Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(9):1542-1551. doi:10.7150/jca.18680 This issue Cite
Research Paper
MicroRNA-765 Enhances the Anti-Angiogenic Effect of CDDP via APE1 in Osteosarcoma
1. Cancer Center, Daping Hospital and Research Institute of Surgery, Third Military Medical University, Chongqing,400042, China;
2. Department of Diagnostic and Therapeutic Ultrasonography, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center of Cancer, Key laboratory of Cancer Prevention and Therapy, Tianjin, China.
* These authors Wei Liang and Xi Wei contributed equally to this work.
Received 2016-12-9; Accepted 2017-2-26; Published 2017-6-2
Abstract
Human osteosarcoma (HOS) is the most common malignancy in children and adolescents and has a heterogeneous presentation and high mortality. Previous studies have shown that microRNAs contribute to RNA silencing and post-transcriptional regulation of gene expression. Here, we showed that significantly increased expression of miR-765 with or without CDDP (Cisplatin) down-regulates APE1 expression and angiogenesis-related markers (VEGF, FGF2, TGFβ, and CD34). Further investigation showed that miR-765 modulates osteosarcoma cell migration and angiogenesis following treatment with cisplatin in vitro and in vivo. MiR-765 increases the anti-angiogenic effect of CDDP in human osteosarcoma. Elucidation of the mechanism of the miR-765-APE1 axis in tumor progression of HOS will be beneficial in identifying biomarkers and therapeutic target of osteosarcoma.
Keywords: MicroRNA-765, APE1, Cisplatin, Anti-angiogenesis, Osteosarcoma.
Introduction
Human osteosarcoma (HOS) is the most common malignancy in children and adolescents and has a heterogeneous presentation and high mortality [1]. HOS pathogenesis involves gene alterations caused by genetic instability and DNA damage repair [2]. Apurinic/apyrimidinic endonuclease1 (APE1) has a dual function in DNA repair and redox gene regulation and maintains DNA binding activity through several cellular pathways [3]. Previous studies have shown that APE1 expression is increased in the variety of cancers, including osteosarcoma, gastric cancer, lung cancer and prostate cancer, and is linked to poor prognosis partly due to resistance to chemotherapeutic agents, such as cisplatin or methyl methanesulfonate [4-6]. APE1 has a pivotal role in DNA repair processes and modulates the base excision repair (BER) pathway, influencing the sensitivity to chemotherapy in tumor cells [3]. Wang et al. [6] reported that inhibition of APE1 enhances the antitumor efficacy of cisplatin in non-small-cell lung cancer. Other researchers showed that an APE1 inhibitor suppressed epithelial-mesenchymal transition (EMT) in lung cancer [7]. Studies have shown that APE1 and osteosarcoma angiogenesis is closely related, and can regulate the angiogenesis of osteosarcoma by regulating TGFβ or FGF2 [8, 9].Thus, we hypothesized that APE1 is involved in apoptosis, angiogenesis, and other biological activities.
MicroRNAs (miRNAs) are single-stranded non-coding RNA molecules containing 18-25 nucleotides that regulate the variety of genes during multiple eukaryotic processes [10]. By binding to the 3' untranslated region (3'UTR) of target genes, miRNAs mediate differentiation, apoptosis and proliferation by inhibiting mRNA translation of related genes [11]. Based on our previous results showing that the miRNA expression profile was altered after transfection with APE1 siRNA in human osteosarcoma, we investigated the role of the miR-765-APE1 axis in the regulation of osteosarcoma following cisplatin treatment [12]. There are currently reported in the literature that cisplatin-induced resistance is mediated by miR-21-3p in ovarian cells [13], and miRNA-21 increases cisplatin sensitivity of osteosarcoma by repressing the expression of Sprouty2 [14]. In addition, miR-765 as a regulatory point in the treatment of prostate cancer patients after treatment of fulvestrant, the impact of prostate cancer generation, migration and invasive ability [15]. However, there is a litter study regarding the association between anti-angiogenic effect of cisplatin and miR-765.
To explore the correlation of APE1 and miRNA, our previous study examined knockdown of APE1 in bone sarcoma HOS cells and differentially expressed miRNAs using high-throughput miRNA chip technology in our laboratory. The results showed that (1) miR-765 was significantly increased (enhanced>5 times); (2) bioinformatics prediction identified miR-765 binding sites in the APE1 3'UTR, and this miRNA may regulate APE1. Clinical osteosarcoma samples showing increased miR-765 expression demonstrated APE1 protein suppression using immunohistological staining. Therefore, we hypothesized that miR-765 regulation of the APE1 pathway in human osteosarcoma mediates cisplatin treatment.
Results
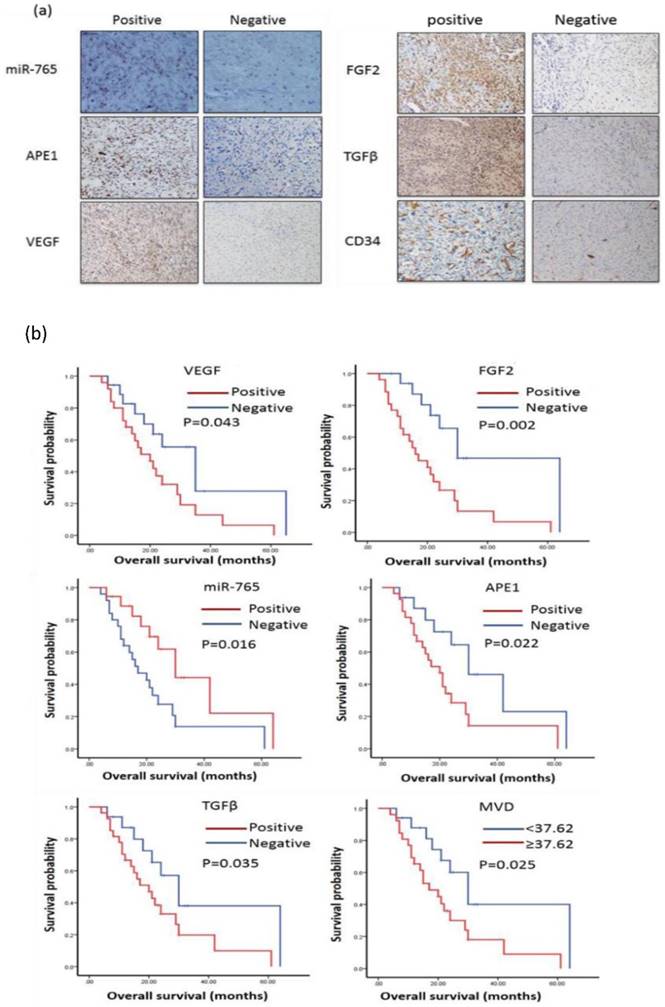
MicroRNA-765 expression is negatively correlated with the expressions of APE1, VEGF, FGF2, TGFβ and CD34 in osteosarcoma tissues
MiR-765 and APE1, VEGF, FGF2, TGFβ, CD34 in order to investigate potential relevance in patients with osteosarcoma by 43 cases (Table 1) of osteosarcoma tissue for immunohistochemistry and in situ hybridization to complete. Images of ISH and IHC staining positive and negative for miR-765, APE1, VEGF, FGF2, TGFB, CD34 expression in osteosarcoma patients are shown in Figure 1. Spearman's rank correlation analysis showed that the expressions of miR-765 and APE1, VEGF, FGF2, TGFβ, CD34 were negatively correlated (P<0.05, Table 2).
MicroRNA-765 expression is associated with good survival in patients with osteosarcoma
The survival analyses showed that 43 cases with positive miR-765 expression had good overall survival by Kaplan-Meier analysis. (MST: 30 m vs. 17 m, P=0.016) (Table 3, Figure 1 b). Additionally, we also found that cases with increased expressions of APE1, VEGF, FGF2, TGFβ and CD34 had poor prognosis (APE1 MST: 30 m vs. 20 m, P=0.022, VEGF MST: 35 m vs. 20 m, P=0.043, FGF2 MST: 30 m vs. 16 m, P=0.002, TGFβ MST: 30 m vs. 20 m, P=0.035 and CD34 MST: 30 m vs. 17 m, P=0.025, respectively) (Table 3, Figure 1 b). Thus, increased miR-765 expression is associated with better outcomes of osteosarcoma patients, indicating that it may be a potential marker or therapeutic target in clinical settings.
The basic characteristics of patients with osteosarcoma
| Parameters | No. of Patients | Percentage (%) |
|---|---|---|
| Age (Range) | 33 (11-77) | |
| ≤33 | 22 | 51.2 |
| >33 | 21 | 48.8 |
| Gender | ||
| Male | 22 | 51.2 |
| Female | 21 | 48.8 |
| TNM Stages | ||
| I~II | 22 | 51.2 |
| III~IV | 21 | 48.8 |
| Histology | ||
| Poor | 10 | 23.3 |
| Moderate | 18 | 41.9 |
| Well | 15 | 34.9 |
Relationship between the miR-765 expression and APE1, VEGF, FGF2, TGFB, MVD in OS patients.
| Variable | MiR-765 | r2 value | P value | ||
|---|---|---|---|---|---|
| Positive | negative | Total | |||
| APE1 | |||||
| Positive | 2 | 24 | 26 | -0.953 | 0.000 |
| negative | 16 | 1 | 17 | ||
| VEGF | |||||
| Positive | 3 | 23 | 26 | -0.809 | 0.000 |
| negative | 15 | 2 | 17 | ||
| FGF2 | |||||
| Positive | 3 | 22 | 25 | -0.857 | 0.000 |
| negative | 15 | 3 | 18 | ||
| TGFB | |||||
| Positive | 1 | 23 | 24 | -0.907 | 0.000 |
| negative | 17 | 2 | 19 | ||
| MVD | |||||
| ≥37.62 | 4 | 22 | 26 | -0.811 | 0.000 |
| <37.62 | 15 | 2 | 17 | ||
| Total | 18 | 25 | |||
MicroRNA-765 in combination with cisplatin inhibits expressions of APE1, VEGF, FGF2, and TGFβ in osteosarcoma cells
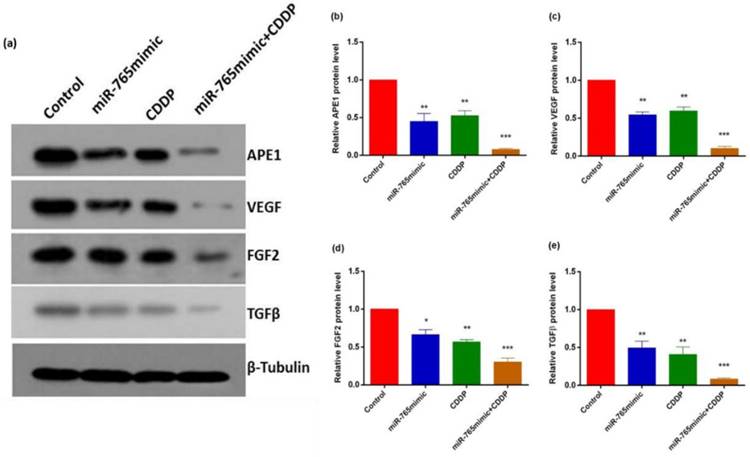
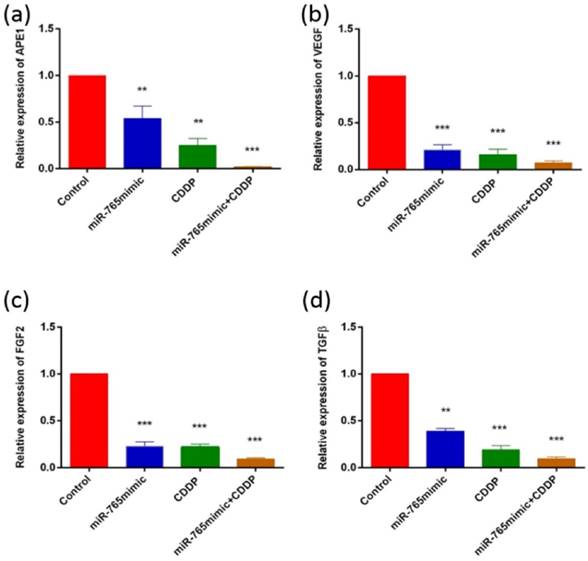
To assess the role of miR-765 in the regulation of various proteins following cisplatin treatment in 9901 cells by Western blot and RT-PCR, we measured the levels of APE1, VEGF, FGF2 and TGFβ and showed that they decreased after transfection with miR-765mimic and treatment with cisplatin compared to the control group. As shown in Fig. 2 and 3, in 9901 cell lines, the protein and RNA expression level of APE1, VEGF, FGF2 and TGFβ in miR-765mimic transfected and cisplatin-treated group was significantly lower than that in miR-765mimic group, simple cisplatin treatment group and blank control group (P < 0.001). The above experiments show that miR-765mimic can significantly inhibit the expression of APE1, so we propose that miR-765 through APE1 can enhance the anti-angiogenic effect of cisplatin in osteosarcoma.
Association between the expressions of miR-765, APE1, anti-angiogenesis markers and overall survival in osteosarcoma patients
| Variable | N (%) | Median OS (months) | P-value |
|---|---|---|---|
| MiR-765 | |||
| Negative | 25(58.1) | 17.0 | 0.016* |
| Positive | 18(41.9) | 30.0 | |
| APE1 | |||
| Negative | 17(39.5) | 30.0 | 0.022* |
| Positive | 26(60.5) | 20.0 | |
| VEGF | |||
| Negative | 17(39.5) | 35.0 | 0.043* |
| Positive | 26(60.5) | 20.0 | |
| TGFβ | |||
| Negative | 19(44.2) | 30.0 | 0.035* |
| Positive | 24(55.8) | 20.0 | |
| FGF2 | |||
| Negative | 18(41.9) | 30.0 | 0.002* |
| Positive | 25(58.1) | 16.0 | |
| MVD | |||
| <37.62 | 17(39.5) | 30.0 | 0.025* |
| ≥37.62 | 26(60.5) | 17.0 |
*p<0.05
The combination of microRNA-765 and cisplatin suppresses the capability of HUVEC migration and tube formation of tumor cells
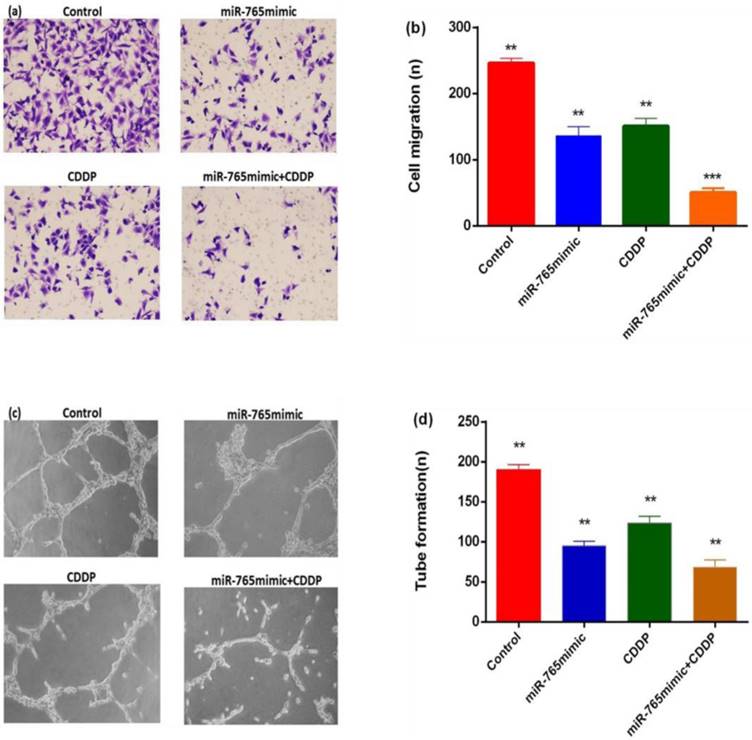
To explore the effects of miR-765 combined with cisplatin in inhibiting tumor angiogenesis, transwell migration and tube formation assays were performed. As shown in Figure 4 a and b, migrations of HUVEC in the miR-765mimic and cisplatin treatment group was significantly less than in the blank control group, with a reduction level of 79.6% (P < 0.001). Secondly, the formation of capillary tube showed that the capillary-like structure formed in miR-765mimic combined with cisplatin treatment group was less than other groups (Figure 4 c and d, P < 0. 01), indicating that miR-765 inhibits HUVEC migration and tube formation in vitro.
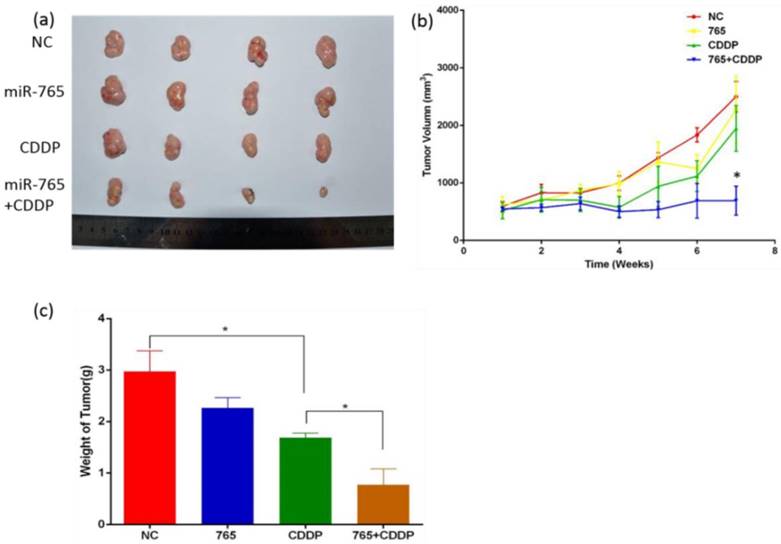
Synergetic treatment of microRNA-765 and CDDP reduces xenograft tumor angiogenesis in vivo
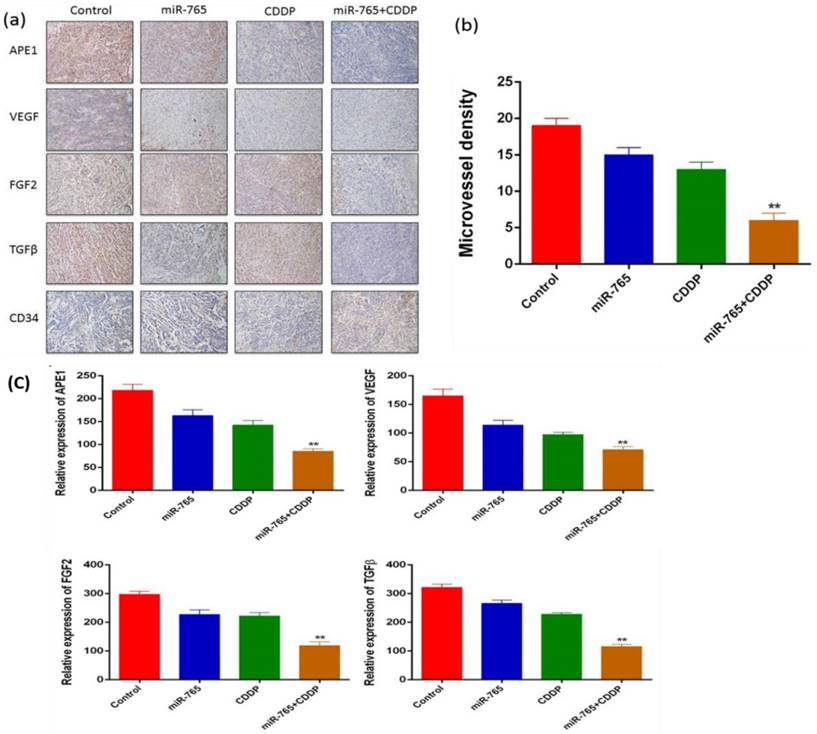
We established 9901 xenograft mice to assess the effects of miR-765 and cisplatin combined treatment in suppression of angiogenesis in vivo. The mice were treated with vehicle alone, miR-765, CDDP plus miR-765 agomir, and CDDP plus scramble agomir. The xenograft tumor size was measured; as time increased, the group treated with miR-765 agomir + CDDP showed inhibited tumor growth compared to the other groups (P<0.05). Additionally, the xenograft tumor weight in the miR-765 agomir + CDDP group was lower than the other groups (P<0.05) (Figure 5 b and c). After the mice were sacrificed, tumor sections were analyzed using immunohistochemistry with a variety of antibodies (Figure 6 a). As expected, the expression levels of APE1, VEGF, FGF2, TGFβ, and CD34 were significantly lower in the miR-765 agomir plus CDDP treatment group (63.6%, 50.9%, 59.7% and 62.9%, respectively) compared to the control group (Figure 6 c) (P<0.05). The density of CD34-positive vessels stained by anti-CD34 antibody was much lower in tumors with miR-765 agomir plus CDDP treatment than that in the control (P<0.05) (Figure 6 b). These results suggest that miR-765 in combination with cisplatin inhibits tumor angiogenesis and growth in vivo by down-regulating APE1, VEGF, FGF2 and TGFβ.
Discussion
MicroRNAs have roles in RNA silencing and post-transcriptional regulation of gene expression. MiRNA profile-based screening assays have been performed in early-stage colorectal cancer and human osteosarcoma to identify their targeted genes for analysis of patient prognoses [16, 17]. Our previous study identified seven up-regulated and six down-regulated miRNAs after analysis of APE1 expression in osteosarcoma [12]. One of up-regulated miRNAs, hsa-miR-765, was selected from the microarray and confirmed by RT-PCR, followed by analysis with bioinformatics methods in APE1 knockdown HOS cells [12]. A combination of surgery and chemotherapy is the current treatment for osteosarcoma, which is most common primary bone malignancy [2]. Resistance to the standard chemotherapies, such as methotrexate, doxorubicin, and cisplatin, is a major challenge in the treatment of osteosarcoma patients [1]. Thus, in this study, we investigated the impact of miR-765 on the anti-angiogenic effect of cisplatin. Several studies have shown the impact of miR-21 on sensitivity of osteosarcoma-derived cells towards chemotherapies [18]. MicroRNAs are critical oncomiRs in tumorigenesis, and their expression has been shown to be up-regulated in breast carcinoma [19]. Our previous study found increased oncomiRs in HOS cells, and these were involved in APE1 pathways related to carcinoma developmental processes, regulation of cellular processes and signaling in cancers [12].
(a) miR-765 expression was negatively correlated with the expressions of APE1, VEGF, FGF2, TGFβ and CD34 in osteosarcoma patient tissues. The samples from patients with osteosarcoma were stained using immunohistochemistry. MiR-765 was inversely correlated with APE1, VEGF, FGF2, TGFβ and CD34 in patients with osteosarcoma tissues. (b) miR-765 positive expression was associated with good prognosis in patients with osteosarcoma by Kaplan-Meier analysis. (MST: 30 m vs. 17 m, P=0.016). APE1 expression was associated with poor prognosis in patients with osteosarcoma by Kaplan-Meier analysis (MST: 30 m vs. 20 m, P=0.022). VEGF expression was associated with poor prognosis in patients with osteosarcoma by Kaplan-Meier analysis. (MST: 35 m vs. 20 m, P=0.043). FGF2 expression was associated with poor prognosis in patients with osteosarcoma by Kaplan-Meier analysis. (MST: 30 m vs. 16 m, P=0.002). TGFβ expression was associated with poor prognosis in patients with osteosarcoma by Kaplan-Meier analysis (MST: 30 m vs. 20 m, P=0.035). The Kaplan-Meier plot showed that CD34 positive expression was associated with poor prognosis in patients with osteosarcoma (MST: 30 m vs. 17 m, P=0.025). MiR-765 expression was associated with good survival, but increased APE1, VEGF, FGF2, TGFβ and CD34 expression was associated with poor prognosis in patients with osteosarcoma.
Increased miR-765 can down-regulate APE1 and angiogenesis-related proteins (VEGF, FGF2 and TGFβ). (a) The APE1, VEGF, FGF2 and TGFβ markers were detected in 9901 cells following incubation with miR-765 mimics, CDDP and miR-765 mimics + CDDP for 48 hours by western blot. (b) Quantitative analysis of western blot. **P < 0.01, ***P < 0.001.
The results of RT-PCR analysis. APE1 (a), VEGF (b), FGF2 (c) and TGFβ (d) expressions were measured by RT-PCR in 9901 cells treated with miR-765 mimics, CDDP and miR-765 mimics + CDDP. **P < 0.01, ***P < 0.001.
MiR-765 mimics and CDDP inhibited the capability of osteosarcoma cells to promote HUVEC migration and tube formation. (a) Typical images of migration. Culture medium from 9901 cells transfected with miR-765 mimics and CDDP results in fewer migrations compared to culture medium from the control group, and the HUVECs in 9901 cells treated with miR-765 mimics +CDDP showed enhanced inhibition of migration. (b) Quantitative analysis of migration. (c) Typical images of tube formation assays. Culture medium of HUVECs transfected with miR-765 mimics and CDDP produced fewer capillary-like structures than culture medium from the control group, and the HUVECs in 9901cells treated with miR-765 mimics +CDDP showed increase inhibition of tube formation. (d) Quantitative analysis of tube formation. **P < 0.01, ***P < 0.001.
In previous studies we found that miR-765 and APE1 have targeted binding sites, miR-765 can inhibit the DNA damage and redox functions of APE1 and furthermore influencing the APE1 regulated chemotherapy sensitivity in osteosarcoma. Based on our data, we hypothesized that miR-765 suppresses the expression of APE1, VEGF, FGF2 and TGFβ in osteosarcoma cells after treatment with cisplatin. Furthermore, the results showed that a combination of miR-765 and cisplatin inhibited osteosarcoma cell migration and microvascular tube formation, indicating that miR-765 enhanced the anti-angiogenic effect of cisplatin in osteosarcoma. Using in vivo experiments, we showed that synergistic treatment of miR-765 and CDDP significantly reduced tumor volume and expressions of APE1, VEGF, FGF2 and TGFβ in vivo. To detect the anti-angiogenic effects on xenograft tumors in vivo, we stained the samples for CD34 expression and revealed that CD34 was lower in tumors compared to the control groups following exposure to a combination of miR-765 and CDDP.
An APE1 inhibitor, AT101, was used to enhance 5-Fu treatment in non-small cell lung cancer in one study [20]. APE1 expression regulation has been shown to be involved in treatment of various cancers [5, 6, 20, 21]. APE1 down-regulation increased chemotherapeutic sensitivity of cancer cells [20, 21]. Other studies indicated that APE1 levels influenced the sensitivity to radiation and chemotherapy in cancer treatment strategies [20, 22]. The dual function of APE1 in DNA repair, and redox has been explored as a mechanistic target of APE1 inhibitors [23]. However, the role of the oncomiR-APE1 axis in treatment of and the effects of microRNA and CDDP combined treatment osteosarcoma has been poorly studied. Thus, our report results provide strong evidence that miR-765 up-regulation enhanced the anti-tumor effect of cisplatin treatment of human osteosarcoma.
In conclusion, exogenous miR-765 induced APE1, VEGF, FGF2 and TGFβ expressions in osteosarcoma cells. Further investigation showed that miR-765 modulates osteosarcoma cell migration and angiogenesis in vitro and in vivo following treatment of cisplatin. The study of the miR-765-APE1 mechanism in tumor progression of HOS will help to identify biomarkers and therapeutic targets of osteosarcoma.
Materials and Methods
Clinical cases
The 43 osteosarcoma patients were treated in Daping Hospital (Chongqing, China) between 2009 and 2013. We assessed the content of the tumor samples by hematoxylin and eosin stain and only evaluated tumor tissue samples containing more than 60%. All of these samples were used after surgery and were fixed in 10% formalin immediately and then stored at room temperature for approximately 24 hours. Tumor samples were fixed, dehydrated and incubated in xylene and then underwent paraffin infiltration and were finally embedded in paraffin. The study was approved by the Ethics Committee of Daping Hospital and Research Institute of Surgery.
Cell lines and cell culture
The 9901 cells were donated by Prof. Qingyu Fan (Fourth Military Medical University, Xian, China) and cultured in RPMI-1640 (HyClone Laboratories Inc., Utah, USA) with 10% FBS. HUVECs (ATCC, Manassas, VA, USA) were grown in DMEM (HyClone) with 10% FBS.
The role of miR-765 and CDDP in the treatment of osteosarcoma in vivo. (a) Xenograft tumor morphology decreased after treatment with miR-765 agomir, CDDP and miR-765 agomir + CDDP compared to the control group. (b) The graph of xenograft tumor size:as time increased, the group treated with miR-765 agomir + CDDP showed inhibited tumor growth compared to the other groups (P<0.05). (c) The graph of xenograft tumor weight: xenograft tumor weight in the group treated with miR-765 agomir + CDDP was lower than the other groups (P<0.05). **P < 0.01.
(a) Immunohistochemical analysis of APE1, VEGF, FGF2, TGFβ, and CD34 in a xenograft mouse model (magnification, ×100); (b) Microvessel density of control and miR-765 agomir, CDDP and miR-765 agomir + CDDP groups. The MVD was significantly lower in the miR-765 agomir + CDDP group compared with the control group (61.1%, P<0.01). (c) As expected, the expression levels of APE1, VEGF, FGF2, TGFβ, and CD34 were significantly lower in the miR-765 agomir plus CDDP treatment group (63.6%, 50.9%, 59.7% and 62.9%, respectively) compared to the control group, **P < 0.01.
RT-PCR analysis
Total RNA was isolated from cells and fresh tissues using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The primer sequences for the genes are as follows:
APE1 forward, 5'-CCGAATTCATGCCGAAGCGTGGGA-3'; reverse, 5'-TCGAGTCACAGTGCTAGGTATAG-3';
VEGF forward: 5'-GCTACTGCCATCCAATCGAG-3'; reverse: 5'-GGTTTGATCCGCATAATCTGCAT-3';
FGF2 forward: 5'-AGAAGAGCGACCCTCACATCA-3'; reverse: 5'-CGGTTAGCACACACTCCTTTG-3';
TGFβ, forward, 5'-CCAAGCTTATGCCGCCCTCCGGGC-3'; reverse, 5'-GCGTCGACCAGCTGCACTTGCAGGAG-3';
GAPDH forward, 5'-GCAGGGGGGAGCCAAAAGGGT-3'; reverse, 5'-TGGGTGGCAGTGATGGCATGG-3'.
Mature miR-765 and the RNU6 endogenous control were analyzed using the TaqMan microRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). The relative expression of miR-765 was normalized against RNU6 expression using the 2-△Ct method.
Immunohistochemical analysis
All immunohistochemical staining was performed in accordance with standard operating procedures. Tumor tissue sections (RM2235; Leica, Solms, Germany) were 4.5 µm per slide. After addition of the primary antibody, 3, 3'-diaminobenzidine was used as a chromogenic substrate with hematoxylin for counterstaining. The results for APE1, VEGF, FGF2 and TGFβ staining were scored based on the percentage of positively stained cells; scores 0 and 1 were categorized as negative expression, and scores>2 were categorized as positive expression [24] (no positive cells, score 0; ≤10% positive cells, score 1; 11%-25% positive cells, score 2; 26%-50% positive cells, score 3; and ≥51% positive cells, score 4). MVD was defined as all CD34-positive endothelial cells separate from nearby microvessels. Staining analysis was performed under the same conditions by two independent experts.
Transwell migration, Matrigel tube formation assay
The transwell migration assay was performed in 24-well plates, and each transwell chamber had an aperture of 8 µm (BD Biosciences, San Diego, CA, USA). Serum-free DMEM media containing 20000 HUVECs were seeded into the upper chamber. The lower compartment was filled with culture medium that contained 9901 cells treated with OptiMEM I medium, blank control, CDDP, miR-765 mimic and miR-765 mimic + CDDP. After 20 hours of incubation, the migrated cells were counted from four randomly stained areas after they were fixed with formaldehyde, treated with crystal violet, and assessed at 200 × magnification by an optical microscope. The Matrigel tube formation assay, supernatant as indicated above was mixed with 20 µL DMEM containing 20000 HUVECs then transferred to the 24-well plate on the Matrigel matrix separately, followed by incubation at 37°C, 5% CO2, for 12 h. And then count the tube formation at 200 × magnification by an optical microscope.
MicroRNA/vector transfections
MiR-765 mimics and its matched miR-NC and agomir-765 and the matched scramble NC were obtained from RiboBio Co., Ltd (Guangzhou, China). Cisplatin was purchased from Sigma-Aldrich (US). Transient transfections of miRNA mimics (RiboBio) were carried out using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol; cells were plated for 24 h without antibiotics before the transfections. All miRNA transfections were for 72 hours.
Western Blot
Western blotting protocol was performed as described previously [25]. Primary antibodies included the mouse monoclonal antibodies anti-APE1 (1:5000), VEGF (1:500), FGF2 (1:1000), TGFβ (1:50) and anti-β-Actin (1:2000), all of the primary antibodies were purchased from Abcam. Visualization was performed using Bio-Rad ChemiDocTM XRS system (Hercules, CA, USA) with enhanced-chemiluminescence substrate and the blots were analyzed using Image Lab 3.0 (BioRad, Hercules, CA, USA). Protein level was normalized to the matching densitometry values of the internal control β-actin.
In situ hybridization (ISH) and analysis
The cellular expression of miR-765 was determined by in situ hybridization, according to the manufacturer's protocol. The 3', 5' DIG-labeled LNA miR-765 antisense probe and LNA scramble control probe were purchased from Exiqo (USA). The DIG blocking buffer and anti-DIG antibody (1:1000) were obtain from Roche (Germany). Scoring was measured by cytoplasmic staining. Based on the scores, patients were further categorized into positive (scores 2 and 3) and negative (scores 0 and 1) expression groups according to ROC curve analysis.
Animal xenograft models
Four groups (n=4 per group) were established with 4-to-6 weeks old BALB/c nude mice. The 9901 cells were cultured at 90% confluence to prepare a cell suspension with 2.0x106 cells/100 μl. After subcutaneously inoculating 9901 cells in the left anterior axilla of nude mice, the mice were injected with agomir in the tumor after 8 days (1 nmol each time, once every 4 days, a total of 8 times). When the tumor size was 10 mm, the mice were injected with cisplatin (CDDP, 4 mg/kg) 1 time every 2 days, 3 times total. The tumor size and weight were measured every 4 days.
Follow-up
Patients were followed up after surgery. The OS was determined from the patient records of survival time. The length of survival was later calculated from the date of surgery until either the time of death or the end of follow-up. The survival data in this study were censored on August 30, 2016.
Ethical approval and informed consent
This study was approved by the Ethics Committee of Daping Hospital and it was in accordance with the principles of the Helsinki Declaration. All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by a named institutional and/or licensing committee. The informed consent was obtained from all subjects.
Statistical analysis
All the data are expressed as the mean ± S.E. Statistical analysis was calculated by ANOVA, χ2 test, or Student's t-test using SPSS 21.0 software. Statistical significance was set at P < 0.05 (*).
Acknowledgements
This work was supported by grants from the National Natural Science Foundations of China (No.81172117). We thank all the people and patients who participated in this study.
Author contributions
W.L, Z-Y.Z and Q.L conceived and designed this study. X.W and W.L were responsible for searching the clinic database. N.D, M-X.L, C-Y.L, J.X., Y.D, X-R.T, X-Y.D and Q.L were jointly involving in western bolt, RT-PCR and animal experiment. C-X.X and D.W finished the Statistical analysis, W.L, Z-Y.Z, X.W and Q.L writing the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015Sep20;33(27):3029-35
2. Moore DD, Luu HH. Osteosarcoma. Cancer treatment and research. 2014;162:65-92
3. Lirussi L, Antoniali G, Vascotto C, D'Ambrosio C, Poletto M, Romanello M. et al. Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Molecular biology of the cell. 2012Oct;23(20):4079-96
4. Qing Y, Li Q, Ren T, Xia W, Peng Y, Liu GL. et al. Upregulation of PD-L1 and APE1 is associated with tumorigenesis and poor prognosis of gastric cancer. Drug design, development and therapy. 2015;9:901-9
5. Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Molecular cancer therapeutics. 2004Jun;3(6):679-86
6. Wang D, Xiang DB, Yang XQ, Chen LS, Li MX, Zhong ZY. et al. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung cancer (Amsterdam, Netherlands). 2009Dec;66(3):298-304
7. Wei X, Li Q, Li Y, Duan W, Huang C, Zheng X. et al. Prediction of survival prognosis of non-small cell lung cancer by APE1 through regulation of Epithelial-Mesenchymal Transition. Oncotarget. 2016May10;7(19):28523-39
8. Ren T, Qing Y, Dai N, Li M, Qian C, Yang Y. et al. Apurinic/apyrimidinic endonuclease 1 induced upregulation of fibroblast growth factor 2 and its receptor 3 induces angiogenesis in human osteosarcoma cells. Cancer science. 2014Feb;105(2):186-94
9. Jiang X, Shan J, Dai N, Zhong Z, Qing Y, Yang Y. et al. Apurinic/apyrimidinic endonuclease 1 regulates angiogenesis in a transforming growth factor beta-dependent manner in human osteosarcoma. Cancer science. 2015Oct;106(10):1394-401
10. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Advances in experimental medicine and biology. 2013;774:1-20
11. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Current genomics. 2010Nov;11(7):537-61
12. Dai N, Zhong ZY, Cun YP, Qing Y, Chen C, Jiang P. et al. Alteration of the microRNA expression profile in human osteosarcoma cells transfected with APE1 siRNA. Neoplasma. 2013;60(4):384-94
13. Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecologic oncology. 2015Apr;137(1):143-51
14. Ziyan W, Yang L. MicroRNA-21 regulates the sensitivity to cisplatin in a human osteosarcoma cell line. Irish journal of medical science. 2016Feb;185(1):85-91
15. Leung YK, Chan QK, Ng CF, Ma FM, Tse HM, To KF. et al. Hsa-miRNA-765 as a key mediator for inhibiting growth, migration and invasion in fulvestrant-treated prostate cancer. PloS one. 2014;9(5):e98037
16. Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M. et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5-6):397-402
17. Lim HJ, Yang JL. Regulatory roles and therapeutic potential of microRNA in sarcoma. Critical reviews in oncology/hematology. 2016Jan;97:118-30
18. Vanas V, Haigl B, Stockhammer V, Sutterluty-Fall H. MicroRNA-21 Increases Proliferation and Cisplatin Sensitivity of Osteosarcoma-Derived Cells. PloS one. 2016;11(8):e0161023
19. Hong XC, Fen YJ, Yan GC, Hong H, Yan CH, Bing LW. et al. Epithelial membrane protein 3 functions as an oncogene and is regulated by microRNA-765 in primary breast carcinoma. Molecular medicine reports. 2015Nov;12(5):6445-50
20. Wei X, Duan W, Li Y, Zhang S, Xin X, Sun L. et al. AT101 exerts a synergetic efficacy in gastric cancer patients with 5-FU based treatment through promoting apoptosis and autophagy. Oncotarget. 2016 Apr 30
21. Xiang DB, Chen ZT, Wang D, Li MX, Xie JY, Zhang YS. et al. Chimeric adenoviral vector Ad5/F35-mediated APE1 siRNA enhances sensitivity of human colorectal cancer cells to radiotherapy in vitro and in vivo. Cancer gene therapy. 2008Oct;15(10):625-35
22. Wang D, Zhong ZY, Li MX, Xiang DB, Li ZP. Vector-based Ape1 small interfering RNA enhances the sensitivity of human osteosarcoma cells to endostatin in vivo. Cancer science. 2007Dec;98(12):1993-2001
23. Ren T, Shan J, Li M, Qing Y, Qian C, Wang G. et al. Small-molecule BH3 mimetic and pan-Bcl-2 inhibitor AT-101 enhances the antitumor efficacy of cisplatin through inhibition of APE1 repair and redox activity in non-small-cell lung cancer. Drug design, development and therapy. 2015;9:2887-910
24. Wei X, Li Y, Zhang S, Ming G. Evaluation of thyroid cancer in Chinese females with breast cancer by vascular endothelial growth factor (VEGF), microvessel density, and contrast-enhanced ultrasound (CEUS). Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014Jul;35(7):6521-9
25. Gu X, Cun Y, Li M, Qing Y, Jin F, Zhong Z. et al. Human apurinic/apyrimidinic endonuclease siRNA inhibits the angiogenesis induced by X-ray irradiation in lung cancer cells. International journal of medical sciences. 2013;10(7):870-82
Author contact
![]() Corresponding author: Professor. Zhao-Yang Zhong MD. PhD. Email: zhongzhaoyang08com; Tel +86-23-68757183; Fax +86-23-68894062
Corresponding author: Professor. Zhao-Yang Zhong MD. PhD. Email: zhongzhaoyang08com; Tel +86-23-68757183; Fax +86-23-68894062

 Global reach, higher impact
Global reach, higher impact