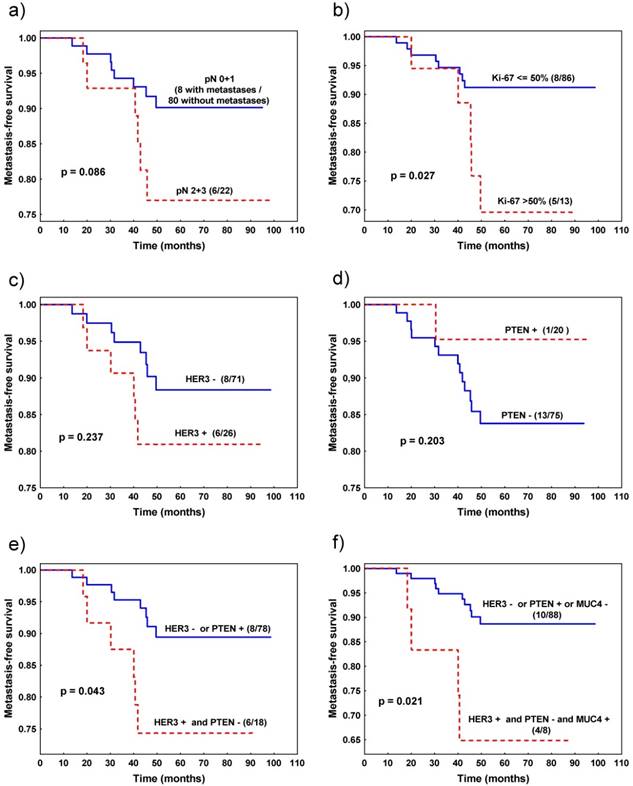
Favourable metastasis-free survival was observed in patients with tumours characterized by Ki-67LI≤50% (vs. Ki-67LI>50%) (p=0.027, Fig. 2b) and the trend was noted for patients with pN0 and pN1 (vs. pN2 + pN3) pathological stage (p=0.086, Fig. 2a) (Table 4).
Metastasis-free survival of patients with invasive ductal breast cancer stratified by: a) nodal status, b) Ki-67 expression, c) HER3 expression, d) PTEN expression, e) combined HER3 and PTEN expression, f) combined HER3, PTEN and MUC4 expression.

Metastasis-free survival according to studied parameters.
| Parameter | Number of patients with metastases /without progression | p value | 5-yr MFS (%) | |
|---|---|---|---|---|
| pT | 1 | 4/37 | 0.500 | 89.5 |
| 2+3 | 9/59 | 84.9 | ||
| pN | 0+1 | 8/80 | 0.086 | 90.1 |
| 2+3 | 6/22 | 77.0 | ||
| G | 2 | 4/29 | 0.934 | 87.7 |
| 3 | 10/70 | 86.6 | ||
| ER/PR* | - | 5/52 | 0.293 | 90.7 |
| + | 9/50 | 83.2 | ||
| Ki-67LI | ≤ 50% | 8/86 | 0.027 | 91.2 |
| > 50% | 5/13 | 69.6 | ||
| HER3 | - | 8/71 | 0.237 | 88.4 |
| + | 6/26 | 80.9 | ||
| MUC4 | - | 8/62 | 0.766 | 87.1 |
| + | 5/31 | 85.5 | ||
| PTEN | - | 13/75 | 0.203 | 83.8 |
| + | 1/20 | 92.5 | ||
| EGFR | - | 10/79 | 0.349 | 87.7 |
| + | 4/16 | 79.9 | ||
| PIK3CA mutations** | - | 10/72 | 0.895 | 87.2 |
| + | 4/14 | 84.4 | ||
| HER3/PTEN | HER3- or PTEN + | 8/78 | 0.043 | 89.4 |
| HER3+ and PTEN - | 6/18 | 74.4 | ||
| HER3/PTEN/MUC4 | HER3- or PTEN + or MUC4- | 10/88 | 0.021 | 88.7 |
| HER3+ and PTEN- and MUC4+ | 4/8 | 64.8 | ||
MFS - metastasis-free survival.
*: “+” identifies ER or PR immunopositive cases, “-“ - ER and PR immunonegative caes; ** “-“ tumours without H1047R or E545K PIK3CA mutations “+” tumours with H1047R or/and E545K PIK3CA mutation.
Other analysed parameters, such as grade, pT, status of ER/PR, EGFR, PTEN, MUC4, HER3 expression and PIK3CA mutation status (analysed separately for each mutation, as well as in combination) did not statistically significantly influence survival of breast cancer patients by itself. In other words, based on expression of only one protein (except Ki-67) we were not able to predict patients' survival. However, for HER3 and PTEN, survival curves were clearly separated (Fig. 2c, d). Therefore, we studied the cumulative effect of HER3 and PTEN and discovered that patients with tumour characterized by HER3 immunonegativity or PTEN immunopositivity survived longer than patients with tumour presenting HER3 expression and lack of PTEN expression (p=0.043, Fig 2e, Table 4). This effect was even more profound when we added MUC4 expression to the model. Patients with tumours characterized by the presence of three potentially negative factors: HER3 and MUC4 immunopositivity and PTEN immunonegativity survived shorter than the rest of the patients (p=0.021, Table 4, Fig. 2f), although the group with poor prognosis consisted only of 12 patients.
Finally, we included Ki-67LI and cumulative effect of HER3 and PTEN to the Cox multivariate analysis. Both variables appeared to be independent negative prognostic factors influencing metastasis-free survival: (i) Ki-67LI>50% (p=0.014, RR=4.6, 95% CI 1.4-15.4), (ii) HER3 immunopositivity with PTEN immunonegativity (p=0.034, RR=3.7, 95% CI 1.1-12.5).
Discussion
MUC4 is a member of transmembrane mucin family. It is expressed in many normal epithelial tissues and secretions (salivary glands, reproductive tract, mammary epithelium, tears, saliva) as well as in many inflammatory diseases and malignancies [16]. MUC4 contains a few functional domains: tandem repeated, NIDO, AMOP, vWD and (at C-terminal) three EGF-like domains [16]. It is postulated that through EGF-like domain, which acts like ligand, MUC4 interacts with HER2 and increases HER2 phosphorylation [7]. Unfortunately, most of the studies concerning MUC4 in aspect of trastuzumab treatment are conducted on cell lines and animal models. Publications where MUC4 is studied in patients cohorts treated with trastuzumab are very scarce [17]. To the best of our knowledge, for the first time we studied MUC4 in HER2-positive breast cancer patients treated with trastuzumab in adjuvant setting. We detected strong/moderate MUC4 expression in 33.3% of cases and did not observe statistically significant differences in MFS according to its expression.
PIK3CA gene encodes one of the 3 forms of p110, which is a catalytic subunit of phosphatidylinositol 3 kinase (PI3K). Alterations of this gene (mutations, increased copy number) were detected in a variety of malignancies [18] and most of them (80%) were located within helical (E542K, E545K) and kinase (H1047R) domains. These activating mutations of PIK3CA are responsible for increased catalytic activity of PI3K (production of phosphatidylinositol [3,4,5] triphosphate [PIP3]) resulting in cell proliferation and survival [19].
Studies concerning relation between mutation status of PIK3CA gene and clinical/biological parameters or its influence on patients' survival/tumour reaction after trastuzumab treatment are difficult to compare. It should be mentioned that these studies were performed in groups of cancer patients with different clinical characteristics or using different material (serum, paraffin-embedded formalin-fixed or frozen tissues). Even more importantly, mutational status of PIK3CA gene was studied using different methods (qPCR, sequencing) and also particular sets of investigated mutations can vary from study to study. In our material, 14 (14.0%) tumours with H1047R mutation and 2 (2.0%) with E545K mutation of PIK3CA gene were detected. This result is within the range of other studies, where mutations were found in 12 - 24% of cases [13, 20-29]. In our series, as in others [24, 25, 27], there was no relation between PIK3CA mutation status and other studied parameters, however, some authors reported associations with grade [13] or nodal status (in ER-positive group) [26]. Influence of PIK3CA mutation status on breast cancer patients' survival is ambiguous, what can be caused by analysing different groups with different clinical characteristics (metastatic breast cancer [22, 29, 30] or patients without distant metastases [25, 31], or various treatment regiments implemented: trastuzumab in neoadjuvant [23, 25, 28] or adjuvant [24, 31] setting). In some studies, patients with tumour characterized by activating PIK3CA mutation had shorter time to progression [29] or had less frequently pathologic complete response [23, 25, 28]. Other studies did not confirm the influence of PIK3CA mutation status on survival (disease/progression/recurrence-free [20, 21, 24], overall [22]) or response to trastuzumab treatment [22]). So far, data suggest that patients with tumours bearing PIK3CA mutation are less likely to have pathological complete response after neoadjuvant trastuzumab treatment and, in case of advanced disease, PIK3CA mutation status might be linked to shorter progression-free survival [32, 33].
Frequently, PIK3CA mutation status and PTEN expression are studied together, because PTEN is a phosphatase and acts as an antagonist of PI3K. In our study, 81.1% of tumours were characterized by low PTEN expression. Other authors noted PTEN loss or weak expression in 15.6-66.2% of cases [11, 12, 17, 21, 22, 23, 29, 34]. Moreover, in our series, survival analysis revealed that only one patient with tumour presenting strong PTEN expression had progression of disease (metastases to liver and adrenal). Nevertheless, we found no statistical significance between groups identified based on PTEN expression (Table 4, Fig 2d). The data concerning prognostic/predictive role of PTEN expression are not consistent. In metastatic breast cancer patients with tumours characterized by PTEN loss, some authors noted shorter survival [12, 22, 29] or shorter time to progression [12, 20]. Moreover, patients who received neoadjuvant chemotherapy with trastuzumab, with reduced PTEN expression in tumour cells, less frequently achieved pathological complete response [23]. Nonetheless, there are also studies which did not confirm relation between PTEN loss in HER2-positive tumours and response to trastuzumab treatment [11, 22], overall [11] and disease-free survival [11, 29].
One of the proposed mechanisms of trastuzumab resistance is heterodimerization of HER2 with other members of EGFR family (EGFR, HER3, HER4) [5, 6, 7, 8]. Data suggest that, out of other heterodimers, HER2/HER3 is the most potent in activation of signalling pathways responsible for proliferation [35]. In our material, EGFR and HER3 positivity were noted in 18% and 31% of cases, respectively. Other authors reported positivity of EGFR in 20 - 66% [11, 12, 20, 36, 37, 38] and of HER3 in 27.2 - 90% of tumours [12, 36, 37, 39]. The above-mentioned discrepancies might, inter alia, result from different scales used for determining of staining immunopositivity. In the present study, status of EGFR did not influence metastasis-free survival. The same was observed by other authors, who reported lack of relation between EGFR expression status and overall/progression-free survival in a group of early stage [20] or metastatic [11, 40, 41] breast cancer patients. However, others noted that EGFR positivity was associated with shorter overall [12] and progression-free survival [12, 20] in a group of metastatic breast cancer patients. In a group treated with neoadjuvant chemotherapy and trastuzumab, negative relation (marginally significant) between pathological complete response and EGFR expression [37] was found, however, contradictory observation - more frequent progression in EGFR-negative group - also was made [36]. For HER3, data are also inconsistent. In some studies, no relations were found between HER3 expression and overall/progression-free survival [41], patients' outcome [36] or pathological complete response [37]. However, other authors reported HER3 expression as an indicator of shorter overall and/or progression-free survival [12, 39, 42]. Also a meta-analysis concerning HER3 overexpression and survival in solid tumours (breast, gastric and ovarian) confirmed HER3 positivity as being associated with worse survival [43].
One of the most valid clues confirming that heterodimerization with other members of EGFR family can affect HER2 signalling pathways and, in the end, impact patients' survival, comes from CLEOPATRA study [44]. It was shown that adding pertuzumab to trastuzumab and docetaxel improved median duration of response by 7.7 months. Pertuzumab is a monoclonal antibody which recognizes 2nd domain of extracellular fragment of HER2, thus blocking dimerization with EGFR, HER3 and HER4 [45]. In our study, we did not find statistical significance when we divided patients according to HER3 expression, however, the survival curves tended to separate quite clearly, and, with larger group, statistical significance probably would be achieved. The similar situation was observed in case of PTEN. There is a clear separation of curves, with only one metastatic event in PTEN positive arm, but the difference is not statistically significant. However, when we combined expression of these two proteins, patients with expression of HER3 and lack of PTEN (two potentially negative factors) had significantly shorter survival than the rest of patients. The similar occurrence was observed by Park and colleagues [12], who reported that patients with expression of HER3 and lack of PTEN were characterized by shorter overall and progression-free survival. Moreover, in this model, differences were statistically more pronounced than in case of expression of only one protein [12]. Our and others' finding concerning HER3 and PTEN confirms that probability of patients' survival after trastuzumab treatment could be influenced not by a single factor, but by a combined effect of two or more molecular events. There are many possible ways in which trastuzumab resistance can be acquired by tumour cells and these mechanisms often overlap, interact and do not exclude each other. Therefore, it is possible that trastuzumab resistance, manifested by poorer patients' outcome, is a cumulative effect of several events. In this respect, we should rather search for a model which includes more than one selected target from a very complicated network of HER2 signalling pathways.
Conclusion
The results of our study suggest that combined analysis of HER3 and PTEN expression might bring information on trastuzumab sensitivity in the group of HER2-positive breast cancer patients treated with trastuzumab in adjuvant setting. Patients with HER3 immunopositivity and PTEN immunonegativity might greatly benefit from additional treatment such as blockers of dimerization of HER2 with HER3 or EGFR (pertuzumab) or PI3K pathway blockers (temsirolimus and everolimus, mTOR inhibitors).
Acknowledgements
We would like to thank Anna Cichocka, MSc for her help during immunohistochemical procedures.
Funding
The study was financed by the National Science Centre, based on the decision numbered DEC-2013/09/B/NZ5/00764.
Competing Interest
The authors have declared that no competing interest exists.
References
1. Slamon DJ, Clark GM, Wong GS. et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177-82
2. Romond EH, Perez EA, Bryant J. et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. New England Journal of Medicine. 2005;353(16):1673-84
3. Vogel CL, Cobleigh MA, Tripathy D. et al. Efficiency and safety of trastuzumab as a single agent in first line treatment of HER 2 overexpresing metastatic breast cancer. J Clin Oncol. 2002;20(3):719-26
4. Nahta R, Esteva FJ. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006;8(6):215
5. Mukohara T. Mechanisms of resistance to anti-human epidermal growth factor receptor 2 agents in breast cancer. Cancer Sci. 2011;102(1):1-8
6. Kruser TJ, Wheeler DL. Mechanisms of resistance to HER family targeting antibodies. Exp Cell Res. 2010;316(7):1083-1000
7. Nahta R, Yu D, Hung MC. et al. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3(5):269-80
8. De P, Hasmann M, Leyland-Jones B. Molecular determinants of trastuzumab efficacy: What is their clinical relevance? Cancer Treat Rev. 2013;39(8):925-34
9. Hammond ME, Hayes DF, Dowsett M. et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med. 2010;134(7):e48-72
10. Wolff AC, Hammond ME, Schwartz JN. et al. American Society of Clinical Oncology/College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131(1):18-43
11. Gori S, Sidoni A, Colozza M. et al. EGFR, pMAPK, pAkt and PTEN status by immunohistochemistry: correlation with clinical outcome in HER2-positive metastatic breast cancer patients treated with trastuzumab. Ann Oncol. 2009;20(4):648-54
12. Park YH, Jung HA, Choi MK. et al. Role of HER3 expression and PTEN loss in patients with HER2-overexpressing metastatic breast cancer (MBC) who received taxane plus trastuzumab treatment. Br J Cancer. 2014;110(2):384-91
13. Beelen K, Opdam M, Severson TM. et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. 2014;16(1):R13
14. Adamczyk A, Niemiec J, Janecka A. et al. Prognostic value of PIK3CA mutation status, PTEN and androgen receptor expression for metastasis-free survival in HER2-positive breast cancer patients treated with trastuzumab in adjuvant setting. Pol J Pathol. 2015;66(2):133-41
15. Niemiec J, Adamczyk A, Ambicka A. et al. BGX-Ki-67 Index as a Supplementary Marker to MIB-1 Index, Enabling More Precise Distinction Between Luminal A and B Subtypes of Breast Carcinoma and Eliminating the Problem of Membranous/Cytoplasmic MIB-1 Staining. Am J Clin Pathol. 2015;143(3):419-29
16. Chaturvedi P, Singh AP, Batra SK. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008;22(4):966-81
17. Koo DH, Lee HJ, Ahn JH. et al. Tau and PTEN status as predictive markers for response to trastuzumab and paclitaxel in patients with HER2-positive breast cancer. Tumour Biol. 2015;36(8):5865-71
18. Jiang BH, Liu LZ. PI3K/PTEN signaling in tumorigenesis and angiogenesis. Biochim Biophys Acta. 2008;1784(1):150-8
19. Mayer IA, Arteaga CL. PIK3CA activating mutations: a discordant role in early versus advanced hormone-dependent estrogen receptor-positive breast cancer? J Clin Oncol. 2014;32(27):2932-4
20. Gallardo A, Lerma E, Escuin D. et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br J Cancer. 2012;106(8):1367-73
21. Wang L, Zhang Q, Zhang J. et al. PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer. 2011;11:248
22. Esteva FJ, Guo H, Zhang S. et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177(4):1647-56
23. Sueta A, Yamamoto Y, Yamamoto-Ibusuki M. et al. An integrative analysis of PIK3CA mutation, PTEN, and INPP4B expression in terms of trastuzumab efficacy in HER2-positive breast cancer. PLoS One. 2014;9(12):e116054
24. Pogue-Geile KL, Song N, Jeong JH. et al. Intrinsic Subtypes, PIK3CA Mutation, and the Degree of Benefit From Adjuvant Trastuzumab in the NSABP B-31 Trial. J Clin Oncol. 2015;33(12):1340-7
25. Loibl S, von Minckwitz G, Schneeweiss A. et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (her2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32(29):3212-20
26. Liedtke C, Cardone L, Tordai A. et al. PIK3CA-activating mutations and chemotherapy sensitivity in stage II-III breast cancer. Breast Cancer Res. 2008;10(2):R27
27. Arsenic R, Lehmann A, Budczies J. et al. Analysis of PIK3CA mutations in breast cancer subtypes. Appl Immunohistochem Mol Morphol. 2014;22(1):50-6
28. Majewski IJ, Nuciforo P, Mittempergher L. et al. PIK3CA Mutations Are Associated With Decreased Benefit to Neoadjuvant Human Epidermal Growth Factor Receptor 2-Targeted Therapies in Breast Cancer. J Clin Oncol. 2015;33(12):1334-9
29. Razis E, Bobos M, Kotoula V. et al. Evaluation of the association of PIK3CA mutations and PTEN loss with efficacy of trastuzumab therapy in metastatic breast cancer. Breast Cancer Res Treat. 2011;128(2):447-56
30. Tolaney S, Burris H, Gartner E. et al. Phase I/II study of pilaralisib (SAR245408) in combination with trastuzumab or trastuzumab plus paclitaxel in trastuzumab-refractory HER2-positive metastatic breast cancer. Breast Cancer Res Treat. 2015;149(1):151-61
31. Oshiro C, Kagara N, Naoi Y. et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat. 2015;150(2):299-307
32. Cescon DW, Bedard PL. PIK3CA genotype and treatment decisions in human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2015;33(12):1318-21
33. Loibl S, von Minckwitz G, Untch M, Denkert C; German Breast Group. Predictive factors for response to neoadjuvant therapy in breast cancer. Oncol Res Treat. 2014;37(10):563-8
34. Chandarlapaty S, Sakr RA, Giri D. et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18(24):6784-91
35. Zaczek A, Brandt B, Bielawski KP. The diverse signaling network of EGFR, HER2, HER3 and HER4 tyrosine kinase receptors and the consequences for therapeutic approaches. Histol Histopathol. 2005;20(3):1005-15
36. Smith BL, Chin D, Maltzman W. et al. The efficacy of Herceptin therapies is influenced by the expression of other erbB receptors, their ligands and the activation of downstream signalling proteins. Br J Cancer. 2004;91(6):1190-4
37. Yonemori K, Tsuta K, Shimizu C. et al. Immunohistochemical expression of HER1, HER3, and HER4 in HER2-positive breast cancer patients treated with trastuzumab-containing neoadjuvant chemotherapy. J Surg Oncol. 2010;101(3):222-7
38. Hudelist G, Köstler WJ, Gschwantler-Kaulich D. et al. Serum EGFR levels and efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. Eur J Cancer. 2006;42(2):186-92
39. Gori S, Foglietta J, Mameli MG. et al. HER-3 status by immunohistochemistry in HER-2-positive metastatic breast cancer patients treated with trastuzumab: correlation with clinical outcome. Tumori. 2012;98(1):39-44
40. Hudelist G, Köstler WJ, Czerwenka K. et al. Her-2/neu and EGFR tyrosine kinase activation predict the efficacy of trastuzumab-based therapy in patients with metastatic breast cancer. Int J Cancer. 2006;118(5):1126-34
41. Robinson AG, Turbin D, Thomson T. et al. Molecular predictive factors in patients receiving trastuzumab-based chemotherapy for metastatic disease. Clin Breast Cancer. 2006;7(3):254-61
42. Berghoff AS, Bartsch R, Preusser M. et al. Co-overexpression of HER2/HER3 is a predictor of impaired survival in breast cancer patients. Breast. 2014;23(5):637-43
43. Ocana A, Vera-Badillo F, Seruga B. et al. HER3 overexpression and survival in solid tumors: a meta-analysis. J Natl Cancer Inst. 2013;105(4):266-73
44. Swain SM, Baselga J, Kim SB, et al; CLEOPATRA Study Group. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724-34
45. Hubalek M, Brantner C, Marth C. Role of pertuzumab in the treatment of HER2-positive breast cancer. Breast Cancer (Dove Med Press). 2012;4:65-73
Author contact
![]() Corresponding author: Agnieszka Adamczyk, Department of Applied Radiobiology, Maria Sklodowska-Curie Memorial Cancer Centre and Institute of Oncology, Cracow, Poland, Cracow Branch, ul. Garncarska 11, 31-115 Cracow, Poland. Phone: +48 12 63 48 371 fax: +48 12 423 1076 e-mail: aa.adamczykcom; z5adamczpl.
Corresponding author: Agnieszka Adamczyk, Department of Applied Radiobiology, Maria Sklodowska-Curie Memorial Cancer Centre and Institute of Oncology, Cracow, Poland, Cracow Branch, ul. Garncarska 11, 31-115 Cracow, Poland. Phone: +48 12 63 48 371 fax: +48 12 423 1076 e-mail: aa.adamczykcom; z5adamczpl.

 Global reach, higher impact
Global reach, higher impact