Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(9):1619-1628. doi:10.7150/jca.18778 This issue Cite
Review
The Emerging Roles of Forkhead Box (FOX) Proteins in Osteosarcoma
1. Department of Orthopaedics, Xi'an Hong-Hui Hospital affiliated to medical college of Xi'an Jiaotong University, Xi'an, Shaanxi, China, 710054;
2. Department of Dermatology, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
* These authors contributed equally to this work.
Received 2016-12-15; Accepted 2017-2-27; Published 2017-6-3
Abstract
Osteosarcoma is the most common bone cancer primarily occurring in children and young adults. Over the past few years, the deregulation of a superfamily transcription factors, known as forkhead box (FOX) proteins, has been demonstrated to contribute to the pathogenesis of osteosarcoma. Molecular mechanism studies have demonstrated that FOX family proteins participate in a variety of signaling pathways and that their expression can be regulated by multiple factors. The dysfunction of FOX genes can alter osteosarcoma cell differentiation, metastasis and progression. In this review, we summarized the evidence that FOX genes play direct or indirect roles in the development and progression of osteosarcoma, and evaluated the emerging role of FOX proteins as targets for therapeutic intervention.
Keywords: forkhead box protein, oncogene, tumor suppressor, osteosarcoma, therapy.
Introduction: Overview of FOX Proteins in Cancer
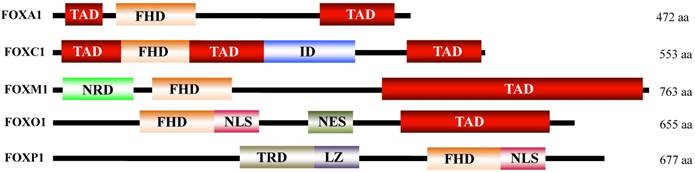
Forkhead box (FOX) proteins are a large family of transcription factors with important functions in multiple biological processes, such as cell cycle control, cell differentiation, proliferation and development [1-4]. Hundreds of FOX genes have been identified in mammals, including 50 members of the human genome and 44 members of the mouse genome [5]. FOX proteins are defined based on the forkhead box (also known as FKH domain), a DNA binding motif containing 80 to 100 amino acids (as shown in Figure 1) [1-4]. Based on similarities in the FKH domain, FOX proteins can be divided into 19 subclasses from FOXA to FOXS [1, 5]. Although FOX proteins share structural similarities in DNA binding motifs, these proteins significantly differ in terms of expression pattern, regulation and function [6].
Genetic analyses indicated that many of these FOX genes are differentially expressed in a variety of diseases, including cancer [1-8]. Several FOX subfamilies, such as FOXA, FOXC, FOXM, FOXO and FOXP, are associated with tumorigenesis and cancer progression, playing roles as either oncogenes or tumor suppressors, depending on the family member and cell type [1, 2, 9]. Three FOX subfamilies, including FOXA, FOXM and FOXO, have been well studied for their functions in cancer [1-9]. Expression of FOXA1 is aberrant in a number of cancer types such as acute myeloid leukemia (AML) [10, 11] lung [12], breast and prostate cancers [10, 13, 14], in which FOXA1 primarily functions as a critical transcription factor to regulate expression of the nuclear hormone receptors, estrogen receptor-α (ER), and androgen receptor (AR) [10, 15]. Most recently, the results from chromatin immunoprecipitation sequencing (ChIP-Seq) revealed that the genes targeted by FOXA1 are most commonly expressed in liver, prostate and breast cancer cells [16]. In addition, cell-specific FOXA1 regulation was also demonstrated that it contributed to unique FOXA1 binding, genetic variations, and/or potential epigenetic regulation [16]. FOXM1 is a proto-oncogene, and its upregulation has been observed in many human cancers, including lung [17], liver [18], prostate [19], breast [20], and pancreatic cancers [21]. The upregulation of FOXM1 promotes tumorigenesis through multiple mechanisms, including the expansion of stem/progenitor cells, modulation of the epigenome, induction of genomic instability, regulation of cell proliferation, progression of the cell cycle and differentiation of cells [17-23]. In mammals, there are four FOXO genes, FOXO1, 3, 4, and 6, which play important roles as tumor suppressors in a variety of cancers through diverse mechanisms, such as initiating apoptosis, prompting cell cycle arrest, and regulating the expression of genes that facilitate DNA repair [24-26]. Members of FOXOs are also regulated by the insulin/PI3K/AKT (phosphorylated by phosphatidylinositol 3-kinase/RAC-alpha serine/threonine-protein kinase) signaling pathway [24-26].
Schematic diagram illustrating the FOX protein domains. All FOX proteins contain a FDH domain, but significantly varying for other domains. FDH, forkhead DNA-binding domain; TAD, transactivation domain; ID, inhibitory domain; NRD, N-terminal repressor domain; NLS, nuclear localization sequence; NES, nuclear export sequence; TRD, transcriptional repressor domain; LZ, leucine zipper.
Expression of FOX genes in osteosarcoma
Emerging evidence has indicated that the expression of FOX genes, including FOXC2 [27], FOXF1 [28], FOXL1 [29], FOXM1 [30, 31], FOXO1 [32, 33] and FOXP2 [34], is disrupted in osteosarcoma cells and primary tumor biopsy samples. As shown in Table 1, according to their expression in osteosarcoma cells and primary tumor biopsy samples, these FOX genes can be divided into two groups: oncogenes and anti-oncogenes. Oncogenes primarily include FOXC2, FOXM1 and FOXP2, and the elevated expression level of these genes has been observed in osteosarcoma cells, which is positively correlated with tumor size, clinical stage, pathological facture, and distant metastasis [27-29]. Anti-oncogenes primarily include FOXF1, FOXL1 and FOXO1, and decreased or absent expression levels of these genes were examined in osteosarcoma cells and primary tumor biopsy samples [30-34].
Expression and functions of FOX family genes in osteosarcoma cells
| Gene | Expression | Osteosarcoma cells | Function | Reference |
|---|---|---|---|---|
| FOXC2 | Upregulated | 143B, U2OS | Regulating osteosarcoma invasion and metastasis | [27] |
| FOXF1 | Downregulated | U2OS, SaOS-2 | Functioning as a target of the p53 family to regulate cancer cell migration and invasiveness | [28, 81] |
| FOXL1 | Downregulated | U2OS, SaOS-2, MG-63 | Regulating osteosarcoma cell growth through induction of cell cycle arrest and apoptosis | [29] |
| FOXM1 | Upregulated | HOS, U2OS, SOSP-9607, MG-63, 143B and SaOS-2 | Functioning as a target of miR-370 to regulate cell growth and metastasis | [31] |
| FOXO1 | Downregulated | U2OS, SaOS-2, MG-63 | FOXO1 inhibits osteosarcoma oncogenesis via Wnt/beta-catenin pathway suppression | [32] |
| FOXP2 | Upregulated | MG-63, 143B, U2OS | Regulating a p21-dependent growth arrest checkpoint | [34] |
Roles of FOX genes in osteosarcoma
Oncogenes
FOXC2
FOXC2 is also called mesenchyme fork head protein 1 (MFH1), and this protein plays a role in early kidney and heart development [35]. In recent years, elevated expression of FOXC2 has been detected in different cancer cells, such as non-small-cell lung cancer [36, 37], colorectal [38], esophageal [39], breast [40] and gastric cancers [41]. Current views on the function and molecular mechanisms of FOXC2 reveal that this protein plays multiple roles in cancer progression and metastasis by affecting different targets [36-41]. For example, FOXC2 directly binds and induces the transcription of some receptors, such as CXCR4 (C-X-C chemokine receptor type 4, also known as fusin or cluster of differentiation 184) and integrin β3 [42]. CXCR-4 is an alpha-chemokine receptor that is specific for stromal-derived-factor-1 (SDF-1), and this factor has been identified as a prognostic marker in various types of cancers [43]. Integrin β3 is critical for tumor invasion, neoangiogenesis, and inflammation [44]. FOXC2 also induces epithelial-mesenchymal transition (EMT) and serves as a link between EMT and stem cell properties in breast cancer [45]. In addition, FOXC2 overexpression induces MET expression and activates the hepatocyte growth factor (HGF)-MET signaling pathway, thereby promoting the invasion and metastasis of colorectal cancer cells [38].
Recently, FOXC2 has been demonstrated to augment tumor propagation and metastasis in osteosarcoma by directly targeting CXCR4 (Figure 2A) [27]. The elevated expression of FOXC2 was also observed in different osteosarcoma cell lines and osteosarcoma samples from patients [27]. Further analysis indicated that FOXC2 overexpression was critical for maintaining osteosarcoma cells in a stem-like state, and anoikis conditions could induce FOXC2 expression [27]. Specific knockdown of FOXC2 expression using short hairpin RNA (shRNA) decreased osteosarcoma cell metastasis and attenuated the anchorage-independent growth and invasion of osteosarcoma cells in vitro [27]. In addition, downregulation of FOXC2 also resulted in decreased CXCR4 mRNA levels and reduced CXCR4-luciferase promoter activity [27]. Interestingly, ectopic expression of CXCR4 restored the invasive capabilities of FOXC2 knockdown cells in vitro and in vivo, indicating that CXCR4 is a downstream target of FOXC2 in osteosarcoma [27].
FOXM1
FOXM1 is a well-known member of the FOX family, and its overexpression has been widely reported in different cancers, such as breast [46], prostate [47], colorectal [48], liver [49], pancreatic [50], and lung cancers [51]. Molecular mechanism studies have revealed that FOXM1 functions in a variety of biological processes, including cell cycle progression, cell proliferation, tumor angiogenesis, apoptosis, cell migration and cell invasion [46-51]. Growing evidence indicates that FOXM1 controls the G1/S to G2/M phase transition and is also involved in M phase progression [52, 53]. FOXM1 has been reported to regulate the expression a number of genes involved in the cell cycle, such as CDC25A and B, Cyclin B, Aurora B, SKP2, CKS1, centromere protein A (CENPA), CENPB and CENPF [54, 55]. In addition, FOXM1 also interacts with B-Myb and MuvB, and the latter gene can be recruited to chromatin through a cell cycle homology (CHR) element [56]. Knockdown of FOXM1 in multiple cancer cells (e.g., lung, liver, colon and breast) leads to the reduction of cell proliferation [23, 57, 58]. FOXM1 can directly bind the promoter of vascular endothelial growth factor (VEGF) and activate its expression, thereby facilitating VEGF-induced angiogenesis in tumor cells [59]. Inhibition of FOXM1 can sensitize human cancer cells to apoptosis induced by proteasome inhibitors, including MG132, bortezomib and thiostrepton [60]. In a number of cancers, such as pancreatic, breast and colorectal cancer, FOXM1 has been reported to regulate the expression of two matrix metalloproteinase (MMP) genes, including MMP-2 and MMP-9, thereby affecting cancer cell migration and invasion [61-64].
Molecular mechanisms of oncogenic FOX proteins in osteosarcoma. (A) FOXC2 directly targets CXCR4, which may further interact with SDF-1, and the SDF-1/CXCR4 signaling axis affects the growth, angiogenesis and metastasis of cancer. (B) A microRNA, known as miR-370, binds to the 3'-UTR of FOXM1 and negatively regulates its expression. The increased expression of FOXM1 in osteosarcoma can lead to decreased E-cadherin, thereby regulating osteosarcoma cell growth and metastasis. (C) Elevated FOXP2 expression in osteosarcoma upregulates p21WAF1/CIP1 and induces osteosarcoma cell growth arrest.
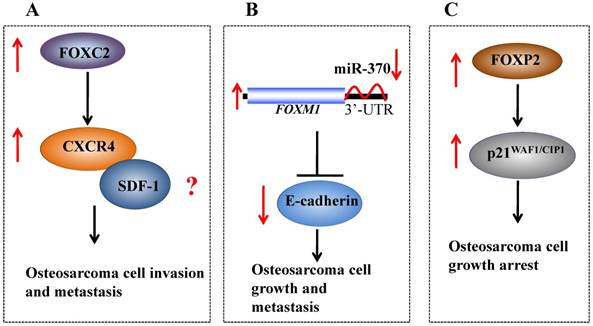
Most recently, FOXM1 was identified as highly expressed in osteosarcoma patient tissues, and overexpression of this protein was correlated with poor prognosis [30, 31]. In addition, FOXM1 expression was also markedly increased in osteosarcoma cells [31]. To elucidate the factors that regulate FOXM1 expression, we examined the microRNA levels in osteosarcoma cells and observed that miR-370 is significantly upregulated and can specifically target the 3'-UTR of FOXM1 [31]. In addition, in osteosarcoma cells overexpressed FOXM1 and miR-370, the decreased E-cadherin expression and increased N-cadherin expression were observed [31]. Thus, our recent studies have revealed that miR-370 regulates the expression of FOXM1 in osteosarcoma cells, thus affecting cell growth and metastasis (Figure 2B) [31].
FOXP2
FOXP2 is a molecule that is required for neurobiology, and its mutation or depletion of expression often results in speech and language disorders [65]. In recent years, its role in cancer has also been evaluated, but the results were controversial. Campbell et al. showed that FOXP2 was overexpressed in lymphoma cells compared to normal counterparts [66]. In a tissue microarray containing samples from more than 11,000 prostate cancer patients, Stumm et al. observed 19% of samples with weak FOXP2 staining, 34% of samples with moderate staining, and 41% of samples with strong staining [67]. However, Cuiffo et al. showed that FOXP2 expression was significantly repressed in breast cancer cells and that its downregulation could trigger tumor initiation and metastasis [68]. In addition, FOXP2 also participates in regulating the expression of a number of genes involved in tumor signaling pathways, including Wnt [69], NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) [70], and IGF-1 (insulin-like growth factor 1) [71]. Importantly, FOXP2 interacts with C-terminal binding protein 1 (CTBP1) [72], a transcriptional corepressor that targets and regulates the expression of tumor suppressors, such as PTEN, p16 and BAX [73].
Recently, Gascoyne et al. identified both FOXP2 mRNA and protein levels were highly expressed in an osteosarcoma cell line (Saos-2) [34]. Treatment with a combination of Vitamin D3 and TGF-β on MG-63 osteosarcoma cells resulted in cell growth arrest at sub-confluence and induction of alkaline phosphatase activity, but not induction of FOXP2 expression [34]. However, the FOXP2 mRNA levels were extremely elevated when MG-63 cells were treated with vehicle [34]. Interestingly, FOXP2 expression was significantly induced when osteosarcoma cell lines with a minimal basal FOXP2 level were cultured to confluency, which was associated with growth arrest and minimal apoptosis [34]. Subsequent analysis indicated that this induction was coincident with strongly induced p21CIP1/WAF1, a critical cell cycle inhibitor (Figure 2C) [34], but the underlying mechanism is still unclear.
Anti-oncogenes
FOXF1
FOXF1 is implicated in a variety of cancers with controversial roles. In human prostate cancer cells, genome-wide array analysis revealed that the FOXF1 mRNA level was significantly reduced compared with that in normal prostate cells [74]. In breast cancer cells, FOXF1 expression is aberrantly repressed through epigenetic mechanisms (DNA hypermethylation in the promoter region) [75]. Overexpression of FOXF1 in breast cancer cells suppresses cell proliferation and tumorigenicity [75]. Further analysis indicated that FOXF1 inhibited the activation of CDK2, thereby affecting the signaling of the CDK2-RB-E2F cascade, resulting in blockage of the G1-S transition [75]. In colon cancer cells, FOXF1 expression was decreased, triggering the ATM/ATR-mediated DNA damage response and activating the p53-p21WAF1 checkpoint pathway [76]. However, elevated FOXF1 levels were also observed in some cancers, such as basal cell carcinoma [77], medulloblastoma [78], rhabdomyosarcomas [78, 79] and Hedgehog (HH)-positive non-small cell lung cancers, compared to healthy cells [78, 80], implying that FOXF1 might function as an oncogene.
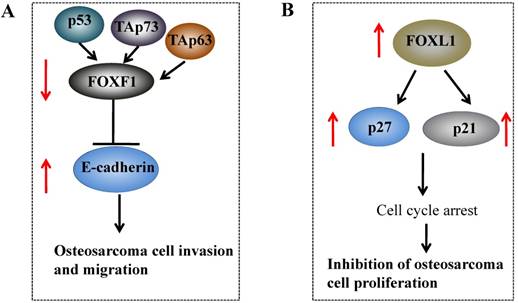
Studies regarding the function of FOXF1 in osteosarcoma cells are poor, and only two studies have shown that FOXF1 expression is decreased in osteosarcoma cells. The first study revealed that FOXF1 is critical for the migration of mesenchymal cells through induction of Integrin-beta3 expression [28], and these authors only used U2OS osteosarcoma cells as a control to determine the FOXF1 level, but did not illuminate its function in osteosarcoma cells [28]. In the second independent study, Tamura et al. showed that FOXF1 knockdown could stimulate cancer cell invasion and migration in Saos-2 osteosarcoma cells [81], and further studies indicated that FOXF1, a target of the p53 family, is upregulated by p53, TAp63 and TAp73 [81]. FOXF1 can regulate the expression of E-cadherin by directly binding to a conserved motif (ATAAATAAATA) located in the E-cadherin promoter region (Figure 3A) [81]. These results strongly suggested that FOXF1 might function as a tumor suppressor in osteosarcoma cells [28, 81].
Molecular mechanisms of FOXF1 and FOXL1 in osteosarcoma. (A) p53, TAp63 and TAp73 upregulates FOXF1 expression, and the activated FOXF1 can further decrease E-cadherin expression by directly binding to its promoter, thereby inhibiting osteosarcoma cell invasion and migration. (B) The overexpression of FOXL1 in osteosarcoma cells induced the expression of p21 and p27, and resulted in cell cycle arrest, eventually leading to inhibition of cell proliferation.
FOXL1
In the mesenchyme of the gastrointestinal tract, FOXL1 was initially implicated in the regulation of the Wnt/APC (Adenomatous Polyposis Coli)/beta-catenin pathway [82]. FOXL1 was also repressed, and this protein plays a role as a tumor suppressor to inhibit cell proliferation and invasion in certain cancers, including colonic [82], pancreatic [83] and gallbladder cancer cells [84]. In colonic cancer, FOXL1 deficiency increased the gastrointestinal tumor load and accelerated tumor initiation in APCMin mice [82]. In pancreatic cancer, lower FOXL1 expression was associated with increased cancer-specific mortality [83]. Overexpression of FOXL1 induced expression of TRAIL (TNF-related apoptosis-inducing ligand) and promoted apoptosis [83]. Moreover, FOXL1 also suppressed ZEB1 (zinc finger E-box-binding homeobox 1) transcription by directly binding to its promoter, thereby affecting cancer cell invasion and aggressiveness, reflecting the fact that ZEB1 is an epithelial-mesenchymal transition (EMT) activator [83]. In gallbladder cancer, decreased expression of FOXL1 was observed in both tissues and cell lines [84]. Overexpression of FOXL1 in gallbladder NOZ cells could trigger mitochondria-mediated apoptosis, thereby inhibiting cell proliferation, migration and invasion in vitro [84]. Additionally, FOXL1 overexpression suppressed ZEB1 expression, induced E-cadherin expression in NOZ cells, and eventually affected cell migration and invasion [84].
Interestingly, recent studies have also indicated that FOXL1 is downregulated in osteosarcoma tissues and cell lines with poor prognosis [29]. Overexpression of FOXL1 in osteosarcoma U2OS cells inhibited cell proliferation and induced expression of p21 and p27, leading to U2OS cell arrest at the G1 phase (Figure 3B) [29]. Moreover, FOXL1 overexpression induced cytochrome c release and disrupted mitochondrial transmembrane depolarization, thereby triggering apoptosis [29].
FOXO1
FOXO1 plays fundamental roles in regulating the reactions of gluconeogenesis and glycogenolysis through insulin signaling [85]. Interestingly, the transcriptional activity of FOXO1 is controlled by its phosphorylation state at the Thr-24, Ser-256, and Ser-319 sites [86]. In the non-phosphorylated state, FOXO1 is localized in the nucleus and binds to the conserved insulin response sequence located in the promoter of glucose 6-phosphatase [85, 86]. However, when it is phosphorylated by the phosphatidylinositol 3-kinase PI3K/AKT (RAC-alpha serine/threonine-protein kinase) pathway, FOXO1 transports from the nucleus to the cytoplasm [85, 86], and this modification subsequently decreases glucose production through a decrease in glucose 6-phosphatase transcription [85, 86]. FOXO1 protein also acts as a tumor suppressor in cancer [87]. Depleted or decreased expression of FOXO1 has been demonstrated in a number of tumors, including breast, pancreatic, prostate, cervical, endometrial and renal cancers [87]. In which, FOXO1 expression can be downregulated by many factors, including miRNAs (miR-9, -27, -96, -153, -182, -183, and -186) [88, 89], estrogen [90] and epidermal growth factor [91]. FOXO1 participates in the regulation of a number of cell cycle regulatory proteins, such as cyclin-dependent kinase inhibitor (p27KIP1, p21WAF1, p130, cyclin G2, cyclin D1 and D2) [92, 93], and this protein can also regulate the apoptosis mediators Bim, Fas ligand and Bcl-6 [94].
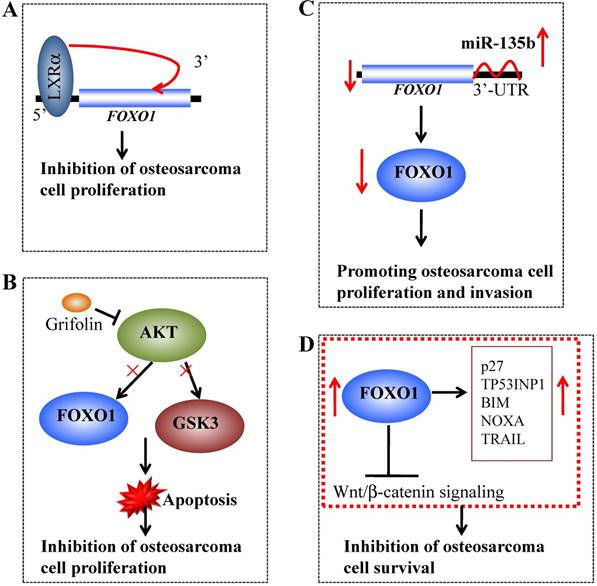
Several recent studies have revealed that FOXO1 expression is suppressed in osteosarcoma tissues and cells. Chang et al. showed that liver X receptor α (LXRα) could directly bind to the promoter and activate the expression of FOXO1, eventually resulting in inhibition of osteosarcoma cell proliferation (Figure 4A) [95]. Jin et al. demonstrated that Grifolin, a natural-derived small molecule, could inhibit AKT activity, thereby suppressing the phosphorylation of its substrates, including FOXO1 and GSK3 (glycogen synthase kinase 3) (Figure 4B), inducing mitochondria-dependent apoptosis [96]. Pei et al. showed that miR-135b directly targeted the 3'-UTR of FOXO1 and that the decreased expression of FOXO1 in osteosarcoma cells was inversely correlated with miR-135b expression (Figure 4C) [97]. Interestingly, FOXO1 knockdown could reverse the effects of miR-135b inhibitor-induced proliferation and invasion in osteosarcoma cells [97]. Recently, Guan et al. showed that FOXO1 at least partially inhibited osteosarcoma oncogenesis by suppression of the Wnt/β-catenin signaling pathway (Figure 4D) [32]. In a study containing 34 osteosarcoma cases, the whole-genome sequencing results demonstrated that the FOXO1 locus had varied copy numbers and showed a loss of heterozygosity, suggesting that chromosomal alterations might contribute to the decrease in FOXO1 expression [32]. Overexpression of FOXO1 in osteosarcoma cells could lead to increased expression of p27KIP1, TP53INP1 (p53-induced-nuclear-protein 1), BCL2L11/BIM (Bcl-2-like protein 11), NOXA and TRAIL as well as decreased expression of Cyclin D1, thereby inhibiting cell proliferation and inducing cell cycle arrest and apoptosis [32]. Importantly, overexpression of FOXO1 significantly impaired Wnt/β-catenin activity. Moreover, inhibition of FOXO1 by its specific inhibitor AS1842856 could inhibit cell differentiation and promote cell survival [32].
Molecular mechanisms of FOXO1 in osteosarcoma. (A) Liver X receptor α (LXRα) can directly bind to the promoter of FOXO1 and activate its expression, eventually resulting in inhibition of osteosarcoma cell proliferation. (B) The small molecule Grifolin inhibits the activity of AKT and causes the suppression of FOXO1 and GSK3 phosphorylation, thereby inducing apoptosis and inhibiting cell proliferation. (B) miR-135b binds to the 3'-UTR of FOXO1 and negatively regulates its expression, thereby promoting osteosarcoma cell growth and invasion. (D) Overexpression of FOXO1 in osteosarcoma cells can increase the expression of p27, TP53INP1 BIM, NOXA and TRAIL and significantly impair Wnt/β-catenin activity, eventually leading to the inhibition of osteosarcoma cell survival.
Targeting FOX genes in osteosarcoma therapy
In recent years, considerable efforts have been made to validate FOX proteins as preferential targets in cancer therapy. However, limited information is available for targeting FOX proteins in osteosarcoma therapy. Collectively, four strategies based on targeting FOX proteins have been developed or are currently under development to inhibit tumor growth. These four promising approaches include (1) small molecules targeting FOX proteins; (2) targeting FOX genes via a CRISPR/Cas9 gene editing tool; (3) developing FOX protein derived peptides for immunotherapy; and (4) targeting molecules or signaling pathways that FOX proteins are involved in (indirect ways).
Small molecules that direct target FOX proteins
The discovery of small molecules as anti-tumor drugs has been significantly developed in the past decade. However, small molecules that directly target FOX proteins have not been extensively reported in cancer therapy, particularly in osteosarcoma therapy. Among the above-mentioned oncogenes (FOXC2, FOXM1 and FOXP2), only FOXM1 was reported to be directly targeted by a number of small molecules. For example, thiostrepton, a natural-derived small molecule, inhibits FOXM1 mRNA and protein levels in breast cancer MCF-7 cells, resulting in induction of apoptosis, primarily by caspase-dependent intrinsic and extrinsic pathways [98]. By applying a high-throughput screen containing a total number of 54,211 small molecules, FDI-6 (NCGC00099374) was shown to block the binding of FOXM1 to its target DNA in MCF-7 cells, resulting in the inactivation of FOXM1 target genes [99]. Siomycin A, an inhibitor of FOXM1, could suppress the expression of FOXM1 at both transcriptional and translational levels in osteosarcoma U2OS cells and inhibit the downstream target genes of FOXM1, such as CDC25B, Survivin, and CENPB [100]. For those anti-oncogenes (FOXF1, FOXL1 and FOXO1), screening strategies to identify molecules that reactivate endogenous FOX proteins are currently being developed. A small molecule known as methylseleninic acid (MSA) in Ewing sarcoma cells increased endogenous FOXO1 expression in a dose- and time-dependent manner [101]. However, it is still absent for small molecules that function to reactivate the expression of FOX genes in osteosarcoma cells.
CRISPR/Cas9 method edits FOX genes
The faster development of the CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated 9) genome editing tool makes it is easy to knockout, knockdown or activate a gene [102]. Currently, CRISPR/Cas9 knockout or knockin kits of these oncogenic and anti-oncogenic FOX genes are commercially available, providing effective methods for researchers to evaluate FOX gene functions in osteosarcoma therapy. Although the oncogenic and anti-oncogenic FOX genes have not been reported to be direct targets via CRISPR/Cas9 methods in cancer therapy, this tool has been suggested to benefit future cancer therapy approaches through direct targeting of interested genes, including FOX genes.
Immunotherapy
The oncogenic FOX genes have the potential to serve as adequate targets for cancer immunotherapy, as these genes are overexpressed in various cancers. Among these genes, FOXM1 has been reported to be a candidate target for anti-tumor immunotherapy in cholangiocarcinoma [103]. The investigation of 23 human FOXM1-derived peptides predicted to bind to human leukocyte antigen (HLA)-A2-restricted T cells revealed three peptides, including FOXM1362-370 (YLVPIQFPV), FOXM1373-382 (SLVLQPSVKV), and FOXM1640-649 (GLMDLSTTPL) could induce HLA-A2-restricted T cells, suggesting that FOXM1 might be a suitable target for immunotherapy against various cancers, including osteosarcoma [103].
Other indirect strategies
Given that FOX proteins participate in or cross-talk with a variety of signaling pathways, these proteins can be regulated by a number of proteins and miRNAs, and the expression of FOX genes is frequently affected when researchers target other molecules in cancer therapy. For example, suppression of PI3K using its specific inhibitor LY294002 resulted in decreased expression of FOXM1 in human U2OS osteosarcoma cells [104, 105]. As previously described, FOXO1 is the target of miR-96 and miR-182, thus treatment with two synthetic antisense inhibitors, including anti-miR-96 and anti-miR-182, in hepatocellular carcinoma and prostate cancer induced FOXO1 expression [106, 107]. In addition, inhibition of the EGFR (epidermal growth factor) signaling pathway using antibodies (such as trastuzumab or cetuximab) or small molecules (such as lapatinib and gefitinib) could induce expression of FOXO3 [2, 108].
Summary and Perspective
Obviously, the deregulation of FOX proteins is frequently associated with tumorigenesis, and these proteins participate in a wide spectrum of cellular processes, including proliferation, differentiation, senescence and apoptosis. Interestingly, both oncogenic and anti-oncogenic FOX proteins are shown in different types of cancers, implying the complexity of these family protein functions. Although FOX proteins are a superfamily comprising 50 members in the human genome, only a few of these proteins have been examined and studied in human osteosarcoma. Additional efforts to explore the precise mechanisms of FOX proteins in tumorigenesis should be conducted in the future, particularly in osteosarcoma.
Emerging evidence suggests that FOX proteins could be intriguing targets for anticancer therapies, including anti-osteosarcoma therapy. Although current views consider that transcription factors are rarely utilized as direct targets, a number of studies are currently investigating FOX proteins as direct or indirect targets to prevent cancer development and progression. The identification of small molecules that selectively block the function of FOX proteins would be a useful step toward designing FOX-specific inhibitors. We expect that FOX-specific inhibitors will be successfully developed and utilized in human cancer therapies. With the rapid development of CRIPSR/Cas9 technology, we propose that this tool will significantly enhance cancer therapy by targeting FOX genes in the coming years.
Acknowledgements
We apologize to all authors whose contributions were not cited due to space limitations.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10(4):233-40
2. Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847-59
3. Zhu H. Targeting forkhead box transcription factors FOXM1 and FOXO in leukemia (Review). Oncol Rep. 2014;32(4):1327-34
4. Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4(11):889-99
5. Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box (FOX) gene families. Hum Genomics. 2010;4(5):345-52
6. Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27(6):224-32
7. van der Heul-Nieuwenhuijsen L, Dits NF, Jenster G. Gene expression of forkhead transcription factors in the normal and diseased human prostate. BJU Int. 2009;103(11):1574-80
8. Zhang YL, Sun FT, Zhang Z. et al. Comprehensive expression analysis suggests functional overlapping of human FOX transcription factors in cancer. Asian Pac J Cancer Prev. 2014;15(23):10475-81
9. Urbanek P, Klotz LO. Posttranscriptional regulation of FOXO expression: microRNAs and beyond. Br J Pharmacol. 2016 DOI: 10.1111/bph.13471
10. Yang YA, Yu J. Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015;2(2):144-151
11. Neben K, Schnittger S, Brors B. et al. Distinct gene expression patterns associated with FLT3- and NRAS-activating mutations in acute myeloid leukemia with normal karyotype. Oncogene. 2005;24(9):1580-8
12. Lin L, Miller CT, Contreras JI. et al. The hepatocyte nuclear factor 3 alpha gene, HNF3alpha (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 2002;62(18):5273-9
13. Hu X, Stern HM, Ge L. et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7(4):511-22
14. Sahu B, Laakso M, Ovaska K. et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962-76
15. Robinson JL, Carroll JS. FoxA1 is a key mediator of hormonal response in breast and prostate cancer. Front Endocrinol (Lausanne). 2012;3:68
16. Zhang G, Zhao Y, Liu Y. et al. FOXA1 defines cancer cell specificity. Sci Adv. 2016;2(3):e1501473
17. Kim IM, Ackerson T, Ramakrishna S. et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153-61
18. Kalinina OA, Kalinin SA, Polack EW. et al. Sustained hepatic expression of FoxM1B in transgenic mice has minimal effects on hepatocellular carcinoma development but increases cell proliferation rates in preneoplastic and early neoplastic lesions. Oncogene. 2003;22(40):6266-76
19. Kalin TV, Wang IC, Ackerson TJ. et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66(3):1712-20
20. Madureira PA, Varshochi R, Constantinidou D. et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281(35):25167-76
21. Wang Z, Banerjee S, Kong D. et al. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67(17):8293-300
22. Teh MT. FOXM1 coming of age: time for translation into clinical benefits? Front Oncol. 2012;2:146
23. Xu XS, Miao RC, Wan Y. et al. FoxM1 as a novel therapeutic target for cancer drug therapy. Asian Pac J Cancer Prev. 2015;16(1):23-9
24. Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011. 1813(11):1938-45
25. Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27(16):2312-9
26. Charitou P, Rodriguez-Colman M, Gerrits J. et al. FOXOs support the metabolic requirements of normal and tumor cells by promoting IDH1 expression. EMBO Rep. 2015;16(4):456-66
27. Gozo MC, Jia D, Aspuria PJ. et al. FOXC2 augments tumor propagation and metastasis in osteosarcoma. Oncotarget. 2016 DOI: 10.18632/oncotarget.11990
28. Malin D, Kim IM, Boetticher E. et al. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Mol Cell Biol. 2007;27(7):2486-98
29. Chen X, Deng M, Ma L. et al. Inhibitory effects of forkhead box L1 gene on osteosarcoma growth through the induction of cell cycle arrest and apoptosis. Oncol Rep. 2015;34(1):265-71
30. Fan CL, Jiang J, Liu HC, Yang D. Forkhead box protein M1 predicts outcome in human osteosarcoma. Int J Clin Exp Med. 2015;8(9):15563-8
31. Duan N, Hu X, Yang X. et al. MicroRNA-370 directly targets FOXM1 to inhibit cell growth and metastasis in osteosarcoma cells. Int J Clin Exp Pathol. 2015;8(9):10250-60
32. Guan H, Tan P, Xie L. et al. FOXO1 inhibits osteosarcoma oncogenesis via Wnt/beta-catenin pathway suppression. Oncogenesis. 2015;4:e166
33. He W, Feng L, Xia D, Han N. MiR-374a promotes the proliferation of human osteosarcoma by downregulating FOXO1 expression. Int J Clin Exp Med. 2015;8(3):3482-9
34. Gascoyne DM, Spearman H, Lyne L. et al. The Forkhead Transcription Factor FOXP2 Is Required for Regulation of p21WAF1/CIP1 in 143B Osteosarcoma Cell Growth Arrest. PLoS One. 2015;10(6):e0128513
35. Mani SA, Yang J, Brooks M. et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069-74
36. Jiang W, Pang XG, Wang Q. et al. Prognostic role of Twist, Slug, and Foxc2 expression in stage I non-small-cell lung cancer after curative resection. Clin Lung Cancer. 2012;13(4):280-7
37. Yu YH, Chen HA, Chen PS. et al. MiR-520h-mediated FOXC2 regulation is critical for inhibition of lung cancer progression by resveratrol. Oncogene. 2013;32(4):431-43
38. Cui YM, Jiao HL, Ye YP. et al. FOXC2 promotes colorectal cancer metastasis by directly targeting MET. Oncogene. 2015;34(33):4379-90
39. Nishida N, Mimori K, Yokobori T. et al. FOXC2 is a novel prognostic factor in human esophageal squamous cell carcinoma. Ann Surg Oncol. 2011;18(2):535-42
40. Pietila M, Vijay GV, Soundararajan R. et al. FOXC2 regulates the G2/M transition of stem cell-rich breast cancer cells and sensitizes them to PLK1 inhibition. Sci Rep. 2016;6:23070
41. Zhu JL, Song YX, Wang ZN. et al. The clinical significance of mesenchyme forkhead 1 (FoxC2) in gastric carcinoma. Histopathology. 2013;62(7):1038-48
42. Hayashi H, Kume T. Foxc2 transcription factor as a regulator of angiogenesis via induction of integrin beta3 expression. Cell Adh Migr. 2009;3(1):24-6
43. Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res. 2014;124:31-82
44. Su X, Esser AK, Amend SR. et al. Antagonizing Integrin beta3 Increases Immunosuppression in Cancer. Cancer Res. 2016;76(12):3484-95
45. Hollier BG, Tinnirello AA, Werden SJ. et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013;73(6):1981-92
46. Yang C, Chen H, Yu L. et al. Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther. 2013;20(2):117-24
47. Cai Y, Balli D, Ustiyan V. et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem. 2013;288(31):22527-41
48. Chu XY, Zhu ZM, Chen LB. et al. FOXM1 expression correlates with tumor invasion and a poor prognosis of colorectal cancer. Acta Histochem. 2012;114(8):755-62
49. Kopanja D, Pandey A, Kiefer M. et al. Essential roles of FoxM1 in Ras-induced liver cancer progression and in cancer cells with stem cell features. J Hepatol. 2015;63(2):429-36
50. Xia JT, Wang H, Liang LJ. et al. Overexpression of FOXM1 is associated with poor prognosis and clinicopathologic stage of pancreatic ductal adenocarcinoma. Pancreas. 2012;41(4):629-35
51. Balli D, Zhang Y, Snyder J. et al. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res. 2011;71(1):40-50
52. Grant GD, Brooks L, Zhang X. et al. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell. 2013;24(23):3634-50
53. Lv C, Zhao G, Sun X. et al. Acetylation of FOXM1 is essential for its transactivation and tumor growth stimulation. Oncotarget. 2016 DOI: 10.18632/oncotarget.11332
54. Wang IC, Chen YJ, Hughes D. et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875-94
55. Davis DB, Lavine JA, Suhonen JI. et al. FoxM1 is up-regulated by obesity and stimulates beta-cell proliferation. Mol Endocrinol. 2010;24(9):1822-34
56. Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26(5):474-89
57. Wu QF, Liu C, Tai MH. et al. Knockdown of FoxM1 by siRNA interference decreases cell proliferation, induces cell cycle arrest and inhibits cell invasion in MHCC-97H cells in vitro. Acta Pharmacol Sin. 2010;31(3):361-6
58. Yang K, Jiang L, Hu Y. et al. Short hairpin RNA- mediated gene knockdown of FOXM1 inhibits the proliferation and metastasis of human colon cancer cells through reversal of epithelial-to-mesenchymal transformation. J Exp Clin Cancer Res. 2015;34:40
59. Karadedou CT, Gomes AR, Chen J. et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2012;31(14):1845-58
60. Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA-damage. PLoS One. 2012;7(2):e31761
61. Zheng Y, Guo J, Zhou J. et al. FoxM1 transactivates PTTG1 and promotes colorectal cancer cell migration and invasion. BMC Med Genomics. 2015;8:49
62. Xue YJ, Xiao RH, Long DZ. et al. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med. 2012;10:200
63. Dai B, Kang SH, Gong W. et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26(42):6212-9
64. Wang IC, Chen YJ, Hughes DE. et al. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283(30):20770-8
65. Lai CS, Fisher SE, Hurst JA. et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519-23
66. Campbell AJ, Lyne L, Brown PJ. et al. Aberrant expression of the neuronal transcription factor FOXP2 in neoplastic plasma cells. Br J Haematol. 2010;149(2):221-30
67. Stumm L, Burkhardt L, Steurer S. et al. Strong expression of the neuronal transcription factor FOXP2 is linked to an increased risk of early PSA recurrence in ERG fusion-negative cancers. J Clin Pathol. 2013;66(7):563-8
68. Cuiffo BG, Karnoub AE. Silencing FOXP2 in breast cancer cells promotes cancer stem cell traits and metastasis. Mol Cell Oncol. 2016;3(3):e1019022
69. Sin C, Li H, Crawford DA. Transcriptional regulation by FOXP1, FOXP2, and FOXP4 dimerization. J Mol Neurosci. 2015;55(2):437-48
70. DeLaughter DM, Christodoulou DC, Robinson JY. et al. Spatial transcriptional profile of the chick and mouse endocardial cushions identify novel regulators of endocardial EMT in vitro. J Mol Cell Cardiol. 2013;59:196-204
71. Vernes SC, Spiteri E, Nicod J. et al. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81(6):1232-50
72. Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24(2):809-22
73. Chinnadurai G. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Res. 2009;69(3):731-4
74. Watson JE, Doggett NA, Albertson DG. et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23(19):3487-94
75. Lo PK, Lee JS, Liang X. et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70(14):6047-58
76. Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cell Signal. 2012;24(1):316-24
77. Wendling DS, Luck C, von Schweinitz D, Kappler R. Characteristic overexpression of the forkhead box transcription factor Foxf1 in Patched-associated tumors. Int J Mol Med. 2008;22(6):787-92
78. Dharmadhikari AV, Szafranski P, Kalinichenko VV, Stankiewicz P. Genomic and Epigenetic Complexity of the FOXF1 Locus in 16q24.1: Implications for Development and Disease. Curr Genomics. 2015;16(2):107-16
79. Armeanu-Ebinger S, Bonin M, Habig K, Poremba C. et al. Differential expression of invasion promoting genes in childhood rhabdomyosarcoma. Int J Oncol. 2011;38(4):993-1000
80. Gialmanidis IP, Bravou V, Petrou I. et al. Expression of Bmi1, FoxF1, Nanog, and gamma-catenin in relation to hedgehog signaling pathway in human non-small-cell lung cancer. Lung. 2013;191:511-21
81. Tamura M, Sasaki Y, Koyama R. et al. Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Oncogene. 2014;33(40):4837-46
82. Perreault N, Sackett SD, Katz JP. et al. Foxl1 is a mesenchymal Modifier of Min in carcinogenesis of stomach and colon. Genes Dev. 2005;19(3):311-5
83. Zhang G, He P, Gaedcke J. et al. FOXL1, a novel candidate tumor suppressor, inhibits tumor aggressiveness and predicts outcome in human pancreatic cancer. Cancer Res. 2013;73(17):5416-25
84. Qin Y, Gong W, Zhang M. et al. Forkhead box L1 is frequently downregulated in gallbladder cancer and inhibits cell growth through apoptosis induction by mitochondrial dysfunction. PLoS One. 2014;9(7):e102084
85. Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27(16):2320-36
86. Zhang X, Gan L, Pan H. et al. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277(47):45276-84
87. Coomans de Brachene A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci. 2016;73(6):1159-72
88. Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284(35):23204-16
89. Myatt SS, Wang J, Monteiro LJ. et al. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70(1):367-77
90. Ahn EH. Regulation of target genes of PAX3-FOXO1 in alveolar rhabdomyosarcoma. Anticancer Res. 2013;33(5):2029-35
91. Ding H, Zhu Y, Chu T, Wang S. Epidermal growth factor induces FoxO1 nuclear exclusion to activate MMP7-mediated metastasis of larynx carcinoma. Tumour Biol. 2014;35(10):9987-92
92. Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011. 1813(11):1978-86
93. Liu P, Kao TP, Huang H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene. 2008;27(34):4733-44
94. Prasad SB, Yadav SS, Das M. et al. Down Regulation of FOXO1 Promotes Cell Proliferation in Cervical Cancer. J Cancer. 2014;5(8):655-62
95. Chang YW, Zhao YF, Cao YL. et al. Liver X receptor alpha inhibits osteosarcoma cell proliferation through up-regulation of FoxO1. Cell Physiol Biochem. 2013;32(1):180-6
96. Jin S, Pang RP, Shen JN. et al. Grifolin induces apoptosis via inhibition of PI3K/AKT signalling pathway in human osteosarcoma cells. Apoptosis. 2007;12(7):1317-26
97. Pei H, Jin Z, Chen S. et al. MiR-135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol Cell Biochem. 2015;400(1-2):245-52
98. Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011;3(9):725-31
99. Gormally MV, Dexheimer TS, Marsico G. et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun. 2014;5:5165
100. Radhakrishnan SK, Bhat UG, Hughes DE. et al. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res. 2006;66(19):9731-5
101. Niedan S, Kauer M, Aryee DN. et al. Suppression of FOXO1 is responsible for a growth regulatory repressive transcriptional sub-signature of EWS-FLI1 in Ewing sarcoma. Oncogene. 2014;33(30):3927-38
102. Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299-311
103. Yokomine K, Senju S, Nakatsura T. et al. The forkhead box M1 transcription factor as a candidate of target for anti-cancer immunotherapy. Int J Cancer. 2010;126(9):2153-63
104. Laoukili J, Kooistra MR, Bras A. et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126-36
105. Wang Z, Ahmad A, Li Y. et al. Forkhead box M1 transcription factor: a novel target for cancer therapy. Cancer Treat Rev. 2010;36(2):151-6
106. Xu D, He X, Chang Y. et al. Inhibition of miR-96 expression reduces cell proliferation and clonogenicity of HepG2 hepatoma cells. Oncol Rep. 2013;29(2):653-61
107. Wallis CJ, Gordanpour A, Bendavid JS. et al. MiR-182 Is Associated with Growth, Migration and Invasion in Prostate Cancer via Suppression of FOXO1. J Cancer. 2015;6(12):1295-305
108. Yang JY, Hung MC. A new fork for clinical application: targeting forkhead transcription factors in cancer. Clin Cancer Res. 2009;15(3):752-7
Author contact
![]() Corresponding authors: caiguo.zhangedu; chenxun2008com
Corresponding authors: caiguo.zhangedu; chenxun2008com

 Global reach, higher impact
Global reach, higher impact