Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(6):1062-1070. doi:10.7150/jca.17080 This issue Cite
Research Paper
The Relation of HPV Infection and Expression of p53 and p16 Proteins in Esophageal Squamous Cells Carcinoma
1. Teaching and Research Institute, Molecular Oncology Research Center, Barretos Cancer Hospital - Pio XII Foundation, Brazil;
2. Department of Endoscopy, Barretos Cancer Hospital - Pio XII Foundation, Brazil;
3. Medical Ambulatory of Specialty Clinics - Barretos, Brazil;
4. Molecular Biology Laboratory, Center for Translational Research in Oncology, Instituto do Câncer do Estado de São Paulo - ICESP, São Paulo, Brazil;
5. Department of Radiology and Oncology, School of Medicine, University of São Paulo, Brazil;
6. Department of Clinical Research - Biohit Oyj, Finland;
7. Medical Laboratory of Medical Investigation (LIM) 14.Department of Pathology, Faculty of Medicine, University of São Paulo, Brazil;
8. Research Institute of Life and Health Sciences (ICVS), University of Minho, Braga, Portugal;
9. ICVS / 3B's - Associated Laboratory to the Government of Portugal, Braga / Guimarães, Portugal.
Received 2016-8-2; Accepted 2016-12-23; Published 2017-4-9
Abstract
GOAL: To investigate the HPV prevalence and characterize the expression of potential molecular surrogate markers of HPV infection in esophageal squamous cell carcinoma.
MATERIALS AND METHODS: The prevalence of HPV in individuals with and without esophageal cancer (EC) was determined by using multiplex PCR; p16 and p53 protein levels were assessed by immunohistochemistry (IHC).
RESULTS: High-risk HPV (hr-HPV) was found in the same frequency (13.8%) in esophageal squamous cell carcinoma (ESCC) and in healthy individuals. The p53 expression was positive in 67.5% of tumor tissue, 20.0% of adjacent non-tumoral tissue and 1.8% of normal esophageal tissue. p16 was positive in 11.6% of esophageal cancer cases and 4.7% of adjacent non-tumoral tissue. p16 was undetectable among control group samples. p53 and p16 levels were not significantly associated with the HPV status.
CONCLUSIONS: These results suggest that hr-HPV types are not associated with the development of ESCC and that p53 and p16 protein expression have no relationship with HPV infection in normal or cancerous esophagus.
Keywords: Esophageal neoplasias, p16, p53, immunohistochemistry, HPV.
Introduction
Esophageal cancer (EC) represents the eighth highest incidence of cancer worldwide (456,000 cases/year), and is usually diagnosed at an advanced stage, showing rapid progression and extremely poor prognosis [1, 2]. For this reason, it is ranked as the sixth most common cause of cancer death (400,000/year) [1]. In Brazil, EC is among the ten most common incident cancers, ranking sixth among men and thirteenth among women, with the higher incidences occurring in southern and southeast Brazil [3], supposedly associated with the high consumption of mate tea [4, 5].
The etiology of esophageal squamous cell carcinoma (ESCC) is complex and the geographical, cultural and ethnic variations observed suggest an association of established risk factors for this disease, such as alcohol consumption, smoking, malnutrition, environmental factors, and infectious pathogenic microorganisms [6, 7].
In the early 80s, Syrjänen et al. (1982) identified for the first time features of HPV cytopathic infection in benign and malignant esophageal tumors [8]. However, studies with populations of varied cultural characteristics, associated with the use of different viral detection methodologies resulted in significantly heterogeneous data concerning HPV prevalence and viral types identified in ESCC [9-11], something that encourages further research in this area and requires knowledge of both molecules activated by the virus, and the expression of surrogate infection markers [12].
During persistent high-risk HPV (hr-HPV) infection, the increased expression of viral oncoproteins E6 and E7 can interfere with cell cycle control and trigger chromosomal instability [13, 14] since they inactivate p53 and pRb tumor-suppressor protein (retinoblastoma protein), respectively, resulting in increased 16INK4A and p53 expressions. Hence, immunohistochemistry (IHC) for p16 and p53 expression is considered as assay for detection of surrogate markers of HPV infection in genital and oropharyngeal carcinomas [15-18].
In HPV positive oropharyngeal, cervical and vaginal tumors [15, 17, 19], the p16 expression is altered, and abnormal p53 expression has also been reported in HPV-positive oropharyngeal carcinoma [19] and penile lesions [20]. Furthermore, clinical development of these HPV-induced tumors, related to the treatment response and overall survival, is favorable when compared to HPV negative tumors [21, 22]. For this reason, the investigation of these proteins in ESCC is appropriate and may be an efficient research tool to find potential tumor markers [23, 24].
Therefore, this study aimed at investigating the prevalence of HPV in ESCC and normal esophageal tissue and also evaluates p16 and p53 expression levels in HPV positive esophageal cancers.
Materials and Methods
From January 2013 to October 2014, patients newly admitted to the Upper Digestive Department at Barretos Cancer Hospital referred for upper digestive endoscopy were screened and a total of 101 patients diagnosed with primary ESCC were enrolled. For the control group, 101 individuals aged 18 years or over, undergoing upper gastrointestinal endoscopy for reasons other than esophageal malignancy and with no microscopic or gross esophageal pathology admitted to the Ambulatory of Medical Specialties (AME, acronym in Portuguese) in Barretos were screened and matched by gender and age (+/- 3 years) with the cases. Sample size of the prospective and controlled study was based on a previously published work [25]. All individuals participating in this study signed an informed consent prior to enrollment and completed a questionnaire regarding their socio-demographical and lifestyle information. Moreover, clinic-pathological information was obtained through medical record review. The protocol was approved by the Ethics Committee at Barretos Cancer Hospital.
Exclusion criteria
Volunteers that had been submitted to adjuvant and/or neoadjuvant therapy, displaying any mental disability or an unfavorable clinical condition that would render them unfit for procedures, or that could provide insufficient amount of sample and/or inadequate sample quality were excluded.
Samples collection
Samples used in this project were collected during endoscopy and processed by the Pathology Department for diagnosis. Of the 101 patients with EC initially recruited for this study, only 87 samples had the enough DNA quantity to be included in the present study. Therefore, 87 controls were matched and included in the analysis. During the endoscopy procedure, biopsies were preceded by visual examination, and sample collection was conducted in the following order: tumor tissue and adjacent non-tumoral tissue for cases; proximal and distal esophageal tissue for controls. Biopsies fixed in 10% buffered formalin and embedded in paraffin were examined by a senior pathologist (CSN).
HPV detection and characterization
Biopsy specimens were digested with proteinase K-SDS 1%; DNA was obtained by organic extraction [26]. HPV DNA was assessed in all samples using a type-specific PCR bead-based multiplex genotyping (TS-MPG) assay that combine multiplex polymerase chain reaction (PCR) and bead-based Luminex technology (Luminex Corp., Austin, TX, USA), as cited in another paper [27]. This methodology is able to identify 21 HPV types [28, 29]. Based on their frequency in cervical cancer and available biological data, 12 of these types, namely types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 59 have been defined as carcinogens (IARC Group 1A)—hereafter referred to as hr-HPV. For eight other types, in the high-risk clade, namely types 26, 53, 66, 67, 68, 70, 73 and 82, the combination of their low frequency, lack of data on their active transcription and their transforming potential in model systems, has led them to be classified as only probable/possible carcinogens (IARC Groups 2A and 2B)—hereafter referred to as possible high-risk (pHR)-HPV types [30].
As a positive control for the quality of the template DNA, primers for β-globin gene were included in the reactions. PCRs were performed with 10µL of template DNA in a 96-well format in 25-μl/well final reaction volume. HPV multiplex PCR was performed with QIAGEN Multiplex PCR Kit (Qiagen, Dusseldorf, Germany), according to manufacturer's instructions. Each reaction consisted of 45 cycles: 94°C for 30 seconds, 63°C for 3 minutes, and 72°C for 90 seconds. The first cycle was preceded by incubation at 95°C for 15 minutes and the last cycle was extended for 10 minutes at 72°C. PCR negative control consisted of a reaction mix without DNA.
Hybridizations were performed according to Schmitt et al (2006) [31]. For each HPV type-specific probe, the mean fluorescence intensity (MFI) values obtained when no PCR product was added to the hybridization mixture was considered as background. The cutoffs were calculated by adding 5 MFI to 1.1 times the value of the median background. MFI values > 20 were considered positive.
Immunohistochemistry
Expressions of p16 and p53 proteins were assessed by IHC using anti-p16 (monoclonal mouse anti-human p16INK4A protein, Clone E6H4TM, ready for use, Ventana, Tucson, AZ, USA) and anti-p53 (monoclonal mouse anti-human p53 protein, Clone DO-7, dilution 1:1200, Cell Marque, Rocklin, CA, USA) in an automated system (Ventana Benchmark ULTRA, Tucson, AZ, USA). Briefly, formalin-fixed, paraffin-embedded tumor specimens (4 μm) were deparaffinized by heating (75°C for 4 minutes), and antigen retrieval was achieved by use of cell conditioning buffer 1 (CC1) at 95°C for 64 minutes. Specific primary antibodies were separately added, and ultraView Universal DAB Detection Kit polymer amplification system (Ventana Medical Systems, Tucson, AZ, USA) was used according to manufacturer's instructions.
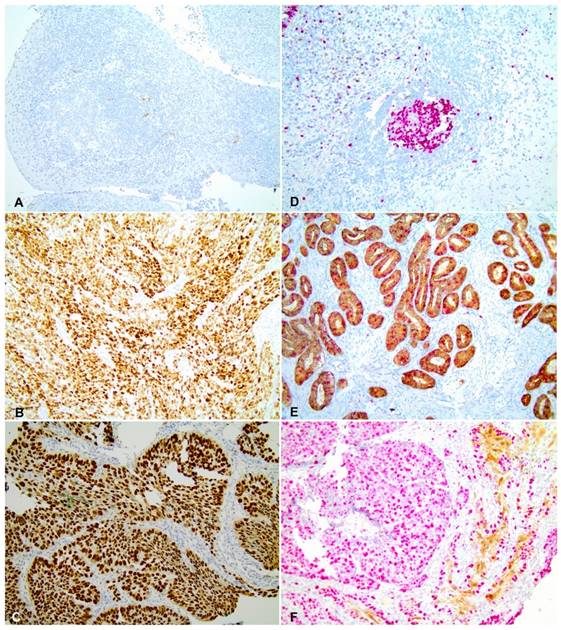
A cervical adenocarcinoma was used as positive control for p16 staining (Figure 2 E) and negative controls were obtained by omitting the primary antibodies (Figure 2 D). Samples with strong and diffuse nuclear and cytoplasmic in more than 70% of the tissue were considered positive [32, 33]. All scorings were conducted blindly by two observers (CSN and ALF). A colon carcinoma sample with a diffuse nuclear p53 staining was used as a positive control for p53 labelling (Figure 2 B) whilst a breast carcinoma sample was used as a negative control (Figure 2 A). The slides were evaluated for nuclear expression, and samples with at least 10% of strong nuclear staining [34] were considered positive for the p53 expression. All scorings were conducted with no knowledge of clinical characteristics or outcome by two observers (CSN and ALF), independently. The scanning of histological images was performed using the Olympus BX43 microscope and the Olympus Soft Imaging Solution GmbH, version 5.2 software (Olympus Corporation, Shinjuku-ku, Tokyo, Japan).
The observers revised all immunohistochemical discordant cases and consensus was achieved.
Statistical Analyses
The description of the study population was made from frequency tables. Chi-square, Fisher's exact, McNemar and Q-Cochran tests were used for comparison between groups (cases x controls; negative HPV x positive HPV). The significance level was 0.05 (5%). All analyses were conducted using the IBM® SPSS® Statistics 21.0 software for Windows (IBM Corporation, Somers, NY, USA).
Results
Figure 1 shows the number of samples included in the HPV analysis and p53 and p16 IHCs. Then, the characteristics of population with 174 volunteers (87 cases and 87 controls) were described. Overall, the majority of the study population was composed by men (78.2%) with an average age of 60 years. However, when lifestyle was investigated, it was shown that the intake of alcohol (90.8%), tobacco smoking (81.6%) and alcohol intake associated with tobacco smoke (75.9%) was significantly higher among individuals in the case group compared to controls individuals (74.7%; 67.8%; 60.9%, respectively) (Table 1).
Characterization of study population
| Variable / Category | Cases | Controls | p* | ||||
|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | ||||
| Gender | |||||||
| Male | 68 | 78.2 | 68 | 78.2 | 1.000 | ||
| Female | 19 | 21.8 | 19 | 21.8 | |||
| Age | |||||||
| ≤ 60 years | 46 | 52.9 | 42 | 48.3 | 0.544 | ||
| >60 years | 41 | 47.1 | 45 | 51.7 | |||
| Alcohol | |||||||
| No | 8 | 9.2 | 22 | 25.3 | 0.005 | ||
| Yes | 79 | 90.8 | 65 | 74.7 | |||
| Tobacco | |||||||
| No | 16 | 18.4 | 28 | 32.2 | 0.036 | ||
| Yes | 71 | 81.6 | 59 | 67.8 | |||
| Alcohol + Tobacco | |||||||
| Never drank or smoked | 3 | 3.4 | 16 | 18.4 | 0.006 | ||
| Drinks and smokes | 66 | 75.9 | 53 | 60.9 | |||
| Drinks or smokes | 18 | 20.7 | 18 | 20.7 | |||
*Used test: Qui-square
Clinical Samples included in the present study *As our study was case-control, only the 87 controls matched with cases were assessed in the work. ** For counting of subjects evaluated for p53 and p16 proteins expression by IHC was considered that individuals with at least one analysis to the tissues for the control group (proximal and/or distal esophageal tissues) and tumor tissue for case group. *** This analysis could not be performed for all individuals included in the study due to lack of material.
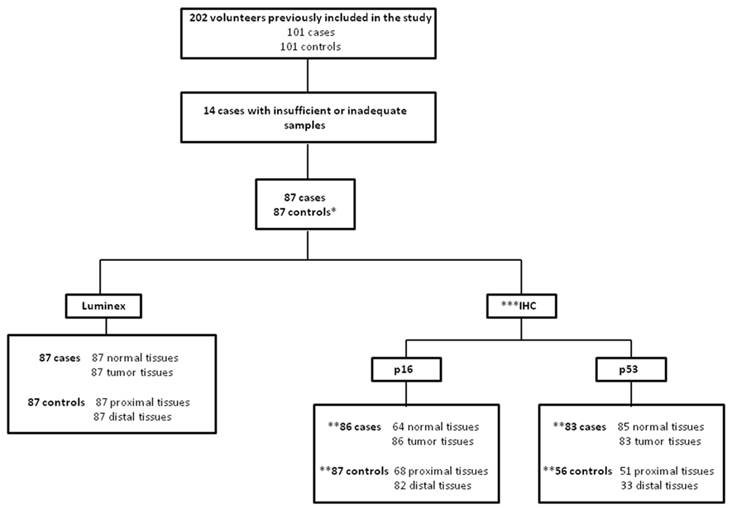
Among the 87 analyzed esophageal tumors, most of them (59%) were moderately differentiated, localized in the middle third of the esophagus (55.2%) and with TNM staging as advanced (73.5%). Moreover, a review of medical records held in May 2016 provided the information that 75.9% of patients in the study died due to the disease and only 14.9% were still alive without ESCC (Table 2).
Characterization of tumor tissue
| Variable | Category | n | % | |
|---|---|---|---|---|
| Degree of differentiation | Well | 11 | 13.3 | |
| Moderate | 49 | 59.0 | ||
| Little | 23 | 27.7 | ||
| Topography | Upper third | 3 | 3.4 | |
| Middle third | 48 | 55.2 | ||
| Lower third | 7 | 8.0 | ||
| More than one location | 29 | 33.3 | ||
| TNM Staging | I and II | 22 | 26.5 | |
| III and IV | 61 | 73.5 | ||
| T | T1/T2 | 10 | 13.0 | |
| T3/T4 | 67 | 87.0 | ||
| N | N0 | 22 | 38.6 | |
| N positive | 35 | 61.4 | ||
| M | M0 | 65 | 79.3 | |
| M1 | 17 | 20.7 | ||
| Patient status | Death due to cancer | 66 | 75.9 | |
| Alive with disease | 8 | 9.2 | ||
| Alive without disease | 13 | 14.9 |
To evaluate HPV DNA prevalence in the esophagus, tissues were initially examined separately in tumor and adjacent non-tumoral tissue for cases and, proximal and distal esophageal tissues for controls. The results obtained were, respectively: 11.5% (10/87), 2.3% (2/87), 10.3% (9/87) and 4.6% (4/87). The overall frequency of HPV in each group was considered as positive when the individual had DNA HPV in at least one tissue. Thus, the frequency of DNA HPV was 13.8% for both of groups.
Table 3 shows the frequency of HPV types detected in each anatomical region analyzed. Regarding the adjacent non-tumoral tissue, two different hr-HPVs (HPV 31 and 66) were detected in equal proportion (1.15%). However, for tumor tissue, HPV 16 (2.3%), 18 (1.15%), 51 (1.15%), 31 (1.15%), 66 (1.15%) types, in addition to coinfections (4.6%) were detected. In controls tissues, HPV 56 (2.3%), 16 (1.15%), 31 (1.15%), 51 (1.15%), 26 (1.15%) types and coinfections (3.4%) were detected in proximal esophagus, whilst in the distal esophagus, only four coinfections (4.6%) were found. Importantly, no low-risk DNA HPV was detected in samples analyzed.
We observed no significant association between the detection of HPV DNA and any of the different variables analyzed (gender, age and consumption of alcohol and tobacco, isolated or combined). In addition, HPV prevalence, clinicopathological characteristics (histological degree and TNM staging) and status of the patient were not significantly related (p=NA*, p=0.502, p=NA*, respectively) (*could not be calculated).
Frequency of the HPV types detected by Luminex
| Variable | Cases | Controls | ||||||
|---|---|---|---|---|---|---|---|---|
| Adjacent normal | Tumor | Proximal | Distal | |||||
| n | (%) | n | (%) | n | (%) | n | (%) | |
| HPV 51 | 0 | 0.0 | 1 | 1.15 | 1 | 1.15 | 0 | 0.0 |
| HPV 31 | 1 | 1.15 | 1 | 1.15 | 1 | 1.15 | 0 | 0.0 |
| HPV 66 | 1 | 1.15 | 1 | 1.15 | 0 | 0.0 | 0 | 0.0 |
| HPV 56 | 0 | 0.0 | 0 | 0.0 | 2 | 2.3 | 0 | 0.0 |
| HPV 18 | 0 | 0.0 | 1 | 1.15 | 0 | 0.0 | 0 | 0.0 |
| HPV 16 | 0 | 0.0 | 2 | 2.3 | 1 | 1.15 | 0 | 0.0 |
| HPV 26 | 0 | 0.0 | 0 | 0.0 | 1 | 1.15 | 0 | 0.0 |
| Coinfecction | 0 | 0.0 | 4* | 4.6 | 3** | 3.4 | 4*** | 4.6 |
| Total | 2 | 2.3 | 10 | 11.5 | 9 | 10.3 | 4 | 4.6 |
Frequencies refer to HPV types detected in each tissue analyzed by Luminex.
*56,66; 39,53,66; 53,66; 16,53.
**16,66; 18,31,33,53; 45,53.
***16,31; 31,59; 16,39,51; 31,45,53.
p53 and p16 immunohistochemistry
Regarding cellular proteins expression, we observed that p16 and p53 levels were significantly higher in tumor tissues (67.5% for p53 and 11.6% for p16) than in control group (1.8% for p53 and 0% for p16) (p <0.001) (Figure 2). p53 expression was considered positive in 67.5% of tumor tissues (Figure 2C), 20.0% of adjacent non-tumoral tissue and 1.8% of normal esophageal tissue. Furthermore, p16 expression was observed in 11.6% of tumor tissues (Figure 2F) and 4.7% of adjacent non-tumoral tissue specimens. No p16 expression was observed in the control group. p53 and p16 expression levels were not significantly associated with HPV status.
Table 4 shows the number of samples that showed agreement and disagreement regarding the p16 expression and the presence of HPV. The majority of samples (80.9%) were double negative concordant (p16-/HPV-). Only one case (0.6%) showed concomitant concordance for positive p16 expression and the presence of HPV. Interestingly, this case did not show characteristics that distinguishes those negative for p16 since was a man aged 73, non-white, with ESCC moderately differentiated type, advanced stage (III), negative expression for p53, was alcohol intake associated with tobacco smoke and died due to cancer. Furthermore, 32 samples were discordant to p16 IHC and HPV results, wherein 9 p16 positive samples (5.2%) were HPV negative whilst 23 (13.3%) samples were negative for p16 expression, but positive for HPV.
Agreement between p16 expression and HPV DNA in esophageal samples
| Agreement | Samples | |
|---|---|---|
| n | (%) | |
| p16- HPV - | 140 | 80.9 |
| p16+ HPV + | 1 | 0.6 |
| p16+ HPV - | 9 | 5.2 |
| p16- HPV + | 23 | 13.3 |
Frequency samples with concordant results between the HPV status and the expression of p16 protein for subjects in the study.
Discussion
The role of HPV in esophageal carcinogenesis is a controversial issue that has been described with significant discrepancies in results, which may be attributed to the different methodologies employed in the HPV DNA detection and, in the many ways, in which sample may be collected and preserved [35] and, maybe, the most important, the characteristics of individuals enrolled, including their sexual practices and social-demographic data [36]. Thus, to our knowledge, this is the first study having a prospective design and controlled in Brazil, which provides important reliability to the results. The goal of this project was to obtain Brazilian data that would help clarify the frequency of HPV in ESCC and the molecular pathways involved in this esophageal cancer development process, besides gathering evidences that could assist in the prognostic evaluation of patients affected by such a malignancy or the prevention of the disease through an HPV vaccine.
We demonstrated that HPV frequencies were identical in case and control groups (13.8%) by using Luminex® technology. The HPV frequency in esophageal tumors we detected is consistent with other studies conducted in Brazil in which the HPV16 and 18 prevalence found was 15.75% by using Nested PCR methodology [37] and 13.0% by using PCR followed by sequencing and in situ hybridization (HPV16 was the most frequent type) [38]. However, this prevalence is higher than others that described HPV prevalence in Brazilian cases of esophageal cancers around 2.5% of low-risk HPV using hybrid capture methodology [39] and 0.0% using nested PCR methodology [40]. Furthermore, in non-cancer volunteers, the HPV frequency of 13.8% was similar to the first Brazilian study carried out with samples from South of the country which found an HPV prevalence of 10% in normal esophageal samples by hybrid capture methodology [39]. However, all other Brazilian studies did not detect DNA HPV in healthy esophageal samples analyzed [37, 38, 40], and this confirms unequivocally that the frequency of HPV in neoplastic and normal esophageal tissues are so variable, not only for any regional differences, but also very important, according to the methodological techniques used to address this goal [35, 36].
Differences in HPV prevalence largely depend on the test used and type of the samples and their conservation. The variation among the groups did not come as a surprise but the real meaning of the findings did. This point was elegantly reported in a recent review of the literature [35]; worldwide HPV-ESCC infection rates ranged from 11.7% to 39.9%, with the high-ESCC-incidence countries relating HPV tumor infections rates significantly higher in relation to low-ESCC-incidence countries. In China, in a high-ESCC-incidence region, for example, HPV infections in ESCC ranged from 32.8% to 63.6%, whereas in North America (low incidence region) rates ranged between 8.7% to 16.6% [35]. Li et al. (2014) also reported that the positive rate for HPV in ESCC was 14.0% for Europe and the Americas, according to our findings [11]. In relation to adjacent non-tumoral esophageal tissue, literature has shown rates as high as 58.9% in some studies, and as low as 0% in others. Such disparities may be justified by the small sample size, making it hard to answer questions of the HPV ESCC prevalence [35].
Global data shown by recent literature indicates that HPV has no significant etiological role in most cases of ESCC [35]. Another recent study showed that HPV infection may play a role in esophageal carcinogenesis only in high-incidence regions [41] and this conclusion corroborates, in part, the results demonstrated by our study, since we found identical HPV frequency in patients without EC group compared to the group with ESCC. To confirm, no correlation with p16 and p53 expression with tumor was observed and they are canonic pathways altered in HPV-cervical cancer.
p53 and p16 immunohistochemistry in ESCC (A) Negative control for p53 staining. (B) Positive control for p53 staining. (C) Infiltrative moderately differentiated ESCC tissue with p53 strong nuclear expression. (D) Negative control for p16 staining. (E) Positive control for p16 staining. (F) Infiltrative poorly differentiated ESCC tissue with strong expression of p16. All images were obtained with 20X magnification.
Despite the HPV frequency be similar, the HPV types detected by us is different from those reported by other Brazilian studies, since the HPV 16 and 18 types were less frequent in our casuistry [39, 40]. However, although less frequent, the HPV types we detected had already been detected in ESCC in different geographic regions of the world [HPV 16 (11.4%), 18 (2.9%), 52 (1.1%), 33 (0.8%) and 31 (0.6%)] [11]. In South America, HPV types most often found in the cervix are HPV 16, 58, 18, 45 and 31, whereas, only HPV types 16, 18 and 31 are cited as frequently in cervical and esophageal cancer. From this information, it is strongly suggested that the main types of HPV that infect the esophageal mucosa in the Brazilian population are different from those that infect the lining of the cervix and esophagus in other regions the world [42].
In addition to detecting HPV presence in normal and tumoral esophageal tissues, we also evaluated the p53 and p16 proteins expression, since it is known that the altered expression (increased or reduced) of these proteins is directly related to tumor progression in several types of cancers, induced or not by HPV. Although there are a significant number of positive HPV samples, they were not associated with p16 and p53 expression in this study. However, the expression of these proteins increases progressively with the severity of esophageal injury, corroborating previous studies [43, 44].
According to p16 expression and HPV status in esophageal samples from patients with and without EC, we found only one sample (0.6%) with concordant results p16+/HPV+. This result corroborates recent systematic review, where it was shown that, unlike the results for cervical and oropharynx cancer, in EC there is slight or no correlation between p16 overexpression and the HPV positivity. By bringing together studies in EC, the authors concluded that the rate of double positivity (HPV+/p16+) is below 5% of cases [35]. Although some studies on head and neck show strong relationship between p16 expression and the HPV presence, it was shown that tests for HPV DNA and p16 immunohistochemistry were not satisfactorily consistent, and that the use of only one of these tests is inadequate to confirm the viral origin of oropharyngeal cancer [45].
We observed that 9 esophageal samples (5.2%) were p16+/HPV-, and similar result obtained by Fonmarty et al. (2015), who analyzed the p16 expression and HPV status of oropharyngeal tumors. The authors identified 11 discordant samples (p16+/HPV-) and according to authors, the p16 overexpression detected by IHC reflects an altered pRb function induced by E7 viral oncoprotein; and this altered function may be secondary to mutation of the Rb gene regardless of HPV persistent infection [45]. Another hypothesis for explain these results can be attributed to PCR's failure to detect the viral DNA, which can be supported by the non-recognition of L1 sequence of the viral genome with complementary primers used in the PCR [45]. Kawakami et al. (2003) showed that 11% of samples were p16+/HPV- and the authors suggested that these result reflect a deregulation in the Rb signaling pathway unrelated to HPV, as noted, for example, in small-cell lung cancer and lymphomas [46].
The inverse correlation between p16-/HPV+ is also questioned by some authors seeking to uncover if there is a chance of HPV playing its carcinogenic role in the esophagus without inducing the overexpression of p16. Among suggested hypotheses, one of them states that esophageal carcinogenesis may involve a high rate of p16 methylation promoters, thus inhibiting its expression in the oncogenic HPV infection [35]. It has been shown that there is loss of p16 expression secondary to promoter methylation in 72% of ESCC cases [47], as it has been shown for other tumor types [48]. The current study also found similar results, since 23 samples (13.3%) were p16-/HPV+. Kawakami et al. (2013) reported that 20% of esophageal tumors were p16-/HPV+, and that most of these tumors showed methylation of the p16 gene promoter [46]. Similar results in head and neck carcinomas also showed p16-/HPV+ cases, although in lower frequencies [49-51].
We concluded that in this particular series, the prevalence of HPV in ESCC and healthy esophagus in the Brazilian population was identical, which suggested that the HPV does not necessarily has a role in the development of esophageal cancer since there was no association between the status of HPV and the expression of p53 and p16.
Abbreviations
ESCC: Esophageal Squamous Cell Carcinoma
hr-HPV: High-risk HPV
PCR: Polymerase Chain Reaction
HPV: Human papillomavirus
EC: Esophageal Cancer
IHC: Imunohistochemistry
PRb: Retinoblastoma protein
AME: Ambulatory Medical Specialties
DNA: Deoxyribonucleic acid
TS-MPG: Type-specific PCR bead-based multiplex genotyping
MFI: Mean fluorescence intensity
CC1: Cell conditioning buffer
TNM: Classification of Malignant Tumours
ICESP: Center for Translational Research in Oncology, São Paulo Cancer Institute
CNPq: Conselho Nacional de Desenvolvimento Científico e Tecnológico
FAPESP: Fundação de Amparo à Pesquisa do Estado de São Paulo
CEP: Ethics committee for research
Acknowledgements
The authors would like to thank the Barretos Cancer Hospital, Ambulatory Medical Specialties (AME) Clinic of Barretos, HPV Teams in the Barretos Cancer Hospital and Molecular Biology Laboratory, Center for Translational Research in Oncology, São Paulo Cancer Institute (ICESP), São Paulo, Brazil. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tencnológico (CNPq) [Grants number 482666/2012-9 to ALF; 573799/2008-3 to LLV]; Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [Grants numbers 13/15968-0 to PRAP; 08/57889-1 to LLV].
Funding statement
CNPq Universal for promoting supplied to the largest study of which this study is part entitled "The role of human papillomavirus (HPV) as the etiologic agent of esophageal cancer. A cross-sectional study, case-control and longitudinal in Barretos Cancer Hospital" (Grant number 482666/2012-9 to ALF); Fundação de Amparo à Pequisa do Estado de São Paulo (FAPESP) (Grant number 13/15968-0 to PRAP); INCT HPV [Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [Grant number 08/57889-1 to LLV]; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Grant number 573799/2008-3 to LLV)].
Ethical approval
This project is part of a larger study approved by the CEP (134 471).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J SI, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. International Agency for Research on Cancer. Lyon, France. 2013
2. Shen ZY, Xu LY, Li EM, Shen J, Zheng RM, Cai WJ. et al. Immortal phenotype of the esophageal epithelial cells in the process of immortalization. Int J Mol Med. 2002;10:641-6
3. Brasil Ministério da Saúde. Estimativa 2016: Incidência de câncer no Brasil. Rio de Janeiro: Instituto Nacional do Câncer. 2015
4. Castellsague X, Munoz N, De Stefani E, Victora CG, Castelletto R, Rolon PA. Influence of mate drinking, hot beverages and diet on esophageal cancer risk in South America. Int J Cancer. 2000;88:658-64
5. Islami F, Malekshah AF, Kimiagar M, Pourshams A, Wakefield J, Goglani G. et al. Patterns of food and nutrient consumption in northern Iran, a high-risk area for esophageal cancer. Nutr Cancer. 2009;61:475-83
6. Syrjanen K. HPV and esophageal cancer. Papillomavirus Research: From Natural History to Vaccines and Beyond. Norwich, UK. 2006:229-53
7. Syrjanen K. Human papillomavirus (HPV) involvement on esophageal carcinogeneses In: Publishing NS, editor. New York, USA. 2010
8. Syrjanen K, Pyrhonen S, Aukee S, Koskela E. Squamous cell papilloma of the esophagus: a tumour probably caused by human papilloma virus (HPV). Diagn Histopathol. 1982;5:291-6
9. Hardefeldt HA, Cox MR, Eslick GD. Association between human papillomavirus (HPV) and oesophageal squamous cell carcinoma: a meta-analysis. Epidemiol Infect. 2014;142:1119-37
10. Petrick JL, Wyss AB, Butler AM, Cummings C, Sun X, Poole C. et al. Prevalence of human papillomavirus among oesophageal squamous cell carcinoma cases: systematic review and meta-analysis. Br J Cancer. 2014;110:2369-77
11. Li X, Gao C, Yang Y, Zhou F, Li M, Jin Q. et al. Systematic review with meta-analysis: the association between human papillomavirus infection and oesophageal cancer. Aliment Pharmacol Ther. 2014;39:270-81
12. World Health Organization International Agency For Research on Cancer. Smokeless Tobacco and Some Tobacco-Specific N-Nitrosamines. Lyon, France. 2007
13. Pinto AP, Degen M, Villa LL, Cibas ES. Immunomarkers in gynecologic cytology: the search for the ideal 'biomolecular Papanicolaou test'. Acta Cytol. 2012;56:109-21
14. Clarke B, Chetty R. Cell cycle aberrations in the pathogenesis of squamous cell carcinoma of the uterine cervix. Gynecol Oncol. 2001;82:238-46
15. Bergeron C, Ronco G, Reuschenbach M, Wentzensen N, Arbyn M, Stoler M. et al. The clinical impact of using p16(INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int J Cancer. 2015;136:2741-51
16. Wang H, Sun R, Lin H, Hu WH. P16INK4A as a surrogate biomarker for human papillomavirus-associated oropharyngeal carcinoma: consideration of some aspects. Cancer science. 2013;104:1553-9
17. Liu SZ, Zandberg DP, Schumaker LM, Papadimitriou JC, Cullen KJ. Correlation of p16 expression and HPV type with survival in oropharyngeal squamous cell cancer. Oral oncology. 2015;51:862-9
18. Holzinger D, Flechtenmacher C, Henfling N, Kaden I, Grabe N, Lahrmann B. et al. Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int J Cancer. 2013;133:1389-99
19. Hellman K, Lindquist D, Ranhem C, Wilander E, Andersson S. Human papillomavirus, p16(INK4A), and Ki-67 in relation to clinicopathological variables and survival in primary carcinoma of the vagina. Br J Cancer. 2014;110:1561-70
20. Mannweiler S, Sygulla S, Winter E, Regauer S. Two major pathways of penile carcinogenesis: HPV-induced penile cancers overexpress p16ink4a, HPV-negative cancers associated with dermatoses express p53, but lack p16ink4a overexpression. Journal of the American Academy of Dermatology. 2013;69:73-81
21. El Hamdani W, Amrani M, Attaleb M, Laantri N, Ennaji MM, Khyatti M. et al. EGFR, p16INK4a and E-cadherin immuno-histochemistry and EGFR point mutations analyses in invasive cervical cancer specimens from Moroccan women. Cell Mol Biol (Noisy-le-grand). 2010;56(Suppl):OL1373-84
22. Sahasrabuddhe VV, Luhn P, Wentzensen N. Human papillomavirus and cervical cancer: biomarkers for improved prevention efforts. Future Microbiol. 2011;6:1083-98
23. Zappacosta R, Colasante A, Viola P, D'Antuono T, Lattanzio G, Capanna S. et al. Chromogenic in situ hybridization and p16/Ki67 dual staining on formalin-fixed paraffin-embedded cervical specimens: correlation with HPV-DNA test, E6/E7 mRNA test, and potential clinical applications. Biomed Res Int. 2013;2013:453606
24. Wang L, Li J, Hou J, Li M, Cui X, Li S. et al. p53 expression but not p16(INK4A) correlates with human papillomavirus-associated esophageal squamous cell carcinoma in Kazakh population. Infectious agents and cancer. 2016;11:19
25. Guo F, Liu Y, Wang X, He Z, Weiss NS, Madeleine MM. et al. Human papillomavirus infection and esophageal squamous cell carcinoma: a case-control study. Cancer Epidemiol Biomarkers Prev. 2012;21:780-5
26. Green MR, Sambrook J. MolecularCloning:ALaboratoryManual,4thed, John Inglis N. 2012.
27. Nunes EM, Sudenga SL, Gheit T, Tommasino M, Baggio ML, Ferreira S. et al. Diversity of beta-papillomavirus at anogenital and oral anatomic sites of men: The HIM Study. Virology. 2016;495:33-41
28. Gheit T, Billoud G, de Koning MN, Gemignani F, Forslund O, Sylla BS. et al. Development of a sensitive and specific multiplex PCR method combined with DNA microarray primer extension to detect Betapapillomavirus types. J Clin Microbiol. 2007;45:2537-44
29. Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24(Suppl 3):1-10
30. Halec G, Schmitt M, Dondog B, Sharkhuu E, Wentzensen N, Gheit T. et al. Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. Int J Cancer. 2013;132:63-71
31. Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead-based multiplex genotyping of human papillomaviruses. J Clin Microbiol. 2006;44:504-12
32. Begum S, Gillison ML, Ansari-Lari MA, Shah K, Westra WH. Detection of human papillomavirus in cervical lymph nodes: a highly effective strategy for localizing site of tumor origin. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9:6469-75
33. Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166-73
34. Keswani RN, Noffsinger A, Waxman I, Bissonnette M. Clinical use of p53 in Barrett's esophagus. Cancer Epidemiol Biomarkers Prev. 2006;15:1243-9
35. Ludmir EB, Stephens SJ, Palta M, Willett CG, Czito BG. Human papillomavirus tumor infection in esophageal squamous cell carcinoma. Journal of Gastrointestinal Oncology. 2015;6:287-95
36. Syrjanen K. Geographic origin is a significant determinant of human papillomavirus prevalence in oesophageal squamous cell carcinoma: systematic review and meta-analysis. Scandinavian journal of infectious diseases. 2013;45:1-18
37. Souto Damin AP, Guedes Frazzon AP, de Carvalho Damin D, Beck Biehl H, Abruzzi de Oliveira L, Auler R. et al. Detection of human papillomavirus DNA in squamous cell carcinoma of the esophagus by auto-nested PCR. Dis Esophagus. 2006;19:64-8
38. Herbster S, Ferraro CT, Koff NK, Rossini A, Kruel CD, Andreollo NA. et al. HPV infection in Brazilian patients with esophageal squamous cell carcinoma: interpopulational differences, lack of correlation with surrogate markers and clinicopathological parameters. Cancer Lett. 2012;326:52-8
39. Weston AC, Prolla JC. Association between esophageal squamous cell carcinoma and human papillomavirus detected by Hybrid Capture II assay. Dis Esophagus. 2003;16:224-8
40. Antunes LC, Prolla JC, de Barros Lopes A, da Rocha MP, Fagundes RB. No evidence of HPV DNA in esophageal squamous cell carcinoma in a population of Southern Brazil. World J Gastroenterol. 2013;19:6598-603
41. Soheili F, Heidary N, Rahbar M, Nikkho B, Fotouhi O, Afkhamzadeh A. et al. Human papillomavirus and its clinical relevance in oesophageal squamous cell carcinoma in a Kurdish population in the west of Iran. Infectious diseases. 2015:1-4
42. Koshiol J, Dunn ST, Walker JL, Zuna RE, Schiffman M, Sherman ME. et al. Reproducibility of linear array for human papillomavirus genotyping. J Clin Microbiol. 2013;51:625-8
43. Bellini MF, Cadamuro AC, Succi M, Proenca MA, Silva AE. Alterations of the TP53 gene in gastric and esophageal carcinogenesis. J Biomed Biotechnol. 2012;2012:891961
44. Tsoumpou I, Arbyn M, Kyrgiou M, Wentzensen N, Koliopoulos G, Martin-Hirsch P. et al. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev. 2009;35:210-20
45. Fonmarty D, Cherriere S, Fleury H, Eimer S, Majoufre-Lefebvre C, Castetbon V. et al. Study of the concordance between p16 immunohistochemistry and HPV-PCR genotyping for the viral diagnosis of oropharyngeal squamous cell carcinoma. Eur Ann Otorhinolaryngol Head Neck Dis. 2015
46. Kawakami H, Okamoto I, Terao K, Sakai K, Suzuki M, Ueda S. et al. Human papillomavirus DNA and p16 expression in Japanese patients with oropharyngeal squamous cell carcinoma. Cancer Med. 2013;2:933-41
47. Salam I, Hussain S, Mir MM, Dar NA, Abdullah S, Siddiqi MA. et al. Aberrant promoter methylation and reduced expression of p16 gene in esophageal squamous cell carcinoma from Kashmir valley: a high-risk area. Mol Cell Biochem. 2009;332:51-8
48. Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42-55
49. Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J. et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:6262-71
50. Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Brandsma J. et al. Molecular classification identifies a subset of human papillomavirus-associated oropharyngeal cancers with favorable prognosis. J Clin Oncol. 2006;24:736-47
51. Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK. et al. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120:1731-8
Author contact
![]() Corresponding author: Adhemar Longatto Filho, M.Sc., PhD, PMIAC, Laboratory of Medical Investigation (LIM) 14, Faculty of Medicine University São Paulo, Av. Dr. Arnaldo, 455 - Cerqueira César 1246-903, São Paulo, Brazil, Phone: + 55 11 3061-7413, FAX: + 55 11 3061-7413, e-mail: longatto16com
Corresponding author: Adhemar Longatto Filho, M.Sc., PhD, PMIAC, Laboratory of Medical Investigation (LIM) 14, Faculty of Medicine University São Paulo, Av. Dr. Arnaldo, 455 - Cerqueira César 1246-903, São Paulo, Brazil, Phone: + 55 11 3061-7413, FAX: + 55 11 3061-7413, e-mail: longatto16com

 Global reach, higher impact
Global reach, higher impact