3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(9):1704-1716. doi:10.7150/jca.16961 This issue Cite
Research Paper
MiR-146b-5p functions as a suppressor miRNA and prognosis predictor in non-small cell lung cancer
1. Department of Lung Cancer Surgery
2. Tianjin key laboratory of lung cancer metastasis and tumor microenvironment, Tianjin Lung Cancer Institute, Tianjin Medical University General Hospital, Anshan Road No.154, Heping District, Tianjin 300052, China
# Contributed equally in this work.
Received 2016-7-24; Accepted 2017-4-8; Published 2017-6-30
Abstract
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death worldwide. However, science has not yet been able to substantially improve the prognosis of lung cancer patients. Accumulating evidence suggests that microRNAs (miRNAs) are key players in the regulation of tumor development and metastasis. Expression of six miRNAs previously shown to play roles in tumor development (miR-146b-5p, miR-128b, miR-21, miR-221, miR-34a, and Let-7a) in other tumor types was examined using real-time RT-PCR in 78 specimens of NSCLC. The results revealed that patients with low expression of miR-146b-5p had significant shorter median and mean survival time than those with high miR-146b-5p expression (33.00 and 30.44 months versus 42.0 and 36.90 months, respectively; log-rank test P=0.048), thus low miR-146b-5p expression level was associated with poor prognosis in NSCLC patients. Univariate Cox hazard regression analysis demonstrated that miR-146b-5p expression levels tended to be a significant prognostic indicator of NSCLC (adjusted hazard ratio=0.482, 95% CI: 1.409- 29.593, P=0.016). Multivariate Cox proportional hazard regression analysis showed that miR-146b-5p expression levels were an independent prognostic factor for NSCLC patients (hazard ratio=0.259, 95% CI: 0.083-0.809, P=0.020). Furthermore, the effects of miR-146b-5p and miR-146b-3p on NSCLC cell growth and invasion in vitro were investigated. Our findings demonstrate that ectopic expression of miR-146b-5p suppressed cell proliferation, clonogenicity, migration/ invasion and also induced G1 arrest in vitro, but did not induce cell apoptosis; whereas enforced expression of miR-146b-3p did not have a significant effect on cell growth and metastasis. Further experiments indicated that miR-146b-5p could reduce mRNA levels of MMP16 and TRAF6 in vitro and was negatively related to the expression of TRAF6 in human NSCLC tissues. In a mouse model, Ago-miR-146b-5p could significantly inhibit the growth of lung cancer xenografts in nude mice. In conclusion, our findings demonstrate that miR-146b-5p functions as a suppressor miRNA and prognosis predictor in NSCLC.
Keywords: miR-146b-5p, non-small cell lung cancer, prognosis, and metastasis
Introduction
Lung cancer, predominantly non-small-cell lung cancer (NSCLC), is the leading cause of cancer-related death worldwide[1]. In spite of the tremendous improvements made in the treatment of NSCLC over the past several decades, the prognosis for lung cancer patients has not substantially improved and the overall five-year survival rate of NSCLC patients has remained unfavorable. This due primarily to initial diagnosis at advanced stages of disease, often accompanied by invasion or lymphatic metastasis[2]. Therefore, early diagnosis is essential for improving lung cancer patients' survival and there is an urgent need for novel diagnostic biomarkers screening.
Mature sequence, miRBase accession number, and proposed clinical relevance for the six chosen miRNAs
| miRNA name | Mature miRNA sequence | miRBase Accession number | Proposed clinical relevance | Reference |
|---|---|---|---|---|
| hsa-miR-146b | UGAGAACUGAAUUCCAUAGGCU | MIMAT0002809 | Down-regulation associated with poor overall survival | [8] |
| hsa-miR-128b | GGGGGCCGAUACACUGUACGAGA | MIMAT0031095 | Decreased expression associated with malignant behavior | [9] |
| hsa-miR-21 | UAGCUUAUCAGACUGAUGUUGA | MIMAT0000076 | High expression associated with poor OS | [10] |
| hsa-miR-221 | ACCUGGCAUACAAUGUAGAUUU | MIMAT0004568 | High expression associated with poor OS | [11] |
| hsa-miR-34a | UGGCAGUGUCUUAGCUGGUUGU | MIMAT0000255 | High expression associated with poor OS | [12] |
| hsa-Let-7a | UGAGGUAGUAGGUUGUAUAGUU | MIMAT0000062 | Low expression correlated with poor prognosis | [13] |
Clinically, NSCLC is characterized by highly variable clinical course with different patients with the same staging and histological types having markedly different clinical outcomes [3]. It raises the necessary to found the molecular signaling which related to diagnosis, classification and prognosis, and so guide the treatment. However, few histology-dependent prognostic biomarkers are available for routine use in clinical practice, especially in resectable patients[4]. Thus, the search for novel and efficient biomarkers for lung cancer, which can be used as prognostic biomarkers, is particularly warranted.
MicroRNAs (miRNAs) are a family of small non-coding RNAs that negatively regulate gene expression at the post-transcriptional level by interacting with the 3'UTR of target mRNAs[5]. A growing number of studies have demonstrated that miRNAs play a fundamental role in the regulation of diverse cellular functions. Hence, deregulation of miRNA expression is often associated with a variety of disorders, including human malignancy. Furthermore, increasing evidence indicates that miRNAs may function as either oncogenes or tumor suppressors and thus can potentially represent prognostic markers[6]. Several studies have already reported miRNAs used as biomarkers for clinical applications[7].
In the present study, we examined gene expression of six tumor-related miRNAs (miR-146b-5p, miR-128b, miR-21, miR-221, miR-34a, and Let-7a)[8-13], which were reported to predict prognosis of NSCLC or other cancers, in 78 specimens of NSCLC using real-time RT-PCR (Table 1). We then correlated the expression of these miRNAs with clinicopathological features and survival with to validate their diagnostic and prognostic potential for NSCLC patients. Our findings revealed that low miR-146b-5p expression levels were associated with poor prognosis in NSCLC patients. Furthermore, we investigated the effects of miR-146b-5p on NSCLC cell growth and invasion in vitro and in vivo. Our findings demonstrate that miR-146b-5p can be used as a prognostic marker in NSCLC.
Materials and Methods
Ethics Statement
This study was approved by the Ethical Review Committee of Tianjin Medical University General Hospital. All biological samples were obtained with patients' written informed consent. All animal procedures and experimental protocols were approved by Laboratory Animal Ethics Committee of Tianjin Medical University.
Patients and Tissue Specimens
Seventy-eight patients diagnosed with NSCLC who underwent surgical resection at the Department of Lung Cancer Surgery, Tianjin Medical University General Hospital from November 2007 to March 2010 were included in the study. The cases were selected based on the following criteria: (1) diagnosis of primary lung cancer clinical stage I to IV (pTNM); (2) undergoing surgical resection without prior chemotherapy or TKI treatment; (3) availability of outcome and follow-up data. Pathologic diagnosis was based on WHO criteria. Lung cancer staging for each patient was performed according to the AJCC Cancer Staging Manual, 7th edition, and was based on findings from physical examination, surgical resection, and computed tomography of the chest, abdomen, pelvis, and brain. The following information was collected from the patients' medical records: age, gender, clinical stage, pathologic diagnosis, differentiation, lymph node status, metastasis, smoking status, and overall survival time. Survival was calculated from the day of resection until Dec 31, 2012. Resected lung tissues were immediately immersed in liquid nitrogen until RNA extraction.
Cell Culture and transfection with miRNA mimics
The H1299 cell line was obtained from SGST (Shanghai, China). The cells were cultured in RPMI 1640 medium (GIBCO, Gaithersburg, MD, USA.) supplemented with 10% fetal bovine serum (FBS) (GIBCO, Grand Island, USA.), penicillin (100 units/mL), and streptomycin (100 units/mL) at 37ºC in an atmosphere of 5% CO2 in a humidified incubator. Before transfection, H1299 cells were replated into six-well plates at a density of 1 × 105 cells per well and incubated overnight. Cells were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. Scrambled miRNA and a miR-146b mimics were purchased from RiboBio Co.,Ltd(Guangzhou, China)and used at a final concentration of 50 nM. At 24 or 48 hours after transfection, cells were harvested for further analysis.
miRNA Extraction
miRNAs were extracted from tissues and cell lines using the miRNeasy mini kit (QIAGEN, Cat. 217004) according the manufacturer's instructions. RNA was quantified using a spectrophotometer (Beckman, USA), and RNA quality was assessed using a denaturing 1.2% agarose gel.
Quantitative Real-Time RT-PCR
For miRNA expression detection, 50 ng RNA was reverse transcribed using the TaqMan MiRNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. TaqMan miRNA assays were used to quantify the expression levels of mature miR-146b, miR-128b, miR-21, miR-221, miR-34a, and Let-7a according to the manufacturer's instructions (Applied Biosystems). The snRNA U6 (Applied Biosystems) was used as a normalization control for all samples.
For quantitative detection of other genes, complementary DNA synthesis was performed using a M-MLV Reverse Transcriptase kit (Promega, USA) followed by a SYBR Green-based real-time PCR analysis as described previously [14]. The primers used are listed as followed: AUF1 forward: 5'-GATCAAGGGGTTTTGGCTTT-3', revered 5'-GTTGTCCATGGGGACCTCTA-3'; TRAF6 forward: 5'-TGCTTGATGGCATTACGAGAA-3', reverse 5'-CATTTGGACATTTCA CCATCAGAG-3'; MMP16 forward: 5'-GTCAGTCGGTGGAAGGTAGC-3', reverse 5'-CTGGAAGACGGTTGGATTTC-3'; GAPDH forward: 5'-GGAGCGAGATCCCT CCAAAAT-3', reverse 5'-GGCTGTTGTCATACTTCTCATGG-3'. Each measurement was performed in triplicate. Real-time RT-PCR was performed using an Applied Biosystems 7900HT Fast Real-Time PCR System instrument and software. PCR data were analyzed using the comparative Ct method (ΔCt), as previously described for determining relative gene expression [15, 16].
Cell proliferation assays
Cell proliferation was assessed using the cell counting kit-8 (CCK-8, Beyotime, China) according to manufacturer's instructions. Twenty-four hours after transient transfection of miRNA mimics or scrambled miRNA, 3×103 cells were seeded into a 96-well plate. Following incubation of cells for 24, 48, or 72h, 10 μL CCK-8 reagents were added to the culture medium for 2 hours. The absorbance of each well was quantified at 450 nm using a microplate reader (SpectraMax M5, Molecular Devices, CA, USA). All data were calculated from triplicate samples.
Cell cycle and apoptosis assays
After transfection with the miRNA mimics or scrambled miRNA for 48h, cells were detached with trypsin and washed 1-2 times with cold PBS. The cells were fixed with cool 70% ethanol at room temperature, and then washed with PBS again. The cells were immediately stained with propidium iodide using the BD Cycletest Plus DNA reagent kit (BD Biosciences, San Jose, CA, USA) following the manufacturer's protocol. To measure cell apoptosis, cells were collected and washed with cold PBS twice, then incubated with Annexin V-FITC and PI for 15 min in the dark. Analyses of cell cycle and cellular apoptosis were performed using a FACS Calibur Flow Cytometer (Beckman Coulter, Atlanta, GA, USA).
Wound healing assay
H1299 cells in 6-well plates were transfected with miRNA mimics or scrambled miRNA. When cells were approximately 90-100% confluent, a 200 µL pipette tip was used to scrap across the confluent cell layer. Then, cells were washed with fresh media to remove floating cells and wound closure was observed at 0 and 24 h under a Nikon TE2000 microscope (Nikon, Tokyo, Japan). Relative migration was assessed at 40× magnification using a TE2000 inverted microscope at 0h and 24h. Wound closure was calculated using the following formula: (0h wound width-24h wound width)/0h wound width.
Colony formation assay
For the colony formation assay, cells were seeded into 6-well plates (500 cells/well) after transfection and incubated at 37℃ for 14 days. Plates were washed twice with PBS and stained with 0.5% crystal violet at the room temperature for 30 min. The number of colonies, defined as a group of > 50 cells, was counted.
Transwell migration and invasion assays
For the cell invasion assay, transwell filters (Corning, NY, USA) were coated with 100 μl 300 µg/mL Matrigel on the upper surface of the polycarbonic membrane. Twenty-four hours after transfection with miRNA mimics or scrambled miRNA, 1 × 105 cells in serum-free medium were seeded in upper chambers and 600 µL medium containing 10% FBS were added to the lower chambers. Cells were cultured at 37℃ in a humidified incubator with 5% CO2. After incubation for 24 h, cells that migrated to the underside of the filter were fixed in methanol and stained using 0.1% crystal violet. Five randomly-selected visual fields were imaged and counted using a Nikon TE2000 microscope (Nikon, Tokyo, Japan) with 100 × magnification. The results were averaged among three independent experiments. For the cell migration assay, the procedure was similar to the cell invasion assay, except that the transwell membranes were not coated with matrigel on the upper surface. Cells were incubated for 12 hr at 37℃ in a 5% CO2 atmosphere. Migrated cells adhering to the lower filter surface were counted the same way as the cell invasion assay.
Xenograft growth evaluation
Female BALB/c athymic nude mice (4-5-weeks old) were purchased from the Experimental Laboratory Animal Center of Beijing University (Beijing, China). For all animal experiments, mice were manipulated and housed according to protocols approved by the Tianjin Medical University Medical Experimental Animal Care Commission. To establish a tumor xenograft model, a total of 1×106 H1299 cells were subcutaneously injected into mice in the left flank. After 2 weeks, when the tumors reached a size of ~62 mm3, mice were randomly distributed into three groups (five mice/group) and injected with either 5 nmol miR-146b-5p agomir (Ago-miR-146b), miRNA agomir scrambled control (Ago-miR-NC, from Ribio, Guangzhou, China), or 0.1 mL PBS buffer into the tumor mass once every 3 days for 3 weeks. Tumor diameters were measured every 3 days. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and calculated using the formula: V =(L ×W2) × 0.5 [17]. After 3 weeks, the mice were sacrificed and the tumors were weighed. The tumors isolated from mice were then preserved in 4% paraformaldehyde at 4°C for future immunohistochemical analysis.
TUNEL assay for apoptotic cells
Tissues were dissected and fixed in 4% paraformaldehyde overnight, deparaffinized, de-hydrated, and embedded in paraffin, then subjected to a TUNEL assay according to the manufacturer's instructions (Roche, New Jersey, USA). In brief, the tissue sections were incubated with Proteinase K (Qiagen, Hilden, Germany) at 37°C for 30 min, then permeabilized with permeabilization solution containing 0.1% Triton X-100 and 0.1% sodium citrate for 8 min. The DNA strand breaks were end-labeled with digoxigenin-conjugated dideoxy-UTP using terminal transferase and the nucleus was counterstained with DAPI (Sigma). The apoptotic and non-apoptotic nuclei were examined using an Olympus fluorescence microscope.
Histological Analysis
Xenografts were fixed in 4% formalin overnight, dehydrated with ethanol, and embedded in paraffin. Sections were first deparaffinized with xylol, and then rehydrated through graded alcohol solutions. After a 10 min blocking step with normal goat serum, the tissues were incubated with anti-Ki67 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at room temperature for 1 h and were subsequently washed with PBS, incubated 60 min with the horseradish peroxidase (HRP)-conjugated secondary antibody (ZSJQ Corp., Beijing, China). Finally, the sections were incubated 3 min at room temperature with 3, 3'-diaminobenzidine (DAB) and counterstained with hematoxylin.
Statistical analysis
Statistical analysis was performed using the software package SPSS version 21.0. Mann-Whitney U and Kruskal-Wallis tests were performed to determine the relationship between miRNA expression and clinicopathological parameters. Survival was estimated using the Kaplan-Meier method, and the differences in survival according to miRNA expression were compared using the log-rank test. Differences between control and miRNA transfected groups were analyzed using the Student's t-test. All P-values obtained in this study were 2-tailed, and the statistical significance level was set at P < 0.05.
Results
Characteristics of the Research Cohort
The study cohort included 52 (66.7%) male and 26 (33.3%) female NSCLC patients, aged 39 to 78 years old (median 63.5 years) (Table 2). Of these 78 patients, 32 (41.0%) had squamous cell carcinoma (SCC), 39 (50.0%) had adenocarcinoma (ADC), and 7 (9.0%) patients had other types of NSCLC. Lymph node metastasis occurred in 47 (60.3%) cases. Finally, 48 patients had stage I or II NSCLC while 30 patients had stage III or IV NSCLC.
Clinicopathological features of 78 NSCLC patients.
| Characteristic | N (%) | |
|---|---|---|
| Age | ||
| ≧63 | 39 (50.0%) | |
| <63 | 39 (50.0%) | |
| Gender | ||
| Male | 52 (66.7%) | |
| Female | 26 (33.3%) | |
| Smoking History | ||
| yes | 49 (62.8%) | |
| no | 29 (37.2%) | |
| Histology | ||
| ADC | 39 (50.0%) | |
| SCC | 32 (41.0%) | |
| Others | 7 (9.0%) | |
| TNM Stage | ||
| I+II | 48 (61.5%) | |
| III+IV | 30 (38.5%) | |
| Lymphatic Metastasis | ||
| Positive | 47 (60.3%) | |
| Negative | 31 (39.7%) | |
| Distant Metastasis | ||
| Positive | 8 (10.3%) | |
| Negative | 70 (89.7%) |
ADC, adenocarcinoma; SCC, squamous cell carcinoma; LN, lymph node
Correlation between miRNA expression and clinicopathological characteristics in NSCLC
We investigated the correlation between the expression level of miR-146b-5p, miR-128b, miR-21, miR-221, miR-34a, and Let-7a and clinicopathological characteristics of NSCLC, as shown in Table 2. As reported previously in the literature, expression of miR-21 was correlated with lymph node metastasis in NSCLC (P< 0.05, Table 3). Higher expression levels of miR-21 were found in lymphatic metastasis-positive tumors than in lymphatic metastasis-negative tumors (P=0.029). Interestingly, expression of miR-221 was correlated with TNM stage (P=0.035) and tended to be lower in advanced tumors (stage III and IV). However, other miRNAs (miR-128b, miR-146b, miR-34a and let-7a) tested were not correlated with age, gender, smoking status, or TNM stage nor lymphatic metastasis status (Table 3).
Association of clinicopathological factors with overall survival
In this study, patient follow up was by telephone call. The postoperative survival time was defined as from the date of operation to the day of follow-up or the date of patients' death. As of December 31, 2012, 26 patients survived, 30 patients had died, and 22 patients were lost for follow-up. The mean survival time was 37 months (range 4-61 months). The clinicopathological factors were selected according to the median value of continuous variables and univariate analysis was performed to identify the factors that were significantly associated with overall survival (Table 4). The results showed that histology type, TNM stage, and lymphatic metastasis were each significant prognostic factor for overall survival of NSCLC patients (log-rank=6.437, 6.126 and 4.278, respectively; P=0.040, 0.013 and 0.039, respectively). Other factors such as age, gender, and smoking status were not statistically correlated with overall survival.
Correlation of miR-128b, miR-146b, miR-21, miR-221, miR-34a, and let-7a expression with clinical characteristics in NSCLC
| Factors | n | miR-128b | miR-146b | miR-21 | miR-34a | miR-221 | Let-7a |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| ≧63 | 40 | 0.0028 | 0.0287 | 1.0049 | 0.0343 | 0.0104 | 1.0788 |
| <63 | 38 | 0.0033 | 0.0260 | 0.7466 | 0.0280 | 0.0090 | 1.0006 |
| P value | 0.842 | 0.368 | 0.150 | 0.242 | 0.368 | 0.936 | |
| Gender | |||||||
| Male | 52 | 0.0027 | 0.0208 | 1.0049 | 0.0308 | 0.0100 | 0.9964 |
| Female | 26 | 0.0033 | 0.0277 | 0.7876 | 0.0426 | 0.0111 | 1.1242 |
| P value | 0.203 | 0.135 | 0.324 | 0.141 | 0.726 | 0.141 | |
| Smoking history | |||||||
| yes | 49 | 0.0029 | 0.0230 | 0.9043 | 0.0274 | 0.0104 | 1.0046 |
| no | 29 | 0.0032 | 0.0257 | 1.0811 | 0.0384 | 0.0090 | 0.9960 |
| P value | 0.723 | 0.492 | 0.691 | 0.155 | 0.285 | 0.873 | |
| Histologic type | |||||||
| Adenoma | 38 | 0.0025 | 0.0263 | 0.7695 | 0.0327 | 0.0084 | 1.0484 |
| Others | 40 | 0.0039 | 0.0208 | 0.9229 | 0.0289 | 0.0108 | 1.0252 |
| P value | 0.395 | 0.920 | 0.826 | 0.569 | 0.054 | 0.484 | |
| TNM stage | |||||||
| I+II | 25 | 0.0034 | 0.0256 | 0.9229 | 0.0324 | 0.0108 | 1.0477 |
| III+IV | 53 | 0.0024 | 0.0229 | 0.8340 | 0.0292 | 0.0084 | 1.0002 |
| P value | 0.234 | 0.742 | 0.853 | 0.172 | 0.035* | 0.615 | |
| Lymphatic metastasis | |||||||
| Positive | 47 | 0.0032 | 0.0209 | 1.1236 | 0.0320 | 0.0090 | 1.0044 |
| Negative | 31 | 0.0029 | 0.0257 | 0.7172 | 0.0315 | 0.0107 | 1.0494 |
| p value | 0.988 | 0.742 | 0.029* | 0.795 | 0.242 | 0.679 |
Factors associated with overall survival in NSCLC
| Factors | n | Mean survival (Months) | Median survival (Months) | Univariate analysis Log-rank P value |
|---|---|---|---|---|
| Age | ||||
| ≧63 | 46 | 37.23 | 42.0 | 2.700 0.100 |
| <63 | 32 | 29.92 | 33.0 | |
| Gender | ||||
| Male | 52 | 33.67 | 38.0 | 0.170 0.681 |
| Female | 26 | 33.65 | 36.5 | |
| Smoking history | ||||
| yes | 49 | 33.16 | 34.0 | 0.753 0.385 |
| no | 29 | 34.52 | 40.0 | |
| Histologic type | ||||
| Adenoma | 38 | 35.71 | 39.0 | 6.437 0.040* |
| SCC | 33 | 33.67 | 38.0 | |
| Others | 7 | 22.57 | 29.0 | |
| TNM stage | ||||
| I+II | 25 | 37.94 | 42.0 | 6.126 0.013* |
| III+IV | 53 | 26.83 | 28.5 | |
| Lymphatic metastasis | ||||
| Positive | 47 | 29.45 | 33.0 | 4.278 0.039* |
| Negative | 31 | 40.06 | 43.0 |
Low levels of miR-146b expression are associated with poor prognosis
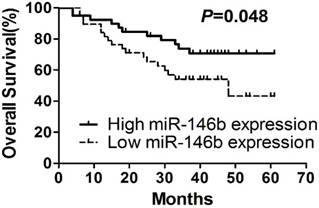
To understand the correlation between expression of those six miRNAs and patient prognosis, the 78 NSCLC patients were assigned to either the high expression group or the low expression group, according to the median expression of each individual miRNA. Kaplan-Meier survival analysis and multi-variable Cox proportional hazards analysis were performed using SPSS software. Kaplan-Meier survival analyses indicated that patients with low expression levels of miR-146b-5p had significantly shorter median and mean survival time than those with high miR-146b expression (33.00 and 30.44 months versus 42.0 and 36.90 months, respectively; log-rank test P=0.048, Figure 1A, Table 5). No significant associations were found when subgroups of all specimens were compared, although there is patients with high miR-146b expression tended to have a better survival time than patients with low miR-146b expression. Expressions of other miRNAs (miR-128b, miR-221, miR-21, miR-34a and let-7a) were not related to the overall survival of NSCLC patients (P>0.05. Suppl. Figure 1).
MiRNA expression levels associated with overall survival in NSCLC
| Factors | n | Mean survival (Months) | Median survival (Months) | Univariate analysis Log-rank P value |
|---|---|---|---|---|
| miR-128b | ||||
| High | 39 | 30.59 | 34.0 | 1.163 0.281 |
| Low | 39 | 36.74 | 42.0 | |
| miR-146b | ||||
| High | 39 | 36.90 | 42.0 | 3.899 0.048* |
| Low | 39 | 30.44 | 33.0 | |
| miR-21 | ||||
| High | 39 | 33.87 | 36.0 | 0.005 0.946 |
| Low | 39 | 33.46 | 38.0 | |
| miR-34a | ||||
| High | 39 | 33.28 | 37.0 | 0.008 0.927 |
| Low | 39 | 34.05 | 37.0 | |
| miR-221 | ||||
| High | 39 | 35.18 | 40.0 | 0.385 0.535 |
| Low | 39 | 32.15 | 34.0 | |
| Let-7a | ||||
| High | 39 | 36.05 | 38.0 | 2.218 0.136 |
| Low | 39 | 31.28 | 34.0 |
In addition, the univariate Cox hazard regression analysis demonstrated that miR-146b expression levels tended to be a significant prognostic indicator of NSCLC (adjusted hazard ratio=0.482, 95% CI: 1.409- 29.593, P=0.016). The multivariate Cox proportional hazard regression analysis also showed that miR-146b expression levels to be an independent prognostic factor for NSCLC patients (hazard ratio=0.259, 95% CI: 0.083-0.809, P=0.020, Table 6). These results indicated that miR-146b is an independent prognostic factor for overall survival and progression-free survival of NSCLC.
Kaplan-Meier survival curves. Kaplan-Meier survival curves for NSCLC patients according to miR-146b-5p expression.
Univariate and multivariate Cox hazard regression analysis.
| Characteristic | Univariate analysis Hazard Ratio (95% CI) | P value | Multivariate analysis Hazard Ratio (95% CI) | P value |
|---|---|---|---|---|
| miR-146b | 0.482 (0.229-1.014) | 0.054 | 0.259 (0.083-0.809) | 0.020* |
| miR-128b | 1.247 (0.608-2.556) | 0.547 | 1.178 (0.500-2.776) | 0.708 |
| miR-21 | 1.035 (0.505-2.117) | 0.926 | 1.755 (0.630-4.886) | 0.282 |
| miR-34a | 0.709 (0.344-1.461) | 0.351 | 1.187 (0.454-3.102) | 0.726 |
| miR-221 | 0.828 (0.404-1.697) | 0.606 | 3.007 (0.530-17.067) | 0.214 |
| Let-7a | 0.716 (0.347-1.477) | 0.366 | 0.332 (0.068-1.614) | 0.172 |
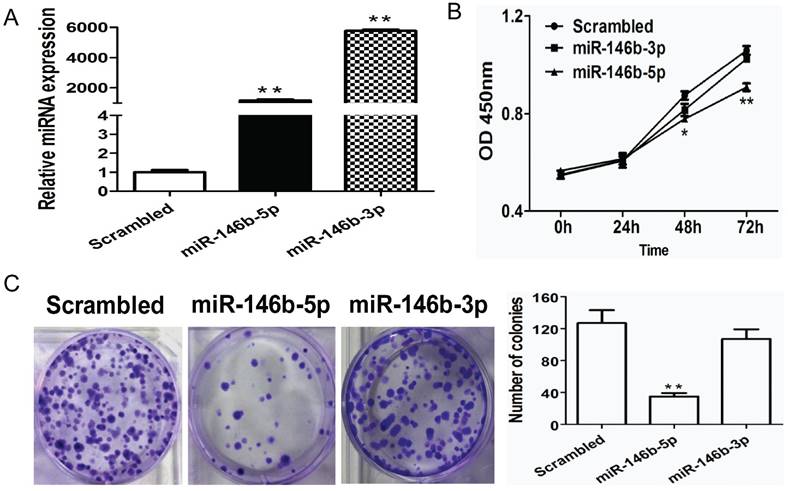
Ectopic expression of miRNA-146b-5p suppresses lung cancer cell growth in vitro
To further investigate the potential function of miR-146b in NSCLC, we evaluated the impact of miR-146b on cell proliferation. Previously, miR-146b-3p was reported to have the opposite function of miR-146b (miR-146b-5p) in NSCLC [18]; therefore, we tested both miR-146b-3p and miR-146b-5p in this study. Human NSCLC H1299 cells were infected with miR-146b-5p or miR-146b-3p mimics or a non-target scrambled oligonucleotide. As shown in Figure 2A, increased levels of both miR-146b-5p and miR-146b-3p were confirmed by real-time PCR (793-fold and 656-fold higher in the miRNA mimic-transfected group as compared to the scrambled group, P < 0.001 and P < 0.001, respectively). Next, the CCK-8 method was used to evaluate the viability of the miR-146-transfected H1299 cells. Compared to the scrambled control, proliferation rates were significantly reduced in miR-146b-5p -transfected cells at 48h and 72h (P < 0.05 and P < 0.001, respectively), whereas the effect of miR-146b-3p on proliferation was not significant (Figure 2B). To assess the functional role of miR-146b in tumor formation, colony formation was measured in H1299 cells. As showed in Figure 2C, a significant increase in the number of colonies was detected in the miR-146b-5p-transfected cells (P < 0.001), but was not affected by miR-146b-3p as compared with the scrambled control-transfected cells.
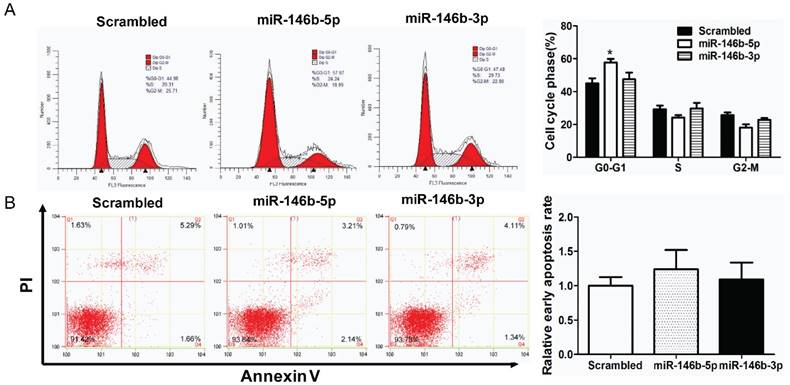
MiR-146-5p induced G0/G1 phase arrest in H1299 cell lines
Furthermore, cell cycle distribution was assessed using PI staining. As illustrated in Figure 3A, miR-146b-5p -transfection significantly increased the percentage of G0/G1-phase cells (44.98% to 57.67%, P < 0.05) indicating G0/G1 phase arrest as compared to the scrambled control-transfected H1299 lung cancer cells, whereas there was no significant difference between miR-146b-3p -transfected cells and the scrambled control-transfected group. In addition, apoptotic changes in H1299 cells after transfected with miR-146b-5p or miR-146b-3p were observed by flow cytometry. Compared to the control group, neither miR-146b-5p nor miR-146b-3p-treated groups displayed differences in the number of Annexin V positive cells (Figure 3B).
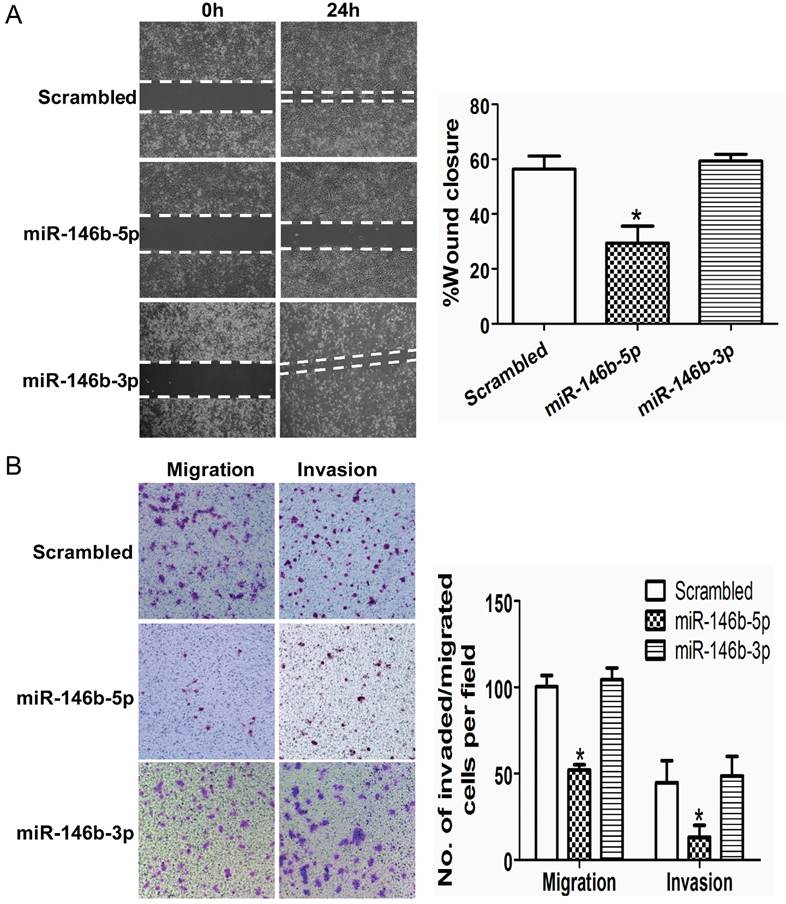
Ectopic expression of miR-146b-5p inhibits lung cancer cell migration and invasion in vitro
We next analyzed the migration and invasion capacities of cells overexpressing miR-146b-5p/3p using a transwell assay and a wound-healing assay, respectively. The results of the wound-healing assay showed that migration ability was significantly decreased with ectopic expression of miR-146b-5p in H1299 cells as compared with control cells ectopically expressing a scrambled oligonucleotide (29.38% vs. 56.34%, P < 0.01, Figure 4A). However, there was no difference between control cells and miR-146b-3p transfected cells (59.36% vs. 56.34%, P = 0.27, Figure 4A). Similarly, results from the transwell assay demonstrated that migration and invasion ability was decreased in miR-146b-5p transfected cells as compared with control cells treated with scrambled oligonucleotide (52.0 vs. 100.3, P < 0.05 for migration assays; 13.1 versus 50.5, P < 0.05 for invasion assays. Figure. 4B). There was no significant difference between the number of miR-146b-3p transfected cells and control cells that penetrated the transwell membrane. Collectively, our data demonstrate that ectopic expression of miR-146b-5p is able to suppress the migration and invasion of NSCLC cells.
MiR-146b-5p inhibits proliferation of H1299 lung cancer cells in vitro. (A) H1299 cells were transfected with miR-146b-5p, miR-146b-3p, or control scrambled oligonucleotides. miRNAs levels were analyzed using qRT-PCR in each group. (B) The effect of miR-146b-5p and miR-146b-3p on cell viability was detected using a CCK-8 assay; (C) Colony formation assays were performed to test the influence of miR-146b-5p and miR-146b-3p on the proliferation of H1299 cells. Data is presented as the mean ± SD. * indicates P < 0.05, ** indicates P <0.01 vs. the scrambled control group.
MiR-146b-5p induced G0/G1 phase arrest in H1299 cell lines. The H1299 cells were transfected with miR-146b-5p, miR-146b-3p, or control scrambled oligonucleotides respectively, and cell cycle distribution (A) and apoptosis (B) were assessed after 48 h. The data is shown as the mean ± SD. * indicates P < 0.05 vs scrambled group.
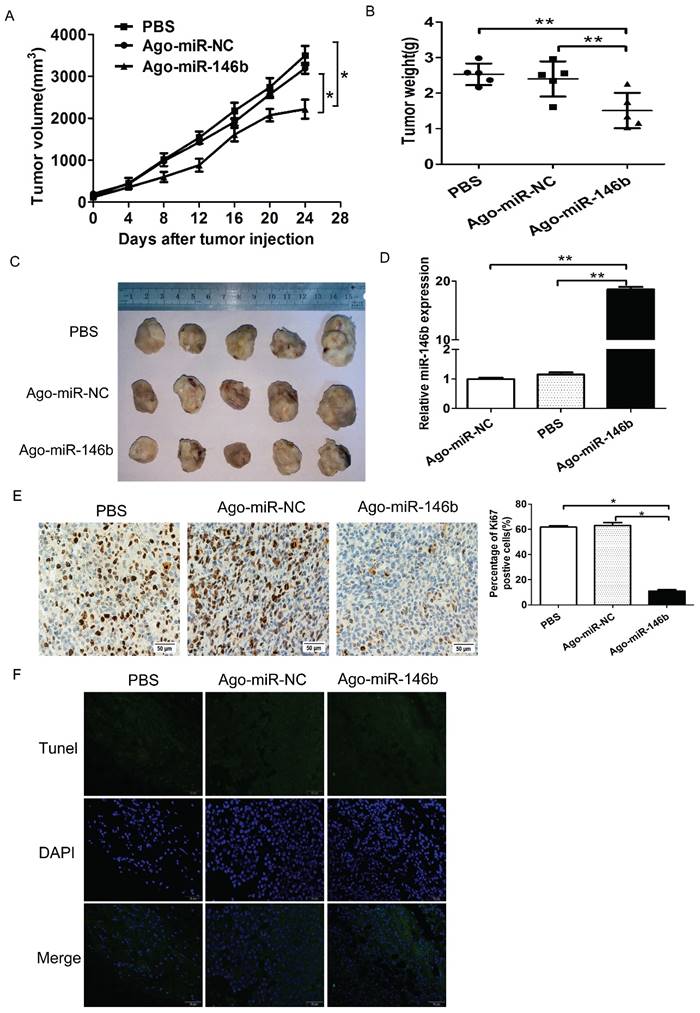
MiR-146b-5p suppresses cell growth of NSCLC cells in vivo
In order to study the effects of miR-146b-5p on the in vivo tumorigenicity of H1299 cells, we established a BALB/c mouse lung neoplasm xenograft model using H1299 cells. Then, Ago-miR-146b-5p, Ago-miR-NC, or PBS were injected into nude mice to examine tumor formation and progression. The tumors were measured every three days. After 4 weeks, the mice were sacrificed and the tumors were weighed. The tumor tissues were excised with a small surgical scissor and photographed. The results showed that mice injected with Ago-miR-146b-5p had significantly smaller in vivo tumor sizes than those injected with Ago-miR-NC or PBS (P < 0.05 and P < 0.05, respectively. Figure 5A). The final weights of tumors with Ago- miR-146b-5p group were also smaller than those from the Ago-miR-NC and PBS groups (Figure. 5B). Photographs of the representative tumors can be observed in Figure 5C. Similar trends were obtained with the weight of tumors (Figure 5C). A qRT-PCR assay confirmed higher levels of miR-145b-5p expression in the Ago-miR-146b-5p injection group (Figure 5D).
To further investigate the underlying mechanism of tumor suppression, xenograft tumor sections were analyzed to verify the expression of the proliferation marker, Ki67, using immunohistochemistry. Representative images were shown in Figure 5E. We found that the expression of Ki67 was reduced in the Ago-miR-146b-5p group compared to the Ago-miR-NC or PBS groups (P < 0.05 and P < 0.05, respectively), which were consistent with the tumor size results described above (Figure 5E). The tumor sections were also analyzed using a TUNEL assay. As shown in Figure 5F, there were no significant difference among the Ago-miR-146b-5p, Ago-miR-NC, and PBS groups. Taken together, the above results indicated that expression of miR-146b-5p inhibited lung cancer cell growth in vivo.
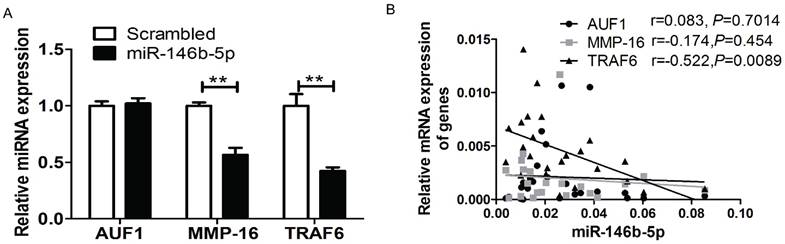
MiR-146b-5p down-regulated the expression of TRAF6, AUF1, and MMP16
It has been reported that miR-146b-5p can directly target AUF1, MMP16, and TRAF6 in several cancer cell types [19-21] including osteosarcoma, glioma, and pancreatic cancer. To verify whether miR-146b downregulates the translation of AUF1, MMP16, or TRAF6 in NSCLC cell lines, real-time PCR analysis was performed to screen the mRNA expression of three genes in both H1299 cell lines and patient tissues. Compared to the control group transfected with scrambled oligonucleotides, ectopic expression of miR-146b-5p mimics significantly reduced the mRNA level of MMP16 and TRAF6, but not AUF1 (P < 0.01, P = 0.0018 and P = 0.058, respectively. Figure 6A). Moreover, we confirmed a linear relationship between the real-time PCR 2-ΔCt values of the three genes and miR-146b-5p levels in patient tumor tissues. And we could conclude that the level of miR-146b-5p was negatively related to the expression of TRAF6 in human NSCLC tissues (r=-0.522, P=0.0089). No significant correlation was found between AUF1and MMP16 with miR-146b-5p levels (r = 0.083, P = 0.7014 and r = -0.174, P = 0.454, respectively. Figure 6B).
MiR-146b-5p inhibits migration and invasion of lung cancer cells in vitro. The H1299 cells were transfected with miR-146b-5p or miR-146b-3p or control scrambled oligonucleotides respectively. (A) A wound healing assay was used to analyze the migration activity in each group. The relative migration activity of cells is plotted on the vertical axis. (B) Transwell migration assays (left) and invasion assays (right) of H1299 cells transfected with miR-146b-5p, miR-146b-3p, or scrambled oligonucleotides. Representative images are shown on the left and quantification of five randomly selected fields is shown on the right. Values are expressed as the mean ± SD. * indicates P < 0.05 when compared with scrambled controls. Data shown are representative of three independent experiments.
MiR-146b-5p suppresses cell growth of NSCLC cells in vivo. H1299 cells were injected into nude mice and either Ago-miR-146b-5p, Ago-miR-NC, or PBS were locally injected into the tumor mass once every 3 days for 3 weeks. At 4 weeks, mice were sacrificed and tumors were removed and weighed. (A-B) Tumor volume (A) and weight (B) of Ago-miR-146b-5p, Ago-miR-NC, and PBS H1299 xenografts in nude mice (n=5 each). *P< 0.05 and **P< 0.001, as compared with the NC and PBS group, respectively. (C) Photographs of tumors isolated from mice by surgical excision on the final experiment day. (D) The expression level of miR-146b-5p in tumors isolated from the three groups of mice was determined by qPCR analysis. **P< 0.001, as compared with the NC and PBS group , respectively. (E) Immunohistochemistry assay for detecting the proliferation marker Ki-67 in xenograft tumor sections. Representative microscopic images showing Ki-67-positive cells in the Ago-miR-146b-5p, Ago-miR-NC, and PBS groups (scale bars, 50 μm.). *P< 0.05, as compared with the NC and PBS group, respectively. (F) Terminal deoxynucleotidyl transferase dUTP nick end-labeling (TUNEL) assay for detecting apoptotic cell death. Representative microscopic images showing TUNEL-positive cells in the Ago-miR-146b-5p, Ago-miR-NC, and PBS groups. Green, TUNEL; blue, DAPI. Scale bars, 50 μm.
MiR-146b-5p down-regulated the expression of TRAF6, AUF1, and MMP16. (A) Relative mRNA expression of TRAF6, AUF1, and MMP16 was detected by qRT-PCR in H1299 cell lines transfected with miR-146b-5p or scrambled control oligonucleotides. ** indicates P < 0.01 vs scrambled group. (B) Relative mRNA expression of TRAF6, AUF1, and MMP16 were detected by qRT-PCR in NSCLC tissues.
Discussion
Although lung cancer is the leading cause of cancer-related death worldwide, the prognosis for NSCLC patients remains poor. The pTNM staging system is still the most useful for predicting outcome for lung cancer patients despite its many limitations[22]. As most patients are diagnosed at a relatively late stage of disease, identifying suitable biomarkers for early diagnosis and prognosis prediction is urgently necessary. In the present study, we analyzed six miRNAs previously reported as dysregulated in cancer for expression level and correlation with clinicopathological factors and survival data from patients with NSCLC. Our results indicated a significant correlation between the expression level of miR-21 and metastasis, which consistent with the literature. In addition, miR-221 showed a significant association with clinical stage. Furthermore, miR-146b-5p was associated with prognosis in lung cancer patients and functioned as an independent prognostic factor for overall survival and progression-free survival of NSCLC. These findings suggested that miR-146b-5p, miR-21, and miR-221 may play a role in the carcinogenesis and metastasis of lung cancer. We also reported that overexpression of miR-146-5p can suppress proliferation, migration, and invasion of lung cancer cells in vitro, suggesting that miR-146b-5p acts as a tumor suppressor gene in NSCLC.
Of the six tumor-related miRNAs assessed in this study, only miR-21 expression was correlated with metastasis of tumor cells. Recent studies have revealed that miR-21 is highly expressed in many cancers, including breast cancer and lung cancer[10, 23]. Recent reports also provide functional evidence that miR-21 targets RECK and participates in cell growth, invasion, and apoptosis in prostate cancer[24]. Here in our present study, we demonstrate that patients with lymph node metastases had a significantly higher expression of miR-21 (P=0.029, Table 2), which suggests that miR-21 functions as a positive regulator of lung cancer metastasis.
Several other studies have reported miR-146b-5p as a prognosis predictor in other tumor types, but the results were contradictory[25, 26]. For example, Raponi et al. reported that mature miR-146b-5p alone was found to have the strongest prediction accuracy for stratifying prognostic groups in non-small cell lung cancer[25]. Chou CK et al. reported that miR-146b-5p is a novel prognostic factor in papillary thyroid carcinoma and patients with higher miR-146b-5p expression levels, which had significantly poorer overall[26]. Conversely, a tissue microarray experiment indicated that miR-146b-5p was a poor prognostic marker for patients with oral squamous cell carcinoma[27]. Wu PY. et al. reported that low expression of miRNA-146b-5p predicts poor outcome of large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone[28]. In this study, we found that a low level of miR-146b-5p expression is associated with poor prognosis. A further Cox proportional hazards regression model analysis indicated that miR-146b-5p was an independent prognostic indicator for patients with NSCLC.
In this study, we demonstrated that mature miR-146b-5p, but not miR-146b-3p can suppress lung cancer proliferation as indicated by decreased colony formation capacity of lung cancer cells in vitro. This conclusion is strongly supported by results from Patnaik and colleagues showing that miR-146b-5p and miR-146b-3p play opposite roles in NSCLC [18]. We further investigated the role of miR-146b-5p on cell growth of lung cancer cells in vivo; the results showed that miR-146b-5p significantly inhibited cell growth in H1299 cells. We also found that ectopic expression of mature miR-146b-5p increased the number of G0/G1-phase cells in human lung cancer cell lines. Furthermore, in vitro assays showed that ectopic expression of miR-146b-5p suppressed the migration and invasion ability of lung cancer cells. These data, corroborating reports by other investigators, indicate that miR-146b acts as a tumor suppressor in NSCLC.
Previous studies had demonstrated that miR-146b-5p inhibits pro-metastatic mesenchymal changes by negatively regulating AUF1, an inducer of mesenchymal features; down-regulation of AUF1 stabilizes the transcription factor, ZEB1, and activates the serine-threonine kinase, AKT, in osteosarcoma cells [24]. Their results indicate that miR-146b-5p may play an important role in carcinogenesis. Liu J et al. reported that TRAF6 is a direct functional target of miR-146b-5p and, further, that miR-146b-5p overexpression significantly decreased phosphorylated TAK1 and IκBα, pivotal downstream effectors of TRAF6 in human glioma[8]. Xia HP et al. reported that miR-146b-5p is involved in glioma cell migration and invasion by targeting MMPs and implicated miR-146b as a metastasis-inhibiting miRNA in glioma[21]. However, little is known regarding how miR-146b-5p regulates lung cancer development. In this study, we demonstrated that ectopic expression of mature miR-146b-5p suppressed mRNA expression of AUF1, TRAF6, and MMP6 in H1299 cell lines in vitro; we also found an inverse correlation between miR-146b-5p and TRAF6 expression in NSCLC tissues. Further investigation will be necessary to determine whether TRAF6 is the direct-target of miR-146b-5p in lung cancer cells and whether miR-146b-5p expression can be used as a novel biomarker to predict disease outcome.
Although the work presented here is limited by being a single-institution study and a relatively small sample size, our results suggest that the expression levels of miR-146b may have potential applications for more useful clinical stratification of NSCLC patients and enable selection of candidates for additional or alternative treatments.
Supplementary Material
Supplementary figure 1.
Abbreviations
NSCLC: non-small cell lung cancer; AJCC: American Joint Committee on Cancer; SGST: Shanghai Guidance of Science and Technology; qRT-PCR: quantitative real-time polymerase chain reaction.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (to Jun Chen, No. 81172233; to Hongyu Liu, No. 81372306) and the Tianjin key project of the Natural Science Foundation (to Jun Chen, 16JCZDJC34200), Tianjin Natural Science Foundation (to Hongyu Liu, 13JCYBJC22600, 16PTSYJC00160), and the Ph.D. Programs Foundation from the Ministry of Education of China (to Jun Chen, 20131202110004).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Shin HR, Bray F. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-17
2. Gadgeel SM, Ramalingam SS, Kalemkerian GP. Treatment of lung cancer. Radiol Clin North Am. 2012;50:961-74
3. Shaw AT, Yeap BY, Mino-Kenudson M. et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247-53
4. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087-92
5. Crea F, Clermont PL, Parolia A. et al. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 2014;33:1-16
6. Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768-71
7. Gilad S, Meiri E, Yogev Y. et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148
8. Liu J, Xu J, Li H. et al. miR-146b-5p functions as a tumor suppressor by targeting TRAF6 and predicts the prognosis of human gliomas. Oncotarget. 2015;6:29129-42
9. Wang P, Guo X, Zong W. et al. MicroRNA-128b suppresses tumor growth and promotes apoptosis by targeting A2bR in gastric cancer. Biochem Biophys Res Commun. 2015;467:798-4
10. Yan LX, Huang XF, Shao Q. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-60
11. Zhang Y, Zhao Y, Sun S. et al. Overexpression of MicroRNA-221 is associated with poor prognosis in non-small cell lung cancer patients. Tumour Biol. 2016;37:10155-60
12. Wei B, Huang QY, Huang SR. et al. MicroRNA34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol Med Rep. 2015;12:5255-61
13. Wang XR, Luo H, Li HL. et al. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol. 2013;15:1491-01
14. Liu H, Liang Y, Li Y. et al. Gene silencing of BAG-1 modulates apoptotic genes and sensitizes lung cancer cell lines to cisplatin-induced apoptosis. Cancer Biol Ther. 2010;9(10):832-40
15. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-8
16. Wang Y, Tang N, Hui T. et al. Identification of endogenous reference genes for RT-qPCR analysis of plasma microRNAs levels in rats with acetaminophen-induced hepatotoxicity. J Appl Toxicol. 2013;33:1330-6
17. Naito S, von Eschenbach AC, Giavazzi R. et al. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109-15
18. Patnaik SK, Kannisto E, Mallick R. et al. Overexpression of the lung cancer-prognostic miR-146b microRNAs has a minimal and negative effect on the malignant phenotype of A549 lung cancer cells. PLoS One. 2011;6:e22379
19. Al-Khalaf HH, Aboussekhra A. MicroRNA-141 and microRNA-146b-5p inhibit the prometastatic mesenchymal characteristics through the RNA-binding protein AUF1 targeting the transcription factor ZEB1 and the protein kinase AKT. J Biol Chem. 2014;289:31433-47
20. Park H, Huang X, Lu C. et al. MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J Biol Chem. 2015;290:2831-41
21. Xia H, Qi Y, Ng SS. et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158-5
22. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471-4
23. Yanaihara N, Caplen N, Bowman E. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189-98
24. Reis ST, Pontes-Junior J, Antunes AA. et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012;12:14
25. Raponi M, Dossey L, Jatkoe T. et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776-83
26. Chou CK, Yang KD, Chou FF. et al. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98:E196-05
27. Scapoli L, Palmieri A, Lo Muzio L. et al. MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int J Immunopathol Pharmacol. 2010;23(4):1229-4
28. Wu PY, Zhang XD, Zhu J. et al. Low expression of microRNA-146b-5p and microRNA-320d predicts poor outcome of large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone. Hum Pathol. 2014;45:1664-73
Author contact
![]() Corresponding authors: Jun Chen and Hongyu Liu. E-mail addresses: huntercj2004com (Jun Chen); liuhongyu123com (Hongyu Liu)
Corresponding authors: Jun Chen and Hongyu Liu. E-mail addresses: huntercj2004com (Jun Chen); liuhongyu123com (Hongyu Liu)

 Global reach, higher impact
Global reach, higher impact