3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(8):1498-1506. doi:10.7150/jca.18626 This issue Cite
Research Paper
Long Noncoding RNA CCAT2 as a Potential Novel Biomarker to Predict the Clinical Outcome of Cancer Patients: A Meta-Analysis
Division of Gastroenterology and Hepatology, Key Laboratory of Gastroenterology and Hepatology, Ministry of Health, State Key Laboratory for Oncogenes and Related Genes, Renji Hospital, School of Medicine, Shanghai Jiao Tong University
Shanghai Institute of Digestive Disease
145 Middle Shandong Road, Shanghai 200001, China.
* Co-first authors, these authors contributed equally to this work.
Received 2016-12-5; Accepted 2017-3-13; Published 2017-6-1
Abstract
Background: Colon Cancer-Associated Transcript 2 (CCAT2) has been demonstrated associated with clinical outcomes in various tumors. However, the results from each study were unfortunately insufficient and not completely consistent. Therefore, we conduct a systematic meta-analysis to evaluate the value for a feasible biomarker for metastasis and prognosis.
Methods: A meta-analysis was performed using data obtained through a systematic search of PubMed, EMBASE, Cochrane Library, China National Knowledge Infrastructure, Wanfang database and VIP database. The pooled odds ratio (OR) and hazard ratio (HR) with 95% Confidence interval (CI ) using random-effect were used to identify the relationship of CCAT2 with clinical outcome of cancer patients. Subgroup analysis and sensitivity analysis were performed.
Results: A total of 867 patients from eight studies were finally included. Patients with high CCAT2 expression underwent an increased risk of lymph node metastasis (LNM) (OR=3.09, 95% CI: 1.53-6.26) and distant metastasis (DM) (OR=7.70, 95% CI: 3.26-18.17). CCAT2 was also significantly correlated with overall survival (OS) (HR=2.19, 95%CI: 1.70-2.82) and progression-free survival (PFS) (HR=2.59, 95% CI: 1.78-3.76). Moderate heterogeneity was observed in meta-analysis for LNM. However, the results remained robust in multiple sensitivity analyses.
Conclusions: High expression of CCAT2 was linked with poor clinical outcome. CCAT2 can serve as a potential molecular marker for prognosis in different types of cancers.
Keywords: lncRNA, CCAT2, cancer, clinical outcome, meta-analysis.
Introduction
The occurrence of cancer continues to increase, and cancer constitutes an enormous burden on society [1]. The current strategies to cancer management have significantly improved some types of cancer patients' overall survival (OS). However, the outcome still remains undesirable. The identification of molecular biomarkers which can use to evaluate potential high-risky cancer patients was therefore of great clinical value. Currently, with the advancements in transcriptome profiling, the number of long noncoding RNAs (lncRNAs) with limited or no protein-coding capacity found in the human genome exceeded ten thousand, but only a tiny fraction of lncRNAs have been functionally characterized [2,3]. Growing evidence has revealed that lncRNAs may play a role in the development and progression of cancer [4-6]. And, some of them could become a new class of cancer biomarkers implicated in diagnosis, prognostication and therapeutic targets, such as HOTAIR and MALAT-1 [7-9]. Therefore, more specific lncRNAs with predictive and prognostic value in management of cancer need to be further teased out.
Colon Cancer-Associated Transcript 2 (CCAT2), a lncRNA that spans the highly conserved 8q24 region harboring the rs6983267 SNP, first discovered by ling et al. [10]. The G allele of the rs6983267 SNP was associated with greater predisposition to colorectal cancer (CRC) than the T allele [11], and the expression and function of CCAT2 were affected by rs6983267 alleles [10]. CCAT2 could induce chromosomal instability and metastases, partly through MYC and Wnt signaling pathway [10]. Recently, CCAT2 was found to reprogram energy metabolism in an allele-specific manner by interacting with the Cleavage Factor I (CFIm) complex to regulate the alternative splicing of glutaminase [12]. Those results suggest that abnormal expression of CCAT2 may contribute to a potential high malignancy degree of cancer.
Abnormal expression of CCAT2 has been reported in various kinds of cancer, including the microsatellite stable CRC, breast cancer, gastric cancer (GC), small cell lung cancer (SCLC), cervical squamous cell cancer (CSCC), ovarian cancer, bladder cancer, oral squamous cell carcinoma (OSCC), non-small cell lung cancer (NSCLC), esophageal squamous cell carcinoma (ESCC), prostate cancer, and hepatocellular carcinoma (HCC) [10, 13-24]. High expression of CCAT2 has been reported associated with metastasis and prognosis in different types of cancer, which indicated the diagnostic or prognostic value of CCAT2 for cancer [13-16, 20, 22, 23]. However, the results from each study were insufficient and not completely consistent in the correlation between CCAT2 and the clinical metastatic factors (as lymph node metastasis (LNM) and distance metastasis (DM)) or prognostic factors (like OS and progression free survival (PFS)). This meta-analysis was conducted to identify the relationship of CCAT2 with cancer patients' clinical outcome, to further validate whether CCAT2 could serve as a feasible biomarker for metastasis and prognosis.
Methods
Literature search strategies
An exhaustive search was performed in PubMed, EMBASE, Cochrane Library, China National Knowledge Infrastructure, VIP database and Wanfang database. The last update of searching time was November 9, 2016. There was no language restriction. The keywords for the search were as follows: “long non-coding RNA CCAT2” or “lncRNA CCAT2” or “CCAT2” or “colon cancer-associated transcript 2” and cancer or carcinoma or tumor or tumour or neoplasms. Besides, the reference lists are manually viewed to get potentially eligible papers and authors contacted for further information where necessary.
Inclusion and exclusion criteria
The criteria for inclusion was as follows: 1) articles evaluating the role of CCAT2 in the development of any type of human cancer, 2) the expression levels of CCAT2 in primary tumor tissue were detected, and 3) patients were grouped based on the expression levels of CCAT2, 4) reporting of clinical characteristics or outcomes, including LNM or DM or OS or PFS, 5) having sufficient data for calculating the odds ratios (OR) or hazard ratios (HR) and corresponding 95 % confidence intervals (CI).Exclusion criteria were the following: 1) nonhuman research, reviews, editorials, expert opinions, letters, and case reports, 2) studies without sufficient data, 3) duplicate publications.
Data extraction and quality assessment
The information was independently extracted and investigated by TJ and HYC from each included studies following the criteria for inclusion and exclusion stated above. A consensus was created by the third investigator (CYX) if there were disagreements. All eligible studies were assessed to be of high quality by using the Newcastle-Ottawa Scale (NOS). For each eligible study, the following information was collected: first author, year of publication, country, tumor type, total number of patients, number of high CCAT2 expression group and low CCAT2 expression group, number of patients with LNM and DM, detection method of CCAT2 expression levels, HRs, and corresponding 95% CIs for OS or PFS. All articles included were assessed the quality with the NOS by two investigators (TJ and HYC) independently.
Statistical methods
All the statistical analyses in this meta-analysis were conducted by using Review Manager version software 5.3.5 (Nordic Cochrane Center) and Stata statistical software version 13.0 (StataCorp, USA). Data of HRs were directly extracted from the eligible studies. If the articles only provided survival curves without offering HR, calculation method introduced by Tierney et al. [25] and Parmar et al. [26] was applied to extract HR with 95%CI using Engauge Digitizer (version 4.1).
The heterogeneity among studies was evaluated by using the Q test (with a significance level of P < 0.10) and I2 statistics [27]. A value of I2 of 0%-30%, 31%-50%, 51%-75%, and 75%-100% each represents an insignificant, low, moderate, and considerable heterogeneity, respectively [27]. A model of random-effect was employed to analyze the data independent of heterogeneity, which renders more precise and authentic results [28]. Begg's test was used to assess the potential publication bias. A two-tailed P values < 0.05 was considered statistically significant.
Results
Characteristics of eligible studies
As shown in Figure 1, a total of 63 published articles were identified from the first attempt to search by using the keywords, of which 23 duplicates were excluded. After screening the title and abstract carefully, 28 articles were excluded. After further inspection of the full articles, 5 articles were excluded, including 3 not relevant to LNM, DM or prognosis, 2 without sufficient data to achieve HR with 95 % CI. Finally, according to the criteria for selection, a total of 8 studies were eligible in this meta-analysis.
Table 1 showed the main characteristics of the included publications. A total of 8 studies involving 867 cancer patients were included. The mean of sample size was 108.4 (range from 48 to 227). Among the eight studies, CCAT2 was tested in 8 types of cancers including prostate cancer, SCLC, ovarian cancer, GC, CSCC, bladder cancer, breast cancer and ESCC. Quantitative real-time PCR (qRT-PCR) was used in all studies to detect CCAT2 expression, of which 7 studies chose GAPDH as a reference gene. In all of the studies, the patients were divided into two groups: high and low expression of CCAT2.
The flow diagram of the meta-analysis
Characteristics of studies in this meta-analysis
| Author | Year | Country | Tumor type | Sample size | CCAT2 expression | CCAT2 assay | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| High expression | High with LNM | High with DM | Low expression | Low with LNM | Low with DM | ||||||
| Zheng JF | 2016 | China | PC | 96 | 59 | 10 | 32 | 37 | 5 | 8 | RT-qPCR |
| Chen SF | 2016 | China | SCLC | 112 | 56 | 45 | 14 | 56 | 22 | 0 | RT-qPCR |
| Huang SY | 2016 | China | OC | 109 | 55 | NA | 36 | 54 | NA | 6 | RT-qPCR |
| Wang CY | 2015 | China | GC | 85 | 44 | 28 | 11 | 41 | 8 | 3 | RT-qPCR |
| Chen X | 2015 | China | CSCC | 123 | 62 | 34 | NA | 61 | 11 | NA | RT-qPCR |
| Li JF | 2016 | China | BC1 | 48 | 28 | 1 | NA | 20 | 2 | NA | RT-qPCR |
| Zhang XL | 2015 | China | ESCC | 227 | 114 | 65 | NA | 113 | 48 | NA | RT-qPCR |
| Cai Y | 2015 | China | BC2 | 67 | 25 | NA | NA | 42 | NA | NA | RT-qPCR |
PC prostate cancer, SCLC small cell lung cancer, OC Ovarian cancer, GC gastric cancer, CSCC cervical squamous cell cancer, BC1 bladder cancer, ESCC esophageal squamous cell carcinoma, BC2 breast cancer, qRT-PCR quantitative real-time PCR, LNM lymph node metastasis, DM distant metastasis, NA not available.
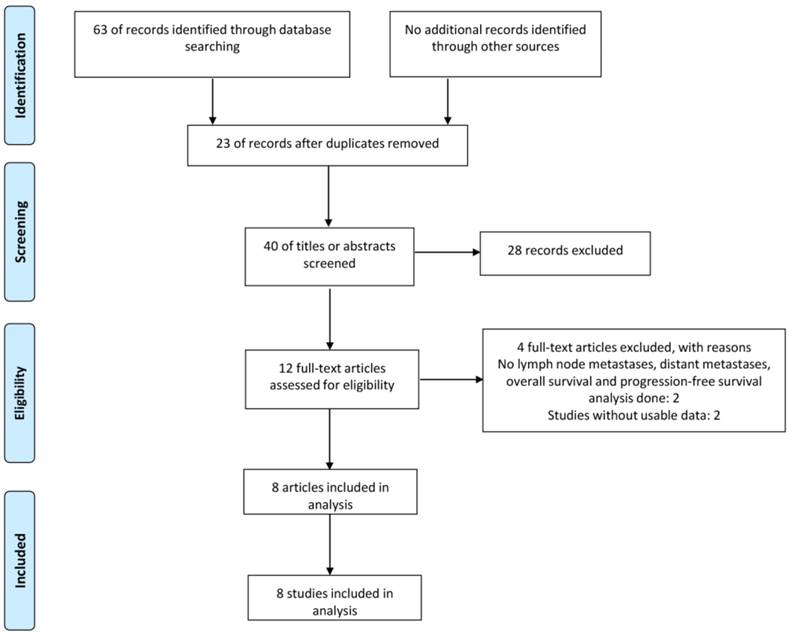
Forest plot for the association between CCAT2 expression levels with LNM
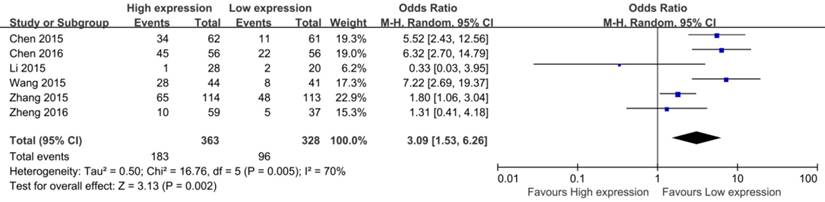
Forest plot for the association between CCAT2 expression levels with DM
Meta-analysis results
Association between CCAT2 and LNM
In the eight eligible studies, six studies reported the LNM of 691 patients based on different CCAT2 expression levels [14, 15, 17, 20, 22, 23]. Four studies demonstrated a statistically significant relationship between high CCAT2 expression and more LNM [14, 15, 20, 22]; the remaining 2 studies did not indicate any association [17, 23]. The OR (the value of high CCAT2 expression group versus low CCAT2 expression group) was 3.09 (95%CI 1.53-6.26, P = 0.002) with a moderate heterogeneity (I2 = 70.0%, Q = 16.76, P bias = 0.005) in a pooled analysis of all 6 studies.
Association between CCAT2 and DM
Among the eight eligible studies, only 4 studies reported the DM of patients, including 402 patients [14, 16, 20, 23]. The result of meta-analysis showed a pooled OR which was 7.70 (95% CI: 3.26 - 18.17, P < 0.01) and low heterogeneity (I2 = 43.0%, P bias = 0.15) (Figure 3). Obviously, patients with high CCAT2 expression were more likely to DM.
Association between CCAT2 and OS
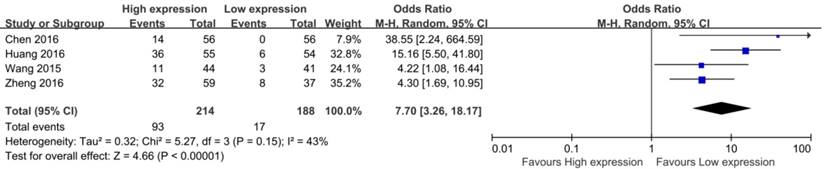
Seven studies including 819 patients were assessed for the correlation between CCAT2 and OS (Table 2) [13-16,20,22,23]. High CCAT2 expression was significantly correlated with poor prognosis (HR 2.19; 95% CI: 1.70-2.82; P <0.01) with a low heterogeneity (I2 = 28%; P bias = 0.21), compared with low CCAT2 expression in a pooled analysis of all seven studies (Figure 4).
Survival data of the eligible studies in this meta-analysis
| Author | Year | Tumor type | Sample size | Survival analysis | Multivariate analysis | HR statistic | Hazard ratios(95% CI) | RNA extraction | Reference control | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OS | PFS | OS | PFS | ||||||||
| Zheng JF | 2016 | PC | 96 | Yes | Yes | Yes | Data in paper | 2.292 (1.370-3.528) | 2.276 (1.199-2.768) | TRIzol(Invitrogen) | GAPDH |
| Chen SF | 2016 | SCLC | 112 | Yes | NA | Yes | Data in paper | 2.034 (1.216-3.402) | NA | RNAiso Plus (TAKARA) | GAPDH |
| Huang SY | 2016 | OC | 109 | Yes | NA | Yes | Data in paper | 2.938 (1.526-5.873) | NA | TRIzol(Invitrogen) | GAPDH |
| Wang CY | 2015 | GC | 85 | Yes | Yes | Yes | Data in paper | 2.405 (1.194-5.417) | 2.315 (1.097-5.283) | TRIzol(Invitrogen) | GAPDH |
| Chen X | 2015 | CSCC | 123 | Yes | Yes | Yes | Data in paper | 2.813 (1.504-6.172) | 3.072 (1.716-8.174) | TRIzol(Invitrogen) | GAPDH |
| Zhang XL | 2015 | ESCC | 227 | Yes | NA | Yes | Data in paper | 1.432 (1.005-2.041) | NA | Total Nucleic Acid Isolation Kit (Ambion) | RNU6B |
| Cai Y | 2015 | BC1 | 67 | Yes | NA | No | Survival curve | 3.87 ( 1.57-9.50 ) | NA | TRIzol(Thermo) | GAPDH |
OS Overall survival, PFS progression free survival, PC prostate cancer, SCLC small cell lung cancer, OC Ovarian cancer, GC gastric cancer, CSCC cervical squamous cell cancer, BC1 breast cancer, ESCC esophageal squamous cell carcinoma, Y Yes, NA not available, 95%CI 95% confidence interval.
Forest plot for the association between CCAT2 expression levels with OS
Forest plot for the association between CCAT2 expression levels with PFS
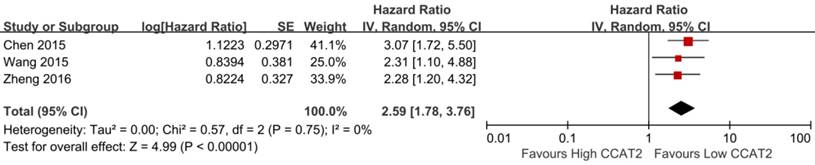
Association between CCAT2 and PFS
As shown in Table 2, three studies reported the PFS of patients [15, 20, 23]. The result indicated that higher expression of CCAT2 might predict poor PFS in various cancers (HR 2.59; 95% CI: 1.78-3.76; P <0.01), with a low heterogeneity (I2 = 0%; P bias = 0.75) (Figure 5).
Subgroup analysis
Subgroup analyses across several different variables were further performed to investigate the heterogeneity of the studies for meta-analysis of CCAT2 and LNM. The LNM related data was stratified into subgroups based on sample size, tumor type, reference control and RNA extraction. The assessment results in each subgroup are also shown in Table 3. Subgroup analysis by sample size explored that high CCAT2 expression status was related to high LNM numbers in big sample size group (n ≥ 100, OR 3.77, 95%CI 1.57 - 9.05, P < 0.01), but not in small size group(n < 100, OR 1.91, 95%CI 0.38 - 9.57, P = 0.43). However, when conducting subgroup analyses on tumor type, we found no significant correlation between high CCAT2 expression and LNM among the studies in Digestive system or Urinary system. According to the results presented in Table 3, when divided by reference control, the subgroup analysis showed that in GAPDH group, up-regulated CCAT2 was associated with more LNM (OR 3.66, 95%CI 1.67-8.03, P < 0.01). As for different RNA extraction method for CCAT2, the relationship between CCAT2 expression and tumor LNM was significant in TRIzol group (OR 2.88, 95%CI 1.01-8.22, P < 0.05), but not in others group (OR 3.22, 95%CI 0.94-11.01, P = 0.06).
Sensitivity analysis
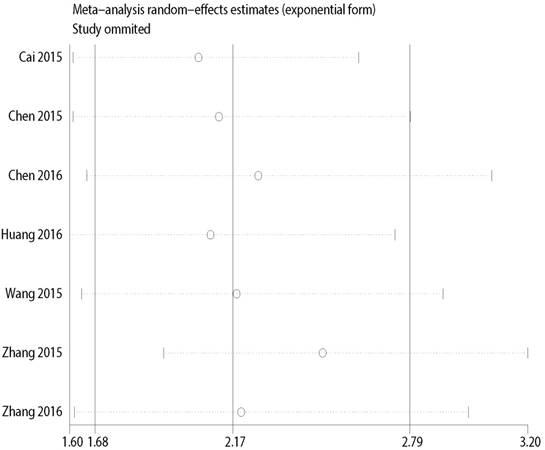
Multiple sensitivity analyses were performed to evaluate whether individual study influenced pooled ORs or HR by excluding one study by turns. It was found that none of the exclusions of a specific study would change the magnitude or direction of the summary effect for the correlation between CCAT2 expression and LNM or OS, which further confirmed the validity of the results (Figure 6/7). The publication bias and sensitivity analysis for DM and PFS were not assessed due to the limited number of studies involved and no severe heterogeneity among studies.
Publication bias
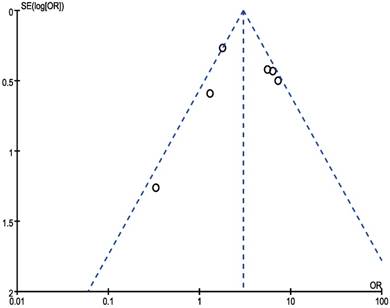
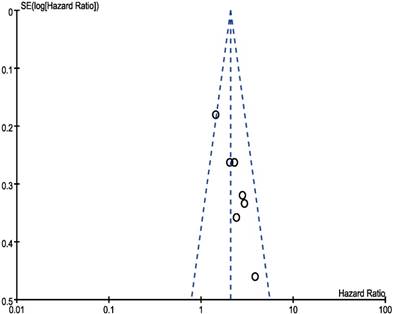
In our meta‑analysis, Begg's tests were introduced to examine potential publication bias. No evidence supporting publication bias was found in analysis between CCAT2 and LNM (P = 1.00) (Figure 8) and analysis between CCAT2 and OS (P = 0.133) (Figure 9). The publication bias, however, cannot be determined on account of the limited number of studies involved in the current meta-analysis.
Subgroup analysis of the role of CCAT2 in LNM in different types of cancer
| Subgroup | No. of studies | No. of patients | Test of Relationship | Test of Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | Q value | P bias | I2, % | ||||
| Overall | 6 | 691 | 3.09 (1.53-6.26) | <0.01 | 15.68 | 0.005 | 70 | |
| Sample size | ||||||||
| < 100 | 3 | 229 | 1.91 (0.38-9.57) | 0.43 | 8.03 | 0.02 | 75 | |
| ≥ 100 | 3 | 462 | 3.77 (1.57-9.05) | <0.01 | 8.72 | 0.01 | 77 | |
| Tumor type | ||||||||
| Digestive system | 2 | 312 | 3.37 (0.87-13.11) | 0.08 | 5.95 | 0.01 | 83 | |
| Urinary system | 2 | 144 | 1.02 (0.36-2.92) | 0.97 | 0.96 | 0.33 | 0 | |
| Others | 2 | 235 | 5.89 (3.26-10.64) | <0.01 | 0.05 | 0.82 | 0 | |
| Reference control | ||||||||
| GAPDH | 5 | 464 | 3.66 (1.67-8.03) | <0.01 | 10.35 | 0.03 | 61 | |
| RNU6B | 1 | 227 | 1.80 (1.06-3.04) | 0.03 | - | - | - | |
| RNA extraction | ||||||||
| TRIzol | 4 | 352 | 2.88 (1.01-8.22) | <0.05 | 9.39 | 0.02 | 68 | |
| Others | 2 | 339 | 3.22 (0.94-11.01) | 0.06 | 6.09 | 0.01 | 84 | |
LNM, lymph node metastasis; OR, odds ratio; 95%CI, 95% confidence interval.
Sensitivity analysis for the association between CCAT2 expression levels with LNM
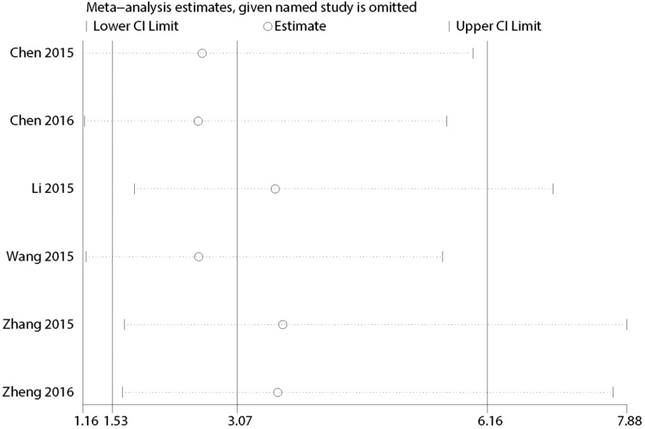
Sensitivity analysis for the association between CCAT2 expression levels with OS
Funnel plot analysis of potential publication bias for LNM
Funnel plot analysis of potential publication bias for OS
Discussion
Evidence from growing publications showed that lncRNAs, similar to protein-coding genes, have oncogenic or tumor-suppressing effects, implicating in various tumorigenesis processes including proliferation, apoptosis and invasion. More and more functional lncRNAs were found to have potential value on predicting disease progression [4-6, 8, 29-31]. CCAT2, a functional lncRNA, first named in 2013, could enhance invasion and metastasis and involve in migration and chemo-resistance in a SNP-independent manner [10, 32]. Recently, CCAT2 was discovered to regulate cancer metabolism in an allele-specific manner by fine-tuning the alternative splicing of glutaminase through interaction with the CFIm complex [12]. The crucial action of CCAT2 inside the molecular carcinogenesis indicated its potential value for diagnosis or prognosis. However, a persuasive support of the CCAT2 in clinical practice is still unavailable, partially due to the uncertainty of the relationship between CCAT2 and metastasis or prognosis implication. Several investigations established a statistically significant relationship between high CCAT2 expression and metastasis (LNM or DM). Nevertheless, some researchers demonstrated an unfavorable impact of CCAT2 abnormality on clinical metastasis. A comprehensive study is therefore demanded.
In the present study, we systematically explored the correlation between CCAT2 and clinic pathological features and prognosis in cancer. Eight eligible studies were summarized quantitatively based on our inclusion and quality assessment criteria. The findings of the current study demonstrated that high CCAT expression was positively correlated with the increase in the risk of LNM and DM in cancer patients. Moreover, we identified that there was a statistical significant positive relationship between CCAT expression and short OS or PFS in cancer patients. In multiple sensitivity analyses, we did not detect any substantial difference in pooled estimates, and there was no excessive influence on the overall results in any individual study.
Among seven studies assessed for the correlation between CCAT2 and OS, six studies used multivariate survival analysis to calculate the HRs and 95% CIs for OS [14-16, 20, 22, 23], and in the included studies for PFS [15, 20, 23], all papers used multivariate survival analysis, suggesting that SPRY4-IT1 expression could probably serve as an independent predictor for OS and PFS. In the included studies from present study, only one study from Huang SY et al. [16], reported the association between increased CCAT2 expression and disease free survival (DFS) in ovarian cancer, thus meta-analysis was not performed for DFS.
Our meta-analysis provided evidence that CCAT2 expression was positively correlated with the clinical metastatic factors (LNM and DM) in patients with various cancer types. Only two studies reported no association between increased CCAT2 expression and LNM in prostate cancer and bladder cancer, respectively, which might be caused by the small size or the tumor type [17, 23]. The mechanism on regulatory activity of CCAT2 in cancer invasion and metastasis has been explored in several cancer types. In a study by Ling et al. [10], cell invasion and metastasis could be promoted by CCAT2 via the MYC-regulated miRNA-17-5p and miRNA-20a. Another study investigated on the function of CCAT2, revealing that CCAT2 might disturb metastasis by regulating EMT-related gene expression (N-cadherin, vimentin and E-cadherin) [23]. Recently, Redis et al. [12] reported that CCAT2 regulates the alternative splicing of GLS, resulting in the preferential expression of the more aggressive splice isoform, which promotes in vivo metastases and in vitro cell migration. These molecular mechanisms of CCAT2 involved cancer progression could elucidate why CCAT2 may serve as a potential biomarker for predicting the patient prognosis.
It is important to explore the sources of heterogeneity in a meta-analysis. In this study, there was insignificant or low heterogeneity among studies analyzing connection between CCAT2 expression and OS or PFS or DM. However, moderate heterogeneity was found during the analyzing correlation between CCAT2 and LNM (I2=70%). Subgroup analyses were conducted according to the majors attributes of primary studies. In present study, different measures, such as reference controls and RNA extractions, were used to extract CCAT2 in different studies. All these variables may influence the heterogeneity. Subgroup analysis showed that reference controls did not change the overall result, but that the sample size, and RNA extractions did. The results showed that big sample size had higher accuracy than small sample size for predicting LNM.
There were also several potential limitations in our study. Firstly, a few clinical studies have focused on CCAT2 at present, and the sample size is therefore small. The data in our analyses were relatively insufficient, which may cause a small-study effect. Secondly, all eligible studies were from China in Asian area, with no Caucasian or African population involved, so the results warranted to be further validated in other population. Thirdly, the cut-off value of high and low CCAT2 expression varied in different studies. It was difficult to reach a same value. Fourthly, our meta-analysis may overestimate the significance of CCAT2 to some extent because studies with null results tend not to be published, even though no conclusive evidence supporting publication bias was observed via visual examination of funnel plot and the Begg test. Fifthly, other parameters that might have a great effect on survival, such as postsurgical treatment and biological types of carcinoma, were not explored, which may result in some heterogeneity.
In conclusion, the expression of CCAT2 was significantly associated with metastasis (LNM, DM) and prognosis (OS, PFS) in different types of cancer. The higher expression of CCAT2, the higher probability of occurrence of LNM and DM patients might suffer with. Meanwhile, shorter OS and PFS may be observed in the patients with high CCAT2 expression. Thus, CCAT2 might be a novel predictive factor for estimating the clinical outcome in different types of cancer. However, further large-scale studies are needed to confirm its value.
Supplementary Material
Table S1. Newcastle-Ottawa Scale for assessment of quality of in included case-control studies. Table S2. Search strategy used in PubMed.
Acknowledgements
This research was supported by the National Key Technology R&D Program (No.2014BAI09B05), the National Natural Science Foundation of China (No. 81572303) & Program of Shanghai Academic/ Technology Research Leader (No. 15XD1502600).
Author Contributions
TJ and HYC contributed to the design of the study. HYC, FLN, WYQ, LQQ collected the data and did the statistical analysis. TJ contributed to the writing of the manuscript. CYX, FJY and XH contributed to the revision of the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Sigova AA, Mullen AC, Molinie B. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110:2876-81
3. Xie C, Yuan J, Li H. et al. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98-103
4. Gomez-Maldonado L, Tiana M, Roche O. et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2015;34:2609-20
5. Zhang X, Gejman R, Mahta A. et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350-8
6. Hu Y, Wang J, Qian J. et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890-902
7. Liu XH, Sun M, Nie FQ. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92
8. Gutschner T, Hammerle M, Eissmann M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-9
9. Wang Y, Xue D, Li Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
10. Ling H, Spizzo R, Atlasi Y. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446-61
11. Tomlinson I, Webb E, Carvajal-Carmona L. et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984-8
12. Redis RS, Vela LE, Lu W. et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell. 2016;61:520-34
13. Cai Y, He J & Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther. 2015;8:2657-64
14. Chen S, Wu H, Lv N. et al. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. Biomed Pharmacother. 2016;82:583-8
15. Chen X, Liu L & Zhu W. Up-regulation of long non-coding RNA CCAT2 correlates with tumor metastasis and poor prognosis in cervical squamous cell cancer patients. Int J Clin Exp Pathol. 2015;8:13261-6
16. Huang S, Qing C, Huang Z. et al. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11:49
17. Li J, Zhuang C, Liu Y. et al. shRNA targeting long non-coding RNA CCAT2 controlled by tetracycline-inducible system inhibits progression of bladder cancer cells. Oncotarget. 2016;7:28989-97
18. Ouyang S, Zhang P, Wang J. et al. [Expression of long non-coding RNA colon cancer associated transcript 2 and its clinicopathologic significance in oral squamous cell carcinoma]. Zhonghua Kou Qiang Yi Xue Za Zhi. 2016;51:286-91
19. Qiu M, Xu Y, Yang X. et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375-80
20. Wang CY, Hua L, Yao KH. et al. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:779-85
21. Wang J, Qiu M, Xu Y. et al. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36:5523-8
22. Zhang X, Xu Y, He C. et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834-9
23. Zheng J, Zhao S, He X. et al. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2016
24. Zhou N, Si Z, Li T. et al. Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12:132-8
25. Tierney JF, Stewart LA, Ghersi D. et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
26. Parmar MK, Torri V & Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815-34
27. Higgins JP, Thompson SG, Deeks JJ. et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-60
28. DerSimonian R & Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-88
29. Ronchetti D, Manzoni M, Agnelli L. et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016;6:e468
30. Jiang CY, Gao Y, Wang XJ. et al. Long non-coding RNA lnc-MX1-1 is associated with poor clinical features and promotes cellular proliferation and invasiveness in prostate cancer. Biochem Biophys Res Commun. 2016;470:721-7
31. Ellinger J, Alam J, Rothenburg J. et al. The long non-coding RNA lnc-ZNF180-2 is a prognostic biomarker in patients with clear cell renal cell carcinoma. Am J Cancer Res. 2015;5:2799-807
32. Redis RS, Sieuwerts AM, Look MP. et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748-62
Author contact
![]() Corresponding author: Ying-Xuan Chen; Email: yingxuanchen71edu.cn
Corresponding author: Ying-Xuan Chen; Email: yingxuanchen71edu.cn

 Global reach, higher impact
Global reach, higher impact