3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(11):2042-2050. doi:10.7150/jca.18852 This issue Cite
Research Paper
Linc00152 promotes Cancer Cell Proliferation and Invasion and Predicts Poor Prognosis in Lung adenocarcinoma
1. Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai 200032, China
2. Department of Pathology, Obstetrics and Gynecology Hospital of Fudan University, Shanghai, 200011, China
3. Department of Respiratory Medicine, Shanghai Changzheng Hospital, Second Military Medical University, Shanghai 200003, China
4. Department of Pathology, Rui Jin Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 20025, China
*Pei-pei Zhang, Yi-qin Wang and Wei-wei Weng shared the first-authorships.
Received 2016-12-21; Accepted 2017-3-25; Published 2017-7-5
Abstract
Background: The long non-coding RNA Linc00152 stimulates tumor progression in cancer. However, its clinical significance and biological functions in lung adenocarcinoma remains unknown. We evaluate the expression of Linc00152 in lung adenocarcinoma and its possible correlation with clinicopathologic features and patient survival to reveal its biological effects in cancer progression and prognosis.
Methods: Total RNA extraction was performed on 110 pairs of lung adenocarcinoma and adjacent normal tissue samples, and then RT-qPCR was conducted. Chi-square test analysis was used to calculate the correlation between pathological parameters and the Linc00152 mRNA levels. Kaplan-Meier and Cox proportional hazards analyses were used to analyze the overall survival (OS) and disease-free survival (DFS) rates. We also detected the potential functional effects of overexpression and knockdown of Linc00152 in vitro cell proliferation, tumor cell invasion and migration, as well as in vivo nude mouse xenograft and metastasis models.
Results: The Linc00152 expression levels were higher in lung adenocarcinoma samples than in the adjacent normal tissues. Linc00152 expression levels tightly correlated with lymph node metastasis station, remote metastasis and TNM staging. The Kaplan-Meier analysis suggested that high Linc00152 expression caused significantly poorer OS and DFS rates, and a multivariate analysis revealed that Linc00152 was an independent risk factor for both DFS and OS. Overexpression of Linc00152 in lung cancer cells stimulated proliferation, tumor cell invasion and migration. Knockdown of Linc00152 inhibited cell growth and cell invasion and migration. Finally, Linc00152 knockdown inhibited lung tumor growth and tumor metastasis in nude mice models.
Conclusions: Our study suggests that Linc00152 independently predicts poor prognosis and promotes tumor progression in lung adenocarcinoma. Linc00152 needs to be considered as a potential molecular target in future cancer pharmacology.
Keywords: Linc00152, lung adenocarcinoma, prognosis, proliferation, invasion
Introduction
Lung cancer (LC) is the leading cause of cancer-related mortality in the world as the top 1 cause of cancer mortality each year in China [1]. The most common histological type of non-small cell lung carcinoma (NSCLC) is lung adenocarcinoma, accounting for 70% of NSCLC and nearly half of all lung carcinoma [2]. Identifying genes that participate in GC biology is critical to improve clinical practice.
The non-coding RNAs with sizes larger than 200 nucleotides (long non-coding RNAs, lncRNAs) are reported to play critical roles in somatic malignancies [3]. Dysregulation of lncRNA expression is associated with lung tumor tumorigenesis, recurrence and metastasis [4, 5]. Exploration of potential lncRNAs related to the pathogenesis of lung adenocarcinoma would be illuminative to future therapeutic strategies of lung cancer [6].
The non-coding RNA Linc00152 is a newly found lncRNA and has been reported to be involved in tumor initiation and progression [7]. Our former study revealed that Linc00152 promotes renal clear cell progression and predicts poor prognosis [8]. Other studies have got similar findings in gastric cancer [9] and hepatic carcinoma [10], which suggests that Linc00152 might serve as a powerful oncogenic lncRNA in somatic malignancies. However, the correlation between Linc00152 expression and tumorigenesis of lung cancer remains elusive.
In this study, we aimed to detect the expression of Linc00152 in lung adenocarcionma and its possible correlation with clinicopathologic features and prognosis to investigate the biological functions of Linc00152 on lung adenocarcinoma.
Materials and Methods
Patient population
A total of 110 patients who underwent surgical resection of primary lung adenocarcinoma at Fudan University Shanghai Cancer Center from 2009 to 2012 were retrospectively analyzed. No patients had received preoperative therapy. The resected cancer tissue and adjacent normal tissue (ANT) samples were frozen in liquid nitrogen immediately and stored at -80 °C until RNA extraction. The related data including age, gender, tumor location, size, histologic stage, lymph node status, remote metastasis and recurrence were collected from the medical record system. All patients were staged based on the TNM classification system. Patient follow-ups were performed every month during the first year after surgery and every 3 months thereafter until August 31, 2016. All patients had complete follow-up information. The Clinical Research Ethics Committee of Fudan University Shanghai Cancer Center approved the study. Written informed consent was obtained from all of the participants for the use of their tissues in the current study.
Cell lines and culture conditions
The human lung cancer cell lines, H1299, H1975, A549 and H1650 were purchased from the Fudan University IBS cell bank, Shanghai, China. All cell lines were cultured in RPMI-1640 or high glucose DMEM (Gibco, Carlsbad, CA, USA) that was supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) and 1% penicillin and streptomycin (Sigma, St. Louis, MO, USA) at 37°C in a humidified atmosphere with 5% CO2. Stable Linc00152-infecting H1299 and H1975 as well as stable Linc00152-knockdown A549 and H1650 cells were maintained with 1 µg/ml puromycin (Sigma, USA).
Plasmids and lenti-virus constructions
The full-length Linc00152 sequence was amplified by PCR from cDNA of NIH: H293 cells and then subcloned into the pcDNA3.1 (+) vector (Transheep, Shanghai, China). pHBLV-IRES-puro lenti-virus was constructed by Hansheng (Shanghai, China). The information of shLinc00152 was listed in the previous article [8].
Proliferation assays
Cells were seeded in 96- or 6-well plates 24 h prior to the experiment. After infected with indicated lenti-virus, proliferation was measured using the CCK-8 (Dojindo, Japan) and the colony-forming assays. The CCK8 assay counts living cells at different time points. Approximately 3.5×103 infected cells in 100 µl were incubated in triplicate in 96-well plates. The CCK-8 reagent (10 µl) was added to each well and incubated at 37 °C for 2 h every 24 h for 4 consecutive days. Then, we measured the optical density at 450 nm using an automatic microplate reader (Synergy4; BioTek, Winooski, VT, USA). For the colony-forming assay, 800 infected cells were seeded in 6-well plate and incubated with according culture for 14 days. Then the cells were fixed with ethanol and then stained by crystal purple solution.
Cell invasion detection
A transwell assay was used to assess cell invasion with a transwell system from Corning co. Ltd., USA. Twenty-four h after transfection, nearly 4.0×104 cells in 100 µl of serum-free medium were added to each upper chamber. Medium containing 10% fetal bovine serum was applied to the lower chamber as chemo-attractant. After a 24 h incubation at 37 °C, the non-invasive cells that were remaining on the chamber were removed with cotton-tipped swabs. The cells that migrated and adhered to the lower surface of the filter were fixed with ethanol, stained with 0.5% crystal violet, photographed at 200×, and counted at 400× magnification (BX51, Olympus, Japan).
Cell Migration Detection
A wound-healing assay was used to assess cell motility. Transfected cells were plated at equal density in 6-well plates and grown to confluent to 90%. Wounds were then scratched with a sterile pipette tip, the cells were washed twice with PBS and serum-free culture medium was added. The wound closing procedure was observed for 48 h, and images were captured every 24 h at 100× magnification (BX51, Olympus, Japan).
Total RNA Isolation and RT-qPCR
Total RNA was extracted from the tumorous and adjacent normal tissues using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Real-time (RT) and quantitative polymerase chain reaction (qPCR) kits were used to evaluate the Linc00152 expression levels from the tissue samples. The RT and qPCR reactions were conducted as previously described [11, 12]. Relative gene expression was calculated using the comparative cycle threshold (CT) (2-ΔΔCT) method. Actin was used as an endogenous control to normalize the data. The utilized Linc00152 primers were designed in a previous study [8]. The Linc00152 mRNA levels were defined as high if they were above the median value [8, 12].
In vivo nude mouse xenograft and tumor metastasis models
The Shanghai Medical Experimental Animal Care Commission approved the animal experiments. Male BALB/c-nu mice (4-5 weeks of age, 18-20 g) were maintained under specific pathogen-free conditions at the Fudan University Experimental Animal Department. All experimental procedures involving animals were undertaken in accordance with the institute's guidelines. For the tumor metastasis models, 1×106 Lenti-shNC and Lenti-shLinc00152 stably infected A549 cells were injected subcutaneously or into tail veins of mice (n=3 per group) and then were kept under observation for 28 days. All the mice were anaesthetized and sacrificed. The livers were excised and measured and fixed followed by H&E sections.
Statistical analyses
Each experiment was repeated three times, and the data are presented as the mean with error bars indicating the standard deviation. All statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). Student's t-test and one-way ANOVA analyses were used for either 2 or multiple group comparisons, respectively, for statistical significance. Correlations among the clinicoparameters and Linc00152 expressions levels were examined using the chi-square or Fisher's exact probability tests. DFS and OS curves were calculated by the Kaplan-Meier method and analyzed with the log-rank test. The DFS rates were calculated from the date of surgery to the date of disease progression (local and/or distal tumor recurrence) or to the date of death. The OS rate was defined as the length of time between the diagnosis and death or last follow-up. Variables with a value of p<0.05 in the univariate analyses were used in the multivariate analysis on the basis of the Cox proportional hazards model. All p values were 2-sided, and statistical significance was established at p<0.05.
Results
Linc00152 was upregulated in lung adenocarcinoma
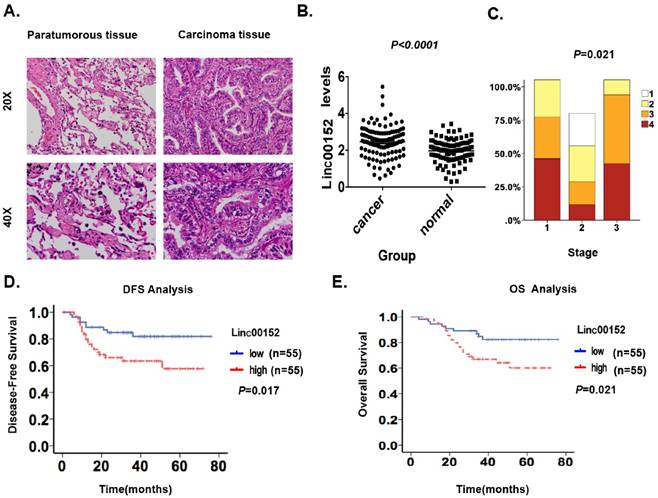
We first detected the Linc00152 levels using RT-qPCR in 110 patients with lung adenocarcinoma to confirm whether its level would be higher in malignant lesions than that in ANT. We observed significantly higher Linc00152 mRNA levels in the 110 malignant lesions than those in the ANTs (p<0.05, Fig. 1A-B), suggesting that Linc00152 could be detected in lung adenocarcinoma and its mRNA level is abnormally upregulated.
Linc00152 upregulation correlated with clinicopathologic characteristics and patient survival of lung adenocarcinoma
Second, we analyzed the correlation between Linc00152 levels and the clinicopathologic status of lung adenocarcinoma (Table 1). The Linc00152 mRNA levels were upregulated in tumors with a higher tumor burden, as defined by further lymph node metastasis station (p=0.005), more remote metastasis (p=0.002) and more advanced TNM staging (p=0.004, Fig. 1C, Table 1). However, no significant correlation was found between the Linc00152 levels and age, tumor grade or T stage only.
Next, we conducted a Kaplan-Meier analysis using the log-rank test to explore the potential influence of Linc00152 expression on patient survival. Linc00152 mRNA levels were divided into low (n=55) and high (n=55) levels based on the median value [11, 13, 14]. The total median follow-up time for the patients who were still alive at the endpoint for analysis was 76 months. The median follow-up time for the patients who were still alive at the endpoint was 37.56 months in the group with high Linc00152 expression and 30.96 months in the group with low expression.
Linc00152 was highly expressed and predicted poor prognosis in lung adenocarcinoma. A: Representative hematoxylin-eosin stained images of tumor and paratumorous tissues selected for the following Linc00152 detections by RT-qPCR. B: Statistical analytical graph of Linc00152 mRNA levels in ANT and lung adenocarcinoma tumor tissues (n=110, p<0.05). C: The correlation graph of Linc00152 with staging of lung adenocarcinoma (n=110, p<0.05). D: Kaplan-Meier disease-free survival (DFS) curve of lung adenocarcinoma patients with different Linc00152 mRNA levels (Low, n=55 vs. High, n=55). E: Kaplan-Meier disease-free survival overall survival (OS) (right) curves of lung adenocarcinoma patients with different Linc00152 mRNA levels (Low, n=55 vs. High, n=55).
Correlation between linc00152 expression and clinicopathological characteristics of lung adenocarcinoma patients
| Characteristics | Number of case | linc00152 | χ2 | P | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | 0.148 | 0.701 | |||
| ≤ 60 | 48 | 23 | 25 | ||
| > 60 | 62 | 32 | 30 | ||
| Gender | 0.587 | 0.444 | |||
| Male | 50 | 23 | 27 | ||
| Female | 60 | 32 | 28 | ||
| T stage | 1.077 | 0.783 | |||
| T1 | 48 | 24 | 24 | ||
| T2 | 52 | 27 | 25 | ||
| T3 | 6 | 3 | 3 | ||
| T4 | 4 | 1 | 3 | ||
| N stage | 12.871 | 0.005* | |||
| N0 | 60 | 36 | 24 | ||
| N1 | 17 | 11 | 6 | ||
| N2 | 32 | 8 | 24 | ||
| N3 | 1 | 0 | 1 | ||
| M stage | 10.523 | 0.002* | |||
| M0 | 57 | 37 | 20 | ||
| M1 | 53 | 18 | 354 | ||
| TNM stage | 10.671 | 0.004* | |||
| Ia + Ib | 52 | 31 | 21 | ||
| IIa + IIb | 22 | 14 | 8 | ||
| IIIa | 36 | 10 | 26 | ||
* Overall p < 0.05
The results showed that patients with high Linc00152 levels (n=55) had significantly shorter DFS (p=0.017; Fig. 1D) and OS rates (p=0.021; Fig. 1E) than those with low levels (n=55). A univariate Cox analysis showed that T stage, TNM stage, N station and Linc00152 levels correlated with the DFS and OS rates (Table 2-3). A multivariate analysis using the Cox proportional hazard model demonstrated that the Linc00152 level was an independent risk factor for DFS (p<0.001, Table 2) and OS (Table 3). Notably, the T stage, TNM staging and Linc00152 were independent risk factors for both DFS and OS (Table 2-3). These results identified that upregulated Linc00152 levels in lung adenocarcinoma predicted poor survival and might be a prognostic biomarker for the disease.
Efficiency identification of Linc00152 overexpression in GC cells
To identify the biological functions of Linc00152 in lung cancer cells, we first detected the baseline mRNA levels of Linc00152 in 5 lung cancer cell lines by RT-qPCR. The RT-qPCR showed that the Linc00152 expression was lower in both the H1299 and H1975 cells but higher in A549 and H1650 cells (Fig. 2A).
Univariate and Multivariate analysis of disease-free survival in lung adenocarcinoma
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |
| T stage | 3.088 | 1.668-5.715 | <0.001* | 2.33 | 1.243-4.365 | 0.008* |
| N stage | 3.3 | 1.861-5.850 | <0.001* | |||
| TNM stage | 4.249 | 2.261-7.988 | <0.001* | 3.514 | 1.845-6.691 | <0.001* |
| Linc00152 expression | 2.331 | 1.318-4.121 | 0.004* | 3.304 | 1.739-6.277 | <0.001* |
HR, hazard ratio; CI, confidence interval; * p <0.05.
Univariate and Multivariate analysis of overall survival in lung adenocarcinoma
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |
| T stage | 4.163 | 1.756-12.118 | 0.002* | 3.504 | 1.312-9.356 | 0.012* |
| N stage | 2.676 | 1.243-5.761 | 0.012* | |||
| TNM stage | 4.229 | 1.719-10.406 | 0.002* | 3.203 | 1.281-8.007 | 0.013* |
| Linc00152 expression | 2.454 | 1.116-5.396 | 0.025* | 3.084 | 1.241-7.666 | 0.015* |
HR, hazard ratio; CI, confidence interval; * p <0.05.
Linc00152 overexpression and knockdown in lung cancer cell lines. A: A statistical analytical graph of the baseline Linc00152 mRNA levels in 5 lung cancer cell lines, as detected by RT-qPCR. Actin was used as an endogenous control. B: A statistical analytical graph of Linc00152 mRNA levels in H1299 and H1975 cells that were transfected with pcDNA3.1-Linc00152 by RT-qPCR. *: p<0.01. C: A statistical analytical graph of Linc00152 mRNA levels in A549 and H1650 cells that were infected with shNC and shLinc00152-1 and -2 by RT-qPCR. *: p<0.01.
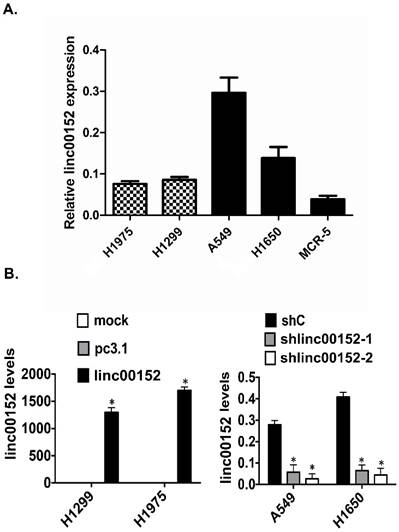
Next, we transfected pcDNA3.1 or pcDNA3.1-Linc00152 constructs into H1299 and H1975 cells for 48 h. We also infected A549 and H1650 cells with shNC and shLinc00152 shRNAs for 48 h. The following RT-qPCR showed that the Linc00152 mRNA levels were significantly increased by Linc00152 overexpression in the H1299 and H1975 cells (Fig. 2B) but decreased by shRNA in A549 and H1650 cells (Fig. 2C).
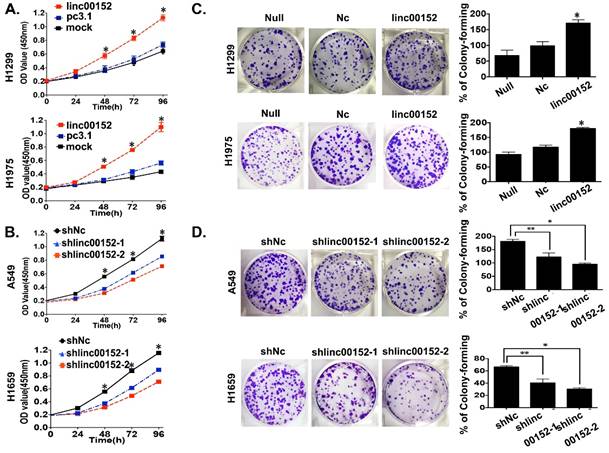
Linc00152 promotes lung tumor cell proliferation
To observe the influence of Linc00152 on tumor cell proliferation, we conducted a panel of experiments. First, the CCK8 assay results suggested that Linc00152 overexpression significantly increased the living H1299 and H1975 cells compared with the controls (p<0.01, Fig. 3A). In contrast, knockdown of Linc00152 significantly reduced the living cells in A549 and H1650 cells compared with controls (p<0.01, Fig. 3B). Next, the colony-forming assay showed more colonies in the Linc00152-overexpressing H1299 and H1975 cells than in the controls (p<0.05, Fig. 3C) but fewer colonies in the Linc00152-knockdown A549 and H1650 cells compared with controls (p<0.05, Fig. 3D). Together, these results suggested that Linc00152 promoted lung cancer cell proliferation in vitro.
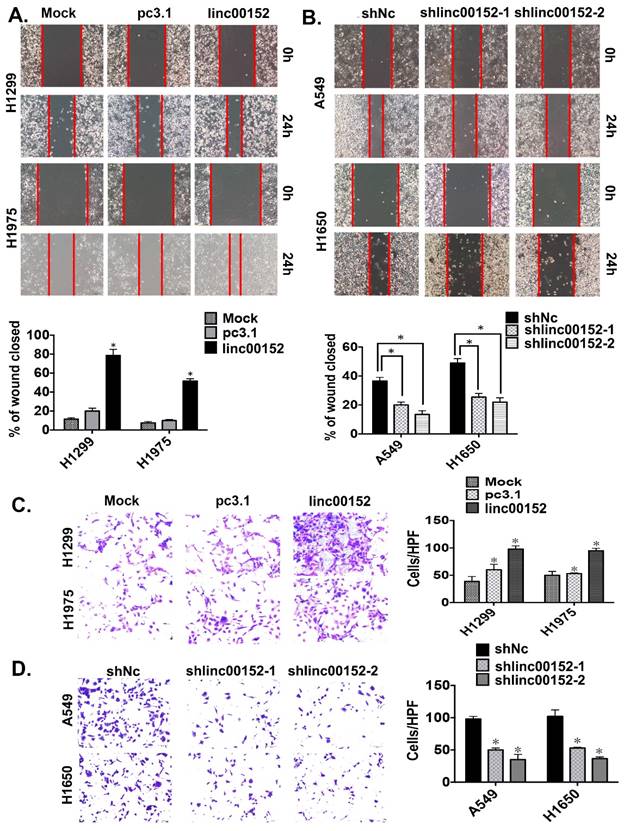
Linc00152 enhanced tumor cell migration and invasion in vitro
To determine whether Linc00152 promoted tumor cell invasion and migration in lung adenocarcinoma, we conducted transwell and wound-healing assays. Linc00152 overexpression in both H1299 and H1975 cells resulted in significantly increased cellular mobility (Fig. 4A, p<0.05). In contrast, knockdown of Linc00152 in A549 and H1650 reduced cell migration compared with controls (Fig. 4B, p<0.05). Consistently, Linc00152 overexpression in both H1299 and H1975 cells resulted in significantly increased cellular invasiveness (Fig. 4C, p<0.05). Meanwhile, knockdown of Linc00152 in A549 and H1650 reduced cell invasion compared with controls (Fig. 4D, p<0.05). Collectively, these data suggest that Linc00152 induces tumor cell migration and invasion in lung cancer cells.
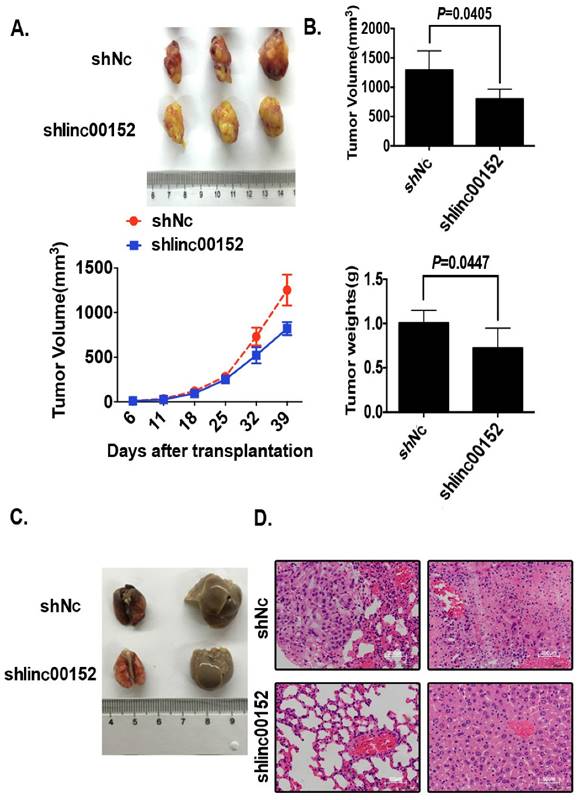
Linc00152 promoted tumor growth and metastasis in nude mice models
Finally, we investigated tumor growth and metastasis by knockdown of Linc00152 in nude mice. Knockdown of Linc00152 decelerated the tumor growth speed in nude mice models compared with controls (Fig. 5A, p<0.05). The final tumor volumes and weights were significantly lower in the group of shLinc00152 compared with controls (Fig. 5B, p<0.05). As to the metastasis models, although there were no conspicuous visible lesions in the samplings of lung and liver in both groups (Fig. 5C), the following H&E slides determined that the metastatic cancer lesions were formed microscopically in the controls but not in Linc00152-overexpressing group (Fig. 5D). Taken together, these data suggest that Linc00152 promotes tumor growth and metastasis in nude mice models.
Discussion
Our study was the first one to report the mRNA level changes and biological functions of Linc00152 in lung adenocarcinoma tissues and cell lines. We found that Linc00152 was abnormally highly expressed in lung tumor tissues compared with ANT. High Linc00152 expression was correlated with increased lung cancer malignancy and poorer prognoses. The in vitro experiments revealed that Linc00152 promoted lung cancer cell proliferation, migration and invasion. Moreover, we identified that knockdown of Linc00152 decelerated tumor growth and reduced the incidence of remote metastasis in the in vivo nude mice models.
Linc00152 is a newly found lncRNA located on 2p11.2[15, 16]. It has been reported to exert oncogenic functions in gastric cancer, breast cancer and hepatic cancer [7, 9, 15, 17]. We formerly also found that Linc00152 predicts poor prognosis and facilitate tumor progression in renal clear cell carcinoma, suggesting that Linc00152 is a general oncogene in somatic malignancies [8]. In our study, we observed abnormally upregulated Linc00152 expression in lung cancer tissues and high level of Linc00152 predicted a poor prognosis in lung adenocarcinoma, reducing both DFS and OS. Moreover, the multivariate analysis found that Linc00152 was an independent risk factor, suggesting that Linc00152 might be a novel prognostic indicator of lung adenocarcinoma. Future studies could explore the serum level of Linc00152 in patients with lung adenocarcinoma to confirm to dig its clinical value in the clinical monitoring of tumor progression and prognosis.
Linc00152 stimulated tumor cell proliferation in lung adenocarcinoma. A: The CCK8 cell counting assays revealed the living cell growth curves of H1299 and H1975 cells that were transfected with the indicated plasmids at the indicated time points. Error bars are shown. *: p<0.01. B: The CCK8 cell counting assays revealed the living cell growth curves of A549 and H1650 cells that were infected with the indicated lenti-viruses at the indicated time points. Error bars are shown. *: p<0.01. C: Representative images of the colony-forming results of H1299 and H1975 cells that were infected with the indicated lenti-virus for 14 days. *: p<0.01. D: Representative images of the colony-forming results of A549 and H1650 cells that were infected with the indicated lenti-virus for 14 days. *: p<0.01. **: p<0.05.
Previous studies have provided evidence that Linc00152 promotes tumor cell proliferation to prompt gastric cancer progression [18]. Consistently, we also found that overexpression of Linc00152 could stimulate tumor cell proliferation in vitro and facilitate tumor growth in nude mice models in vivo. What was more important was that the statistical analysis showed that Linc00152 was correlated with lymph node metastasis station, remote metastasis and TNM staging. Notably, the expression of Linc00152 was not correlated with T stage only in our study, suggesting that the tight correlation between Linc00152 expression and TNM staging in lung adenocarcinoma might be mainly due to its contribution to tumor invasion and metastasis. These clinical analyses were also supported by the in vitro and vivo experiments in our study, as overexpression of Linc00152 promoted tumor cell invasion and migration while knockdown of Linc00152 suppressed tumor cell invasion as well as metastasis in nude mice models, suggesting that Linc00152 might contribute to tumor metastasis majorly in lung adenocarcinoma. Future studies could focus on the potential mechanisms by which Linc00152 drives tumor invasion and metastasis in lung cancer.
Conclusions
Our study suggests that Linc00152 predicts poor prognoses and promotes tumor progression in lung adenocarcinoma. Linc00152 might be applied as a prognostic predictor for the prediction of patient risk.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81572254, 81602269, 81601988 and 81602078), and Shanghai Health Development Planning Commission General Project (201540280).
Linc00152 induced tumor cell migration and invasion in lung adenocarcinoma. A: Representative images (left) and quantification (right) of wound-healing assays for H1299 and H1975 cells. (Scale bars, 400 µm). *: p<0.01. B: Representative images (left) and quantification (right) of wound-healing assays for A549 and H1650 cells. (Scale bars, 400 µm). *: p<0.01. C: Representative images (left) and quantification (right) of transwell invasion assays for the H1299 and H1975 cells. (Scale bars, 50 µm). *: p<0.01. D: Representative images (left) and quantification (right) of transwell invasion assays for the A549 and H1650 cells. (Scale bars, 50 µm). *: p<0.01.
Linc00152 promoted xenograft growth and metastasis in nude mice models. A: Representative xenograft images (top) of the Lenti-shNC and Lenti-shLinc00152 H1299 groups, and the statistical analytical graph (bottom) of the xenograft tumor growth speed. The error bars are shown. *: p<0.01. B: A statistical analytical graph of the xenograft tumor weights and volumes. The error bars are shown. *: p<0.01. C: The representative gross photos of lung and liver tissues from Lenti-shNC and Lenti-shLinc00152 A549 groups. D: The representative H&E photos of lung and liver tissues from Lenti-shNC and Lenti-shLinc00152 A549 groups. Top: The scale bars=200µm. Bottom: The scale bars=50µm.
Competing Interests
The authors have declared that no competing interests exist.
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016
2. Li HM, Guo K, Yu XY, Yu Z, Xu P. Expression and clinical significance of A-kinase anchor protein 4 in lung adenocarcinoma tissue. Thorac Cancer. 2016;7:273-278
3. Yang ZG, Gao L, Guo XB, Shi YL. Roles of long non-coding RNAs in gastric cancer metastasis. World J Gastroenterol. 2015;21:5220-5230
4. Wu Y, Yu DD, Hu Y, Yan D, Chen X, Cao HX, Yu SR, Wang Z, Feng JF. Genome-wide profiling of long non-coding RNA expression patterns in the EGFR-TKI resistance of lung adenocarcinoma by microarray. Oncol Rep. 2016;35:3371-3386
5. Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, Wei M, Xu C, Wu C, Zhang Z, Gao X, Liu Z, Hou J, Huang J, Sun Y. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949-2959
6. Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z, Zhang LW. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20:2285-2295
7. Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA Linc00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441-5447
8. Wu Y, Tan C, Weng WW, Deng Y, Zhang QY, Yang XQ, Gan HL, Wang T, Zhang PP, Xu MD, Wang YQ, Wang CF. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6:285-299
9. Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112-3123
10. Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, Sun B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687-696
11. Linardou H, Kalogeras KT, Kronenwett R, Kouvatseas G, Wirtz RM, Zagouri F, Gogas H, Christodoulou C, Koutras AK, Samantas E, Pectasides D, Bafaloukos D, Fountzilas G. The prognostic and predictive value of mRNA expression of vascular endothelial growth factor family members in breast cancer: a study in primary tumors of high-risk early breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Breast Cancer Res. 2012;14:R145
12. Wang YQ, Zhang QY, Weng WW, Wu Y, Yang YS, Shen C, Chen XC, Wang L, Liu KJ, Xu MD, Sheng WQ. Upregulation of the Non-Coding RNA OTUB1-isoform 2 Contributes to Gastric Cancer Cell Proliferation and Invasion and Predicts Poor Gastric Cancer Prognosis. Int J Biol Sci. 2016;12:545-557
13. Xu MD, Qi P, Weng WW, Shen XH, Ni SJ, Dong L, Huang D, Tan C, Sheng WQ, Zhou XY, Du X. Long non-coding RNA LSINCT5 predicts negative prognosis and exhibits oncogenic activity in gastric cancer. Medicine (Baltimore). 2014;93:e303
14. Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122
15. Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol Ther. 2016
16. Yang T, Zeng H, Chen W, Zheng R, Zhang Y, Li Z, Qi J, Wang M, Chen T, Lou J, Lu L, Zhou T, Dai S, Cai M, You W, Pan K. Helicobacter pylori infection, H19 and Linc00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147-153
17. Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. Linc00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813-42824
18. Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J, Wang J, Zhang Q, Xu Z. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135
Author contact
![]() Corresponding authors: Dr. Professor C-F Wang, Department of Pathology, Rui Jin Hospital, Shanghai Jiao Tong University School of Medicine, 197, Rui Jin Er Road, Shanghai 200025, China. E-mail: wang_chaofucom; Tel: +86-21-64175590; Fax: +86-21-64174774. And Dr. M-D Xu, Department of Pathology, Fudan University, Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, China. Email: xumd27202003com; Tel: +86-21-64175590.
Corresponding authors: Dr. Professor C-F Wang, Department of Pathology, Rui Jin Hospital, Shanghai Jiao Tong University School of Medicine, 197, Rui Jin Er Road, Shanghai 200025, China. E-mail: wang_chaofucom; Tel: +86-21-64175590; Fax: +86-21-64174774. And Dr. M-D Xu, Department of Pathology, Fudan University, Shanghai Cancer Center, 270 Dong'an Road, Shanghai 200032, China. Email: xumd27202003com; Tel: +86-21-64175590.

 Global reach, higher impact
Global reach, higher impact