3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(7):1284-1291. doi:10.7150/jca.17635 This issue Cite
Research Paper
Preferential Association of Lissencephaly-1 Gene Expression with CD133+ Glioblastoma Cells
1. “Carol Davila” University of Medicine and Pharmacy, 37 Dionisie Lupu Street, 020021 Bucharest, Romania;
2. "Bagdasar-Arseni" Clinical Hospital, Neurosurgery Clinic, 10-12 Berceni Street, 041915 Bucharest, Romania;
3. Institute of Cellular Biology and Pathology, "Nicolae Simionescu", 8 B. P. Hasdeu Street, 050568 Bucharest, Romania;
4. “ Victor Babes” National Institute of Pathology, 99-101 Splaiul Independentei Street, 055096 Bucharest, Romania.
* These authors contributed equally to the research in this paper.
Received 2016-9-19; Accepted 2017-1-12; Published 2017-5-11
Abstract
Lissencephaly-1 (Lis1) protein is a dynein-binding protein involved in neural stem cell division, morphogenesis and motility. To determine whether Lis1 is a key factor in glioblastoma, we evaluated its expression and function in CD133+ glioblastoma cells. Global, Lis1 gene expression is similar in glioblastoma and normal samples. Interestingly, immunohistochemistry data indicate increased Lis1 expression colocalized with CD133 in a subset of glioma cells, including the tumor cells with perivascular localization. Lis1 gene expression is increased up to 60-fold in CD133 positive cells isolated from primary cultures of glioblastoma and U87 glioblastoma cell line as compared to CD133 negative cells. To investigate the potential role of Lis1 in CD133+ glioblastoma cells, we silenced Lis1 gene in U87 cell line obtaining shLis1-U87 cells. In shLis1-U87 cell culture we noticed a significant decrease of CD133+ cells fraction as compared with control cells and also, CD133+ cells isolated from shLis1-U87 were two times less adhesive, migratory and proliferative, as compared with control transfected U87 CD133+ cells. Moreover, Lis1 silencing decreased the proliferative capacity of irradiated U87 cells, an effect attributable to the lower percentage of CD133+ cells. This is the first report showing a preferential expression of Lis1 gene in CD133+ glioblastoma cells. Our data suggest a role of Lis1 in regulating CD133+ glioblastoma cells function.
Keywords: Glioblastoma, Lis1, CD133, U87 cell line.
Introduction
Lissencephaly-1 (Lis1) protein, also known as platelet activating factor acetylhydrolase 1b regulatory subunit 1 (Pafah1b1), is a dynein-binding protein involved in neural stem cell division, morphogenesis and motility [1, 2]. Mutations or deletion of the gene encoding this protein may cause lissencephaly type I [3, 4]. Recent evidence shows a key role of Lis1 in cellular processes such as polarity establishment, fate determinants localization and mitotic spindle orientation by anchoring to the cellular cortex [5-7]. Data indicate that Lis1 is a major player in hematopoietic stem cells function, but also in leukemogenesis by regulating the asymmetric division and self-renewal of normal and malignant hematopoietic stem cells [8, 9]. Glioma stem cells represent a small subpopulation in glioblastoma that is of crucial importance in gliomagenesis, radio resistance, recurrences and survival [10-13]. One of the most known glioma stem cell marker is CD133 [14-17]. CD133 is not just a surface marker, but it is also involved in glioblastoma stem cell maintenance [18]. The working hypothesis of our paper is that Lis1 could play an important role in glioma stem cells function. To test our hypothesis, we evaluated Lis1 expression and function in CD133+ glioblastoma cells.
Material and Methods
Cell culture, transfection and CD133+ cell isolation
U87 cells (U87-MG, HTB14) obtained from ATCC were grown in culture medium (CM), represented by DMEM supplemented with 10% fetal bovine serum (FBS). U87 cells were transfected with a mix of three plasmids, each containing specific shRNA for Lis1 using Plasmid Transfection Reagent in Plasmid Transfection Medium (Santa Cruz, CA). Stably transfected U87 cells in which Lis1 gene was silenced were denoted shLis-U87. Mock transfected cells were obtained using control plasmid A, containing non-specific shRNA (Santa Cruz, CA, USA). Selection of stably transfected cells was done with 5 µg/ml puromycin for three weeks. Primary cell culture, obtained from glioblastoma specimens by trypsinization, were grown in DMEM/F12 supplemented with 10% FBS and used at passage three. In order to increase CD133+ cell fraction, control U87 and shLis1-U87 cells were incubated in stem cell conditioned medium (SM), containing DMEM/F12 supplemented with 10 ng/ml βFGF, 20 ng/ml EGF, and growth factors N2 and B27 (Sigma) at the concentrations recommended by the supplier.
CD133 positive cells were isolated using CD133 MicroBead Kit (Miltenyi Biotec, AC-133/1), as previously described [13]. Briefly, control U87, shLis1-U87 and primary glioblastoma cultures, grown for five days in DMEM supplemented with 10% FBS or in SM, were detached using 33.3 µg/ml Liberase (Roche). The cells were counted, washed successively with DMEM and phosphate-buffered saline containing 0.5% bovine serum albumin (PBS-BSA) and passed through a 70 µm mesh filter. To the cell suspension, 20 µl FcR Blocking Reagent and then 20 µl CD133 MicroBeads were added. After 30 min incubation on ice, cells were washed in 2 ml PBS-BSA and centrifuged at 300 g for 10 min. Cells were resuspended in 500 µl PBS-BSA and applied onto the pre-equilibrated MS column, on the magnetic stand. After three washes with 500 µl PBS-BSA each, the column was removed from the magnetic field and the cells were collected in 1 ml PBS-BSA. Positive and negative CD133 cells were collected, counted and prepared for further analysis.
Gene expression
Lis1 gene expression was evaluated by Real-Time PCR in 36 glioblastoma samples (WHO grade IV glioma [19]) and in 5 normal samples obtained by open surgical procedures in 36 patients with glioblastoma operated in ''Bagdasar-Arseni'' Clinical Hospital, with written informed consent. Normal tissue samples were obtained when anterior temporal lobectomy was performed concomitant with tumor resection in 5 patients from the cohort with severe grand-mall temporal seizures, according to our institutional neurosurgical protocols. The research protocol was approved by the Ethics Committee of ''Bagdasar-Arseni'' Clinical Hospital in accordance with current national and European ethics legislation in medical research. The samples were homogenized using 0.5 mm diameter-zirconium beads in the homogenization solution of Maxwell® 16 LEV simplyRNA kit (Promega, Madison, WI) using a SpeedMill (Analytik-Jena, Germany), in two cycles of 30 sec each, with 30 sec cooling in between. After centrifugation (1 min at 5000 g), 200 μl supernatant were mixed with 200 μl lysis buffer, placed in the Maxwell 16 cartridge (Promega) and further processed according to the manufacturer's instruction. After reverse transcription, gene expression was analyzed by Real Time PCR using TaqMan Gene Expression Assays and specific probes Hs00181182_m1 (labelled with FAM) for Lys1 and Hs02758991_g1 (labelled with VIC) for GAPDH (for normalization), in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Qiagility instrument (Qiagen) was used to pipet the 384 well PCR plates. The results were analyzed with DataAssist V3.01 Software (Applied Biosystems).
Lis1 immunodetection and colocalization with CD133 in human glioblastoma samples
Lis1 protein expression was assessed by immunohistochemistry in eight random glioblastoma samples obtained from the specimen repository of “Bagdasar-Arseni'' Clinical Hospital. CD133 colocalization was performed in five Lis1 positive samples. Formalin-fixed, paraffin-embedded material was available in each case as paraffin blocks. Two µm - thick sections of each sample were obtained from the most representative paraffin block, avoiding large necrotic areas or nervous tissue existing in the main tumor mass. Each section was deparaffinized and dehydrated in gradually increased ethanol concentrations. After heat-induced antigen retrieval, the slides were treated with 3% hydrogen peroxide for 20 min at room temperature, to block endogenous peroxidase activity. Tissue slides were incubated with anti-Lis1 antibody (Abcam: ab2607, Cambridge, UK) diluted 1:2000, overnight at room temperature. After washing, detection was performed with Poly-HRP-GAM/R/R IgG kit and 3, 3'-diaminobenzidine HRP substrate (Immunologic, Duiven, and The Netherlands). Counterstaining was performed with hematoxylin. Then, anti-CD133 antibody (AA: 20-36, GGQPSSTDAPKAWNYELc) from MyBioSource (San Diego, California, USA), diluted 1:500 was applied overnight at room temperature, followed by alkaline phosphatase-labelled secondary antibody. After washing with PBS, the reaction was visualized with the chromogen Fast Red from BioGenex (Milmont Drive, Fremont, CA, USA).
Assessment of Cell Adhesion, Proliferation and Migration
Control U87 or shLis-U87 cells were X-ray irradiated with doses from 5 to 50 Gy (RS2000 apparatus, Rad Source Technologies, Inc). Then, cells were seeded at 1x104 cells/ well, in quadruplicates or triplicates in E-plates and placed in xCelligence RTCA instrument (Roche), and followed-up for 100 hours. Alternatively, irradiated or non-irradiated cells were seeded in 24-well plates and cell proliferation assessed by DNA amount per well was determined using Hoechst 33342 (Molecular Probes), at day 2 and 9 post-irradiation.
To test cell adhesion and proliferation, CD133+ cells isolated from control U87 or from shLis-U87 were seeded on E-plates (104 cells/well), which were introduced in xCELLigence RTCA and followed-up for 3 hours (for cell adhesion tests) or 160 hours (for proliferation tests). To assess the capacity to migrate toward serum, CD133+ cells isolated from U87 or shLis-U87 were seeded at a density of 4x104 cells per well in the upper chamber of CIM plates; in the lower compartment, DMEM (control) or DMEM containing 20% FBS was added. The plate was placed in xCELLigence instrument and followed-up for 3 hours.
Statistical analysis
Real-Time PCR and xCELLigence experiments were performed in triplicates or quadruplicates. The experiments were performed three times. The results were statistically analyzed using One-way ANOVA. The data were expressed as means ± standard deviations.
Results and Discussion
Lis1 expression and its colocalization with CD133 in glioblastoma
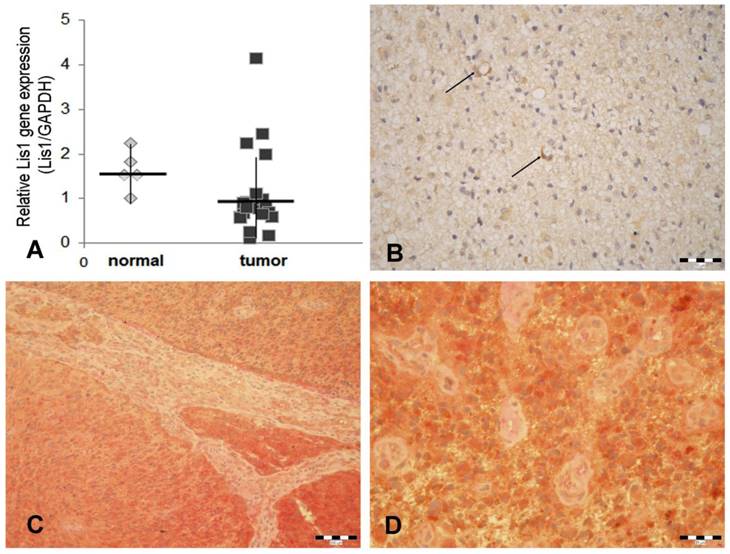
Our data obtained by Real Time PCR showed that Lis1 gene is similarly expressed in normal and glioblastoma tissues (Figure 1A). These data are in agreement with those obtained by Suzuki et al. (2007) which reported that Lis1 protein levels (assessed by Western blot) were not consistently higher in glioblastoma samples as compared to normal brain tissues [20]. Considering the critical role of Lis1 in regulating the asymmetric division and self-renewal of both normal and malignant hematopoietic stem cells [8], we hypothesized that in glioblastoma, elevated Lis1 levels are associated with the small population of CD133+ cells. To determine whether differences exist in Lis1 expression among distinct glioblastoma cells, we investigated Lis1 distribution in tumor tissues. Lis1 immunostaining in glioblastoma samples displayed a broad distribution from negative to positive. In the positive specimens, a subset of glioma cells expressed Lis1 protein (Figure 1B). Colocalization of Lis1 and CD133 was noticed in certain cells, some of them distributed in the perivascular tumor area (Figure 1C, D). Previous reports showed that the main population found in the perivascular niche in glioblastoma is represented by glioma stem cells [21-23]. These cells may be found among general tumor population either as CD133+ perivascular niches or as single cells [24]. However, CD133 immunostaining results should be analyzed with caution, as a former report demonstrated inconsistent immunohistochemical expression patterns among different CD133 antibody clones [25].
Lis1 and CD133 gene expression in neurosphere-like U87 cells
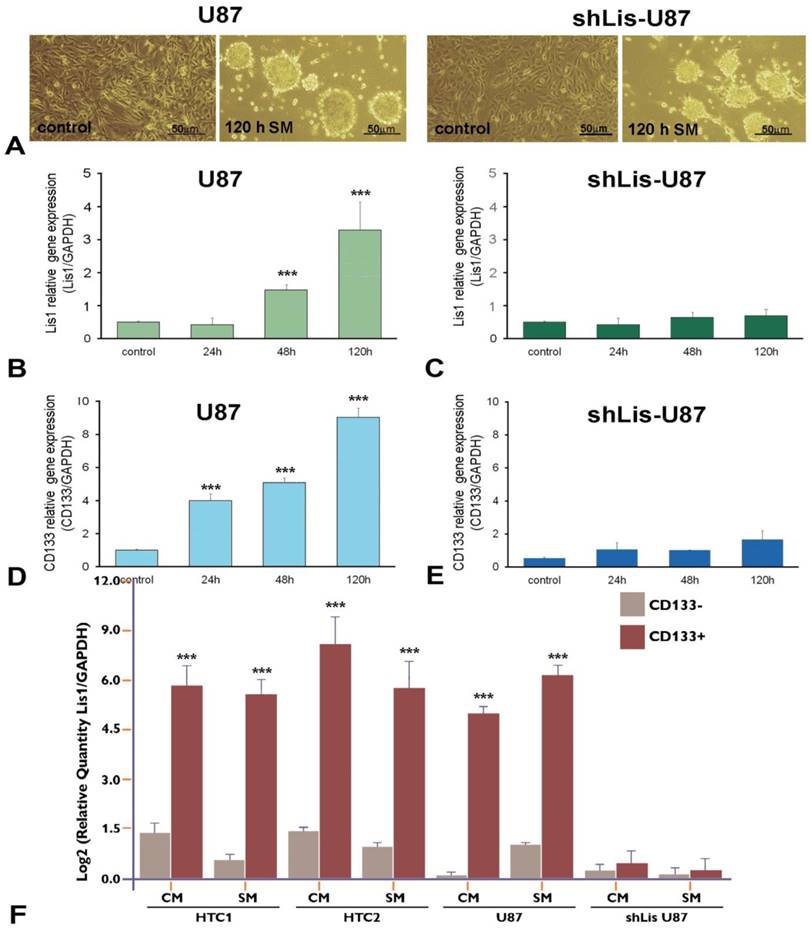
To verify the correlation between CD133 and Lis1, we silenced Lis1 gene in U87 cells by stable transfection with shRNA specific for Lis1, and we obtained shLis-U87cells, that did not express Lis1 either in normal medium (Figure 2C, control) or after exposure to serum-free, neural stem cell conditioned-medium (SM) for various periods (Figure 2C, columns 24h, 48h and 120h). Incubation of U87 cells with SM induced phenotypic changes resulting in neurosphere-like morphology, as illustrated in Figure 2A left. The same morphology was noticed for U87 cells in which Lis1 was silenced (Figure 2A right). Interestingly, we found that both Lis1 and CD133 expression increased progressively within five days of U87 cells exposure to SM (Figure 2B and D, respectively), suggesting a possible correlation between Lis1 and CD133 expression. shLis-U87 cells expressed less CD133 and this could not be induced even after exposure to stem-conditioned medium (Figure 2E). As expected, Lis1 expression could not be induced in U87 cells transfected with plasmids containing specific shRNA for Lis1 (Figure 2C).
Lis1 gene expression and Lis1 colocalization with CD133 in glioblastoma. (A) Five normal and 36 glioblastoma samples were analyzed for Lis1 expression by Real-Time PCR. Lis1 expression normalized to GAPDH expression in normal (grey rhombuses) and tumor samples (black squares) are not statistically different. (B) In Lis1 positive glioblastoma samples, some intensely stained cells present a perivascular distribution (arrows). (C) Colocalization of Lis1 and CD133 in a Lis1 positive case of glioblastoma. Lis1 positive cells (brown color), and CD133 (red staining) are widely spread. The microvascular structure, separating tumor areas, is negative. (D) Details of the case presented in (C). The majority of tumor cells express both antigens. The small tumor vessels are negative. (Bars, 50 µm in B, 100 µm in C, and 25 µm in D).
Lis1 gene is overexpressed in CD133+ glioblastoma cells
Next, we evaluated Lis1 gene expression in CD133+ and CD133- cells isolated from primary cultures obtained from glioblastoma tumors as well as from U87 cultures incubated in CM or SM for five days. Lis1 expression was increased up to 60 fold in CD133+ fraction obtained from primary glioblastoma cells grown in culture medium (Figure 2F, HTC1/CM and HTC2/CM columns) or up to 32 fold in SM (Figure 2F, HTC1/SM and HTC2/SM columns). Lis1 gene was overexpressed 32 fold in CD133+ cells compared with CD133- cells isolated from U87 culture (Figure 2F, U87/CM columns) or up to 35 fold in SM (Figure 2F, U87/SM columns). As negative control we used CD133+ cells isolated from shLis-U87 incubated in CM or SM, when Lis1 was not increased as compared with CD133- cells (Figure 2F, shLis-U87/CM and SM columns).
Proliferation of irradiated U87 and shLis-U87 cells
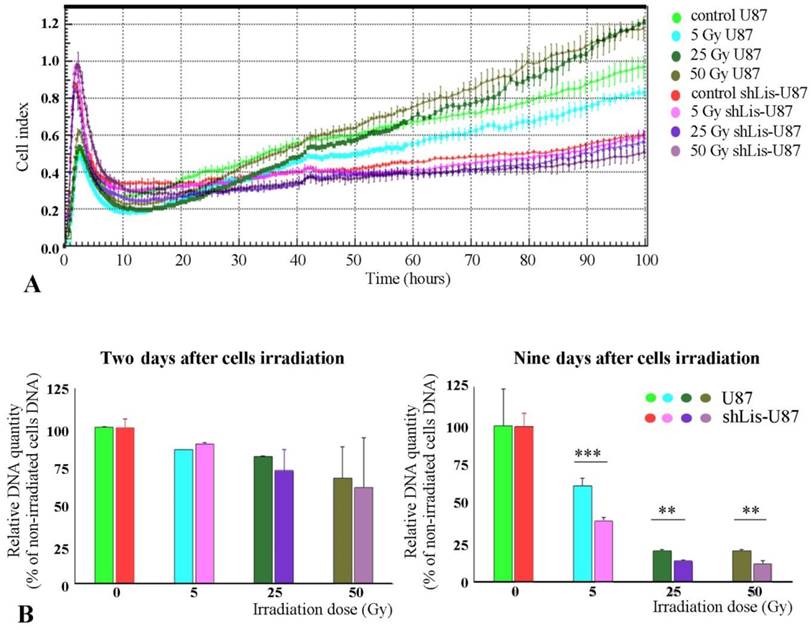
As previously known, CD133+ cells are resistant against tumor radiation therapy (11). Interestingly, we found that irradiated U87 cells recovered better their proliferative capacity than shLis-U87 cells, whether assessed by the electrical properties of the cells, as in the xCELLigence system (Figure 3A) or by quantification of the DNA amount (Figure 3B). The proposed explanation is that U87 culture contains a higher number of CD133+ stem cells resistant to irradiation compared with shLis-U87 culture. Our data confirm this hypothesis and show that the percentage of CD133+ cells in control U87 cultures exposed for five days to SM was ~2%, data in agreement with results from the literature demonstrating an increase of CD133+ cell fraction in U87 cells cultivated in SM [26]. However, the number of CD133+ cells found in shLis1-U87 under the same condition dramatically decreased to ~0.3% CD133+ (data not shown).
Lis1 and CD133 gene expression in neurosphere-like U87 cells. (A) Exposure of U87 glioma cells to stem-conditioned medium (SM) induces phenotypic modifications resulting in neurospheres after five days, in both regular U87 cells (left) and shLis1-U87 cells (right). The expression of Lis1 (B) and CD133 (D) is induced in U87 cells exposed to SM, with the highest level at day 5 of incubation. (C) As expected, silencing Lis1 gene in U87 cells (shLis-U87) inhibits its induction in cells incubated with SM. CD133 induction is almost abrogated in shLis-U87 cells (E) compared with U87 cells (D). Lis1 is highly expressed in CD133+ cells isolated from U87 cell line and primary glioblastoma (HTC1 and HTC2) cell cultures in normal culture medium (CM) or SM (F). Data show enrichment up to 60 fold in Lis1 expression in CD133 + fractions for cells grown in CM and up to 32 fold in cells incubated in SM. Lis1 expression in CD133+ fraction isolated from U87 cells grown in CM is 32 times higher than that of CD133- fraction (U87 columns); in CD133+ fraction isolated from U87 incubated in SM Lis1 expression is 35 times higher than in CD133 negative cells from the same culture. The negative control is represented by CD133+ cells isolated from shLis-U87 incubated in CM or SM for which Lis1 was not increased as compared with CD133- cells (shLis-U87/CM and SM columns).
Proliferation of irradiated U87 and shLis-U87 cells. Cells having Lis1 silenced or not were irradiated with X-ray doses from 5 to 50 Gy. Cells seeded at a density of 1x104 cells/well, in quadruplicates or triplicates in E-plates and placed in xCelligence RTCA instrument, were followed-up for 100 hours (A). Alternatively, irradiated or not irradiated cells were seeded in 24-well plates and the DNA amount per well was determined using Hoechst 33342 (B). Both methods showed that irradiated U87 cells recovered better their proliferative capacity than shLis-U87 cells.
Lis1 impact on cell adhesion, migration and proliferation of CD133+cells
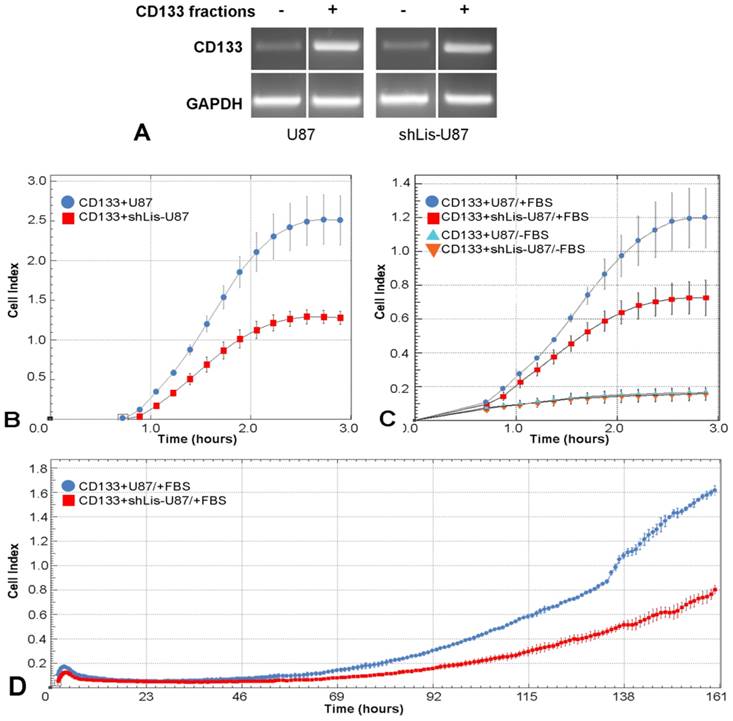
To explore Lis1 function in CD133+ glioblastoma cells, we tested cell adhesion, migration and proliferation of CD133+ cells isolated from U87 and shLis-U87 cells, following their behavior in real time in xCELLigence system. The purity of the fractions was assessed by CD133 expression determined by RT-PCR. Data showed a good enrichment of CD133+ cells in the positive fractions, as showed in Figure 4A.
The cell index for CD133+ cells isolated from shLis-U87 was half of that obtained for CD133+ cells isolated from control U87 cells (Figure 4B), indicating that Lis1 silencing delayed the adhesion of CD133+ cells. The recorded cell index for migratory test was two times lower for shLis-U87 CD133+ cells as compared with that obtained for control U87 CD133+ cells (Figure 4C). Proliferation test showed that after five days, control U87 CD133+ cells presented a cell index double than that obtained for CD133+ cells isolated from shLis-U87 cells (Figure 4D).
Taken together, these data suggest that Lis1 silencing led to a deregulation of the mechanisms involved in the maintenance of the CD133+pool in glioblastoma cell population.
Conclusions
Our results demonstrate that Lis1 gene is preferentially expressed in CD133+ glioblastoma cells and suggest a role of Lis1 in maintaining the CD133+ glioblastoma cell population. To our knowledge, this is the first report evaluating Lis1expression in CD133+ glioblastoma cells. Future studies are required to fully elucidate the role of Lis1 in glioblastoma stem cells function, and to determine if Lis1 may become a molecular target for glioblastoma therapy.
Cell adhesion, migration and proliferation of CD133 + cells isolated from U87 and shLis-U87 cells. CD133+ cells were isolated from control U87 and shLis-U87 cells. The purity of the fraction is revealed by higher CD133 expression (assessed by RT-PCR) in CD133+ fraction as compared with CD133- fraction (A). CD133+ cells isolated from control U87 (blue circles) or from shLis-U87 (red squares) cultures were subjected to functional assays using xCELLigence Real-Time Cell Analysis instruments. The data representing the recorded cell index at different times show that CD133+ cells isolated from shLis-U87 culture present two times lower (B) adherence to the surface, (C) migratory potential and (D) proliferative rate, as compared with the those isolated from control U87 culture.
Abbreviations
Lis1: Lissencephaly-1 protein; CM: culture medium, SM: stem-conditioned medium; RT-PCR: reverse transcription polymerase chain reaction; WHO: World Health Organization; DMEM: Dulbecco's Modified Eagle Medium; FBS: Fetal Bovine Serum, HTC- human tumor cells.
Acknowledgements
We thank Dr. Madalina Fenyo for help with irradiating cells, and Dr. Ovidiu Croitoru for graphical design.
Funding
This work was supported by a grant of the Romanian National Authority for Scientific Research, CNCS - UEFISCDI, project number PN-II-RU-TE-2012-3-0235 and a grant of the Romanian National Authority for Scientific Research and Innovation, CCCDI - UEFISCDI, project number 46 M/13.07.2016.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. The Journal of Cell Biology. 2005;170(6):935-945
2. Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10(8):970-979
3. Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein betasubunit-like repeats. Nature. 1993;364(6439):717-721
4. Lo Nigro C, Chong CS, Smith AC, Dobyns WB, Carrozzo R, Ledbetter DH. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum Mol Genet. 1997;6(2):157-164
5. Britta W, Ulrich S. Stem cell fate regulation by dynein motor protein Lis1. Nature Genetics. 2014;46(3):217-218
6. Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319(1):1-9
7. Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132(3):474-486
8. Zimdahl B, Ito T, Blevins A, Bajaj J, Konuma T, Weeks J, Koechlein CS, Kwon HY, Arami O, Rizzieri D, Broome HE, Chuah C, Oehler VG, Sasik R, Hardiman G, Reya T. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nature Genetics. 2014;46(3):245-252
9. Chen X, Zhang J, Zhao J, Liu M, Sun X, Zhao M, Liu X. Lis1 is required for the expansion of hematopoietic stem cells in the fetal liver. Cell Research. 2014;24(8):1013-1016
10. Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396-401
11. Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756-760
12. Shibahara I, Sonoda Y, Saito R, Kanamori M, Yamashita Y, Kumabe T, Watanabe M, Suzuki H, Watanabe T, Ishioka C, Tominaga T. The expression status of CD133 is associated with the pattern and timing of primary glioblastoma recurrence. Neuro-Oncology. 2013;15(9):1151-1159
13. Ardebili SY, Zajc I, Gole B, Campos B, Herold-Mende C, Drmota S, Lah TT. CD133/prominin1 is prognostic for GBM patient's survival, but inversely correlated with cysteine cathepsins' expression in glioblastoma derived spheroids. Radiol Oncol. 2011;45(2):102-115
14. Dahlrot RH, Hermansen SK, Hansen S, Kristensen BW. What is the clinical value of cancer stem cell markers in gliomas? Int J Clin Exp Pathol. 2013;6(3):334-348
15. Beier D1, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010-5
16. Zhang M, Song T, Yang L, Chen R, Wu L, Yang Z, Fang J. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. Journal of Experimental & Clinical Cancer Research. 2008;27:85
17. Wu A, Oh S, Wiesner SM, Ericson K, Chen L, Hall WA, Champoux PE, Low WC, Ohlfest JR. Persistence of CD133+ cells in human and mouse glioma cell lines: detailed characterization of GL261 glioma cells with cancer stem cell-like properties. Stem Cells Dev. 2008;17(1):173-84
18. Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31(5):857-69
19. Cavenee WK, Furnari FB, Nagane M, Huang HJS, Newcomb EW, Bigner DD. Diffusely infiltrating astrocytomas. In: (ed.) Kleihues P, Cavenee CW. Pathology and Genetics of Tumours of the Nervous System. World Health Organization Classification of Tumours. 2nd ed. Lyon, France: IARC Press. 2000:10-21
20. Suzuki SO, McKenney RJ, Mawatari SY, Mizuguchi M, Mikami A, Iwaki T, Goldman JE, Canoll P, Vallee RB. Expression patterns of LIS1, dynein and their interaction partners dynactin, NudE, NudEL and NudC in human gliomas suggest roles in invasion and proliferation. Acta Neuropathol. 2007;113(5):591-599
21. Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell. 2007;11(1):69-82
22. Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Stem Cell-like Glioma Cells Promote Tumor Angiogenesis through Vascular Endothelial Growth Factor. Cancer Res. 2006;66(16):7843-8
23. Gilbertson RJ, Rich JN. Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nature Reviews Cancer. 2007;7(10):733-736
24. Christensen K, Schrøder HD, Kristensen BW. CD133+ niches and single cells in glioblastoma have different phenotypes. Journal of Neuro-Oncology. 2011;104(1):129-143
25. Hermansen SK, Christensen KG, Jensen SS, Kristensen BW (2011). Inconsistent Immunohistochemical Expression Patterns of Four Different CD133 Antibody Clones in Glioblastoma. Journal of Histochemistry & Cytochemistry. 2011;59(4):391-407
26. Annabi B, Laflamme C, Sina A, Lachambre MP, Béliveau R. A MT1-MMP/NF-κB signaling axis as a checkpoint controller of COX-2 expression in CD133(+) U87 glioblastoma cells. Journal of Neuroinflammation. 2009;6:8
Author contact
![]() Corresponding author: Felix Mircea Brehar, "Bagdasar-Arseni" Clinical Hospital, Neurosurgery Clinic, 10-12 Berceni Street, Bucharest, 041915, e-mail: felixbreharcom, phone: +40724257549, fax: +40213347350
Corresponding author: Felix Mircea Brehar, "Bagdasar-Arseni" Clinical Hospital, Neurosurgery Clinic, 10-12 Berceni Street, Bucharest, 041915, e-mail: felixbreharcom, phone: +40724257549, fax: +40213347350

 Global reach, higher impact
Global reach, higher impact