Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(13):2561-2574. doi:10.7150/jca.19972 This issue Cite
Research Paper
LHX6, An Independent Prognostic Factor, Inhibits Lung Adenocarcinoma Progression through Transcriptional Silencing of β-catenin
1. Institute of Toxicology, College of Preventive Medicine, Third Military Medical University, Chongqing 400038, PR China;
2. Department of emergency, Yan'an Hospital, Kunming Medical University, Kunming 650500, PR China.
Received 2017-3-7; Accepted 2017-6-10; Published 2017-8-2
Abstract
Introduction: Our previous study identified LIM homeobox domain 6 (LHX6) as a frequently epigenetically silenced tumor-suppressor gene in lung cancer. However, its clinical value has never been evaluated, and the in-depth anti-tumor mechanism remains unclear.
Methods: Public database was used for lung cancer, lung adenocarcinoma and lung squamous carcinoma patients and tissue microarray data was used for lung adenocarcinoma patients to study prognostic outcome of LHX6 expression by Kaplan-Meier and Cox-regression analysis. In vitro proliferation, metastasis and in vivo nude mice model were used to evaluate the anti-tumor effect of LHX6 on lung adenocarcinoma cell lines. The mechanisms were explored using western blot, TOP/FOP flash assays and luciferase reporter assays. LHX6 expression and clinical stages data were collected from The Cancer Genome Atlas database (TCGA).
Results: Expression of LHX6 was found to be a favorable independent prognostic factor for overall survival (OS) of total lung adenocarcinoma patients (P=0.014) and patients with negative lymph nodes status (P=0.014) but not related the prognostic outcome of lung squamous cell carcinoma patients. The expression status of LHX6 significantly correlated to histological grade (P<0.01), tumor size (P=0.026), lymph node status (P=0.039) and clinical stages (P<0.01) of lung adenocarcinoma patients. Functionally, LHX6 inhibited the proliferation and metastasis of lung adenocarcinoma cells in vitro and in vivo. Furthermore, LHX6 suppressed the Wnt/β-catenin pathway through transcriptionally silencing the expression of β-catenin, and the promoter region (-1161 bp to +27 bp) was crucial for its inhibitory activity.
Conclusions: Our data indicate that the expression of LHX6 may serve as a favorable prognostic biomarker for lung adenocarcinoma patients and provide a novel mechanism of LHX6 involving in the tumorigenesis of lung adenocarcinoma.
Keywords: LHX6, lung adenocarcinoma, prognostic factor, β-catenin.
Introduction
Lung cancer, one of the most common malignancies, is the primary cause of cancer-related death worldwide [1, 2]. Among the different types of lung cancers, non-small cell lung cancer (NSCLC), primarily comprising lung adenocarcinoma and lung squamous cell carcinoma, constitutes over 80% of lung cancer patients [3, 4]. Lung adenocarcinoma is the most frequent type of NSCLC and accounts for approximately 40% of lung cancer [5]. Although numbers of novel therapies have been applied in the clinic, the five-year survival rate of lung cancer patients is lower than 20% [6, 7]. The undesirable prognosis of lung cancer is largely due to its high mortality rate and aggressive phenotypes. Recently, various genetic and epigenetic alterations have been shown to drive this process in lung cancer and even one gene may indicate adverse prognostic outcomes in lung adenocarcinoma and lung squamous cell carcinoma patients [8-10]. Thus, identification of new functional genes and biomarkers in different types of lung cancer may elucidate the mechanism of lung tumorigenesis and provide alternative approaches to improve cancer therapy and diagnosis.
The human genome contains at least 12 LIM homeobox (LHX) genes encoding LIM homeodomain transcription factors. These genes usually have a LIM domain in addition to a homeo domain [11]. Studies have shown that LHX6 plays important roles in organ development and oncogenesis [12-17]. Hypermethylation of LHX6 serves as a sensitive biomarker in early diagnosis of head and neck carcinomas and cervical cancer [11, 18]. We have previously reported that LHX6 is a novel tumor suppresser which is epigenetically silenced by DNA methylation in lung cancer, but whether it is related to the clinical progression has yet to be evaluated [17]. A recent study has demonstrated that LHX6 suppresses breast cancer progression by interfering the Wnt/β-catenin pathway. However, the in-depth regulatory mechanism has not been investigated [16].
It is well established that Wnt/β-catenin signaling helps maintain cancer stem cells and is a putative stem cell marker [19, 20]. Abnormal activation of this pathway is a crucial oncogenic step in tumor initiation and progression [21, 22]. Wnt/β-catenin signaling is normally over-activated in the tumorigenesis process through accumulation of β-catenin (the key mediator of this signaling pathway) present in the cytoplasm which is then subsequently translocated into the nucleus where it forms a complex with members of the T-cell factor (TCF)/Lymphoid enhancer-binding factor (LEF) family of transcription factors, leading to the activation of downstream targets genes, such as the oncogenes Cyclin D1, MMP7 and c-Myc [19, 23, 24]. Previous studies have indicated that Wnt/β-catenin signaling is frequently activated with promoted lung cancer cell proliferation and metastasis [20, 25-27]. However, the mechanisms leading to the abnormal activation of the Wnt/β-catenin signaling still remains unclear in lung cancer.
In the present study, we found that LHX6 expression was an independent favorable prognostic factor and tightly associated with clinical parameters, which indicated that LHX6 might be a potential prognostic biomarker for lung adenocarcinoma patients. Furthermore, LHX6 suppressed tumor growth and metastasis by transcriptional silencing the expression of β-catenin, and this provided a novel mechanism of the tumorigenesis process of lung adenocarcinoma.
Materials and Methods
Cell lines and patient samples
The human lung cancer cell lines SPC-a-1, 95D, LTEP-a-2, A549, H358 and an immortalized HBE cell line were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China). All cells were cultured in the corresponding medium (recommended by the suppliers) supplemented with 10% fetal bovine serum, and maintained at 37 °C in an incubator with 5% CO2. For expression assay, 15 matched tumor and adjacent non-tumor lung cancer patients' samples were obtained from the Southwest Hospital in Chongqing, China. For the tissue microarray analysis, a total of 88 primary lung adenocarcinoma tissues were obtained from cancer patients who had undergone surgical resection with curative intent between 2004 and 2009 at the Southwest Hospital in Chongqing, China. The clinicopathological information was retrieved from the patients' electronic medical records including age, gender, tumor size, histological grade, lymph node status (negative or positive) and clinical stage (defined according to American Joint Committee on Cancer. 7th edition) and follow-up information (5-10 years) for overall survival rates. This study was approved by the ethics committee of the Southwest Hospital Affiliated with Third Military Medical University, and all experiments were carried out in accordance with approved guidelines of Third Military Medical University. Informed consent was signed by all of the recruited patients.
Reverse-transcription polymerase chain reaction (RT-PCR) and real-time qPCR
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RT-PCR and real-time quantitative PCR analyses were performed as previously described [28], and primer sequences are listed in Supplementary Table 1.
Antibodies and western blot
WB was performed as previously described [17]. After incubation with the secondary antibody, the proteins were detected by chemiluminescence (Millipore Germany). The primary antibodies used in this study were anti-LHX6 (1:1000; Santa Cruz Biotechnology), anti-CTNNB1 (1:700; Santa Cruz Biotechnology), anti-MMP7 (1:1000; Abcam), anti-c-Myc (1:1000; Santa Cruz Biotechnology), anti-CCND1 (1:1000; Santa Cruz Biotechnology) and anti-GAPDH (1:2000; Beyotime China).
Tissue microarray analysis (TMA) generation
All samples from lung adenocarcinoma patients were reviewed histologically after hematoxylin and eosin staining. To construct the TMA slides, two cores were taken from each representative tumor tissues (within a distance of 20 mm). The tissues were stained with hematoxylin-eosin and then reviewed histologically by at least two pathologists. Finally, the TMAs were constructed (in collaboration with Shanghai Biochip Company Ltd, Shanghai, China) as previously described [8].
Immunohistochemical (IHC) analysis and scoring
IHC staining was performed using an antibody against LHX6 (1:500; Abcam) as described previously [29]. A double-blind method was used to analyze IHC results by randomly choosing three visual fields from different areas of each specimen. The immunostaining was considered positive when ≥10% of the tumor cells was immunoreactive, and samples were scored as previously described [8]. Comprehensive score = staining percentage × intensity. The LHX6 expression ≤4 was denoted as low expression, and >4 were scored as high expression, according to the median of 88 lung adenocarcinoma patients.
Construction of LHX6 mi-RNA expression vectors
Negative control-miRNA and targeting LHX6-miRNA were generated by Invitrogen and the sequences are: LHX6-miRNA forward strand 5'-3' TGCTGAGAAGCAGGCGAAGCAGGCCAGTTTTGGCCACTGACTGACTGGCCTGCCGCCTGCTTCT (The italic indicated mature mi RNAi sequence targeting the open reading frame region of human LHX6); LHX6-miRNA reverse strand 5'-3' CCTGAGAAGCAGGCGGCAGGCCAGTCAGTCAGTGGCCAAAACTGGCCTGCTTCGCCTGCTTCTC; Negative control-miRNA forward strand 5'-3' TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT (The italic sequence indicated mature control-mi-RNA); Negative control-miRNA reverse strand 5'-3' CCTGAAATGTACTGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCGCAGTACATTTC. Each miRNA oligo was cloned into the pcDNA6.2™-GW/ EmGFP-miR expression vector as previously described [30, 31] and then sequenced for following experiments.
Cell viability assay
Briefly, cells were seeded in 96-well plates at a density of 4000 cells/well and transfected with LHX6-overexpression or Vector control and LHX6-miRNA or Control-miRNA respectively. The absorbance was determined one day after cells were plated to confirm an identical number of cells within groups. After 1-4 days of transfection, cell viability was evaluated by with a Cell Proliferation Reagent MTS (Promega) according to the manufacturer's instructions.
Generation of stable cell lines
The LHX6 over-expression or knockdown cell lines were generated as previously described [28]. The stably transfected cells were screened with G418 (Calbiochem, La Jolla, CA, USA) or Blasticidin (Invitrogen Preservation). Single clones were obtained by the cylinder method. Several positive clones were confirmed by WB and then mixed for subsequent experiments.
Cell migration and invasion assay
For the migration assay, cells (4×104/ chamber) were resuspended in 300 µL of serum-free medium to avoid the effect of proliferation and seeded in the upper Transwell chamber (8 µm pore size, BD Biosciences). For the invasion assay, cells in serum-free medium were placed into the upper chamber of an insert coated with Matrigel (BD Biosciences). For each assay, medium supplemented with 20% fetal bovine serum was added into the lower chamber. After incubation for 24 h at 37 °C, non-migrated or non-invaded cells on the upper membrane were removed with a cotton swab. Cells that had migrated or invaded through the membrane were stained with 0.1% crystal violet. The experiment was performed in triplicate wells.
Colony formation assay
The colony formation assay was conducted as previously described [28]. Briefly, LTEP-a-2 and SPC-a-1 cells were plated in 6-well plates at a concentration of 2.5×105 cells per well. For knockdown, LTEP-a-2-LHX6 stably cells were plated in 6-well plates at a concentration of 2×105 cells per well. After culturing for 24 h, the cells were transfected with LHX6-overexpression or Vector control and LHX6-miRNA or Control-miRNA, respectively. After 48 h of transfection, cells were collected, diluted 1:3, plated in 6-well plates and selected with 0.8 mg/mL of G418 or 2 μg/mL of Blasticidin for 14 days to establish stable clones in which the plasmids had stably integrated into genomic DNA. Surviving colonies (>50 cells per colony) stained with crystal violet (Sigma-Aldrich, St Louis, MO, USA) were counted.
Animal experiments
BALB/c-nude mice (4 weeks of age, 18-20 g) were purchased from the Center of Experimental Animals of Third Military Medical University, China and housed in a sterile environment. Eight 4-week-old male BALB/c-nude mice (half males and half females) were randomly divided into two groups (n=4/group). LTEP-a-2 cells (8×106) stably expressing LHX6 or control vector were injected subcutaneously into the right flanks of the nude mice. The tumor volume was determined using the equation V = 0.5 × D × d2 (V, volume; D, longitudinal diameter; d, latitudinal diameter) every 3 days. The mice were sacrificed after 28 days for the following experiments. For the tumorigenesis experiments, tumors were dissected and weighed. To evaluate metastasis, livers were dissected to observe the metastatic sites after H&E staining and to quantify the metastasis of the human cancer cells with qPCR. All experimental animal procedures were approved by the Institutional Animal Care and Use Committee of Third Military Medical University, China.
Luciferase and TOP/FOP flash reporter assays
The previously reported promoter region of full-length human β-catenin (CTNNB1) promoter (Catenin-promoter-full: -2760/+27) [32] and two overlapping regions, Catenin-promoter-1 (-2760/-1161) and Catenin-promoter-2 (-1952/+27), were amplified and cloned into a pGL3-basic vector (Promega, Madison, WI, USA). LTEP-a-2 and SPC-a-1 cells were transfected with the pGL3-CTNNB1-promoters together with pIRES2-EGFP-LHX6 or control vector and pRLTK-Luc (Renilla-TK-luciferase vector, Promega) to normalize the transfection efficiency. After 36 h, the activities of firefly luciferase and renilla luciferase were measured in triplicate using the fluorescence microplate reader measurement system Varioskan LUX (Thermo Fisher, Waltham, MA, USA) with a Dual-luciferase reporter kit (Promega) as previously described [9]. For the TOP and FOP flash assay, LTEP-a-2 and SPC-a-1 cells were plated into 24-well plates at a concentration of 2.0×104 cells per well. Cells were co-transfected with 300 ng of either TOP flash (T-cell factor reporter plasmid) or FOP flash (mutant T-cell factor reporter plasmid) expression plasmids (Millipore, Temecula, CA, USA) and 300 ng of pIRES2-EGFP-LHX6 or control vector and 20 ng pRL-TK. Luciferase activity was detected as described above. The experiment was performed in triplicate wells for three times.
Analysis of publicly available datasets
To analyze the effect of LHX6 expression on prognosis of lung cancer patients, we generated Kaplan-Meier survival curves of lung cancer patients with low or high expression of LHX6 using Kaplan-Meier Plotter (www.kmplot.com/analysis) [33]. Specifically, “Lung cancer” at the upper-right corner of the home page of www.kmplot.com/analysis was selected, and “LHX6” was entered into the Gene symbol. The 2015 release of the database was used, auto select best cut-off was chosen in the analysis by clicking “Auto select best cutoff”, and then, “Draw Kaplan-Meier plot” was clicked. The patients were automatically split into low LHX6 and high LHX6 groups according to the LHX6 expression values determined by the microarray.
To analyze the expression status of LHX6 in lung cancer patients and normal samples and the expression correlation between LHX6 and CTNNB1, we first used The UCSC Cancer Genomics Browser (https://genome-cancer.soe.ucsc.edu/proj/site/hgHeatmap/) to analyze the results and further downloaded the raw data from TCGA data base. The TCGA lung cancer (LUNG) RNAseq (IlluminaHiSeq; n=1124), TCGA lung adenocarcinoma (LUAD) RNAseq (IlluminaHiSeq; n=571) and TCGA lung squamous carcinoma (LUSC) RNAseq (IlluminaHiSeq; n=553) data sets were downloaded and analyzed.
Statistical analysis
Statistical analyses were performed using SPSS 16.0 software (SPSS, Inc., Chicago, IL). Data are expressed as the means ± standard deviation (SD). Student's t-test or Mann-Whitney U test was used to compare means between groups. Kaplan-Meier survival plots and the Cox regression methods were used to compare the survival outcome between the two LHX6 expression groups. Pearson χ2 test was applied to analyze the relationship between LHX6 expression and clinical-pathological parameters, except the histological grade and clinical stages classification which were analyzed by Mann-Whitney U test. Pearson correlation was used to analyze the expression relation. P-value <0.05 was considered to denote statistical significance.
Results
LHX6 expression is significantly down-regulated in lung cancer tissues
Our previous work identified LHX6 as an epigenetically silenced gene in lung cancer [17]. To further investigate its clinical values in lung cancer, we first analyzed the expression status of LHX6 in lung tumor tissues. The results showed that LHX6 was down regulated in tumor tissues compared with adjacent non-tumor tissues (Fig. 1A). To further confirm this observation, the expression pattern of LHX6 was subsequently analyzed in lung cancer patients from The Cancer Genome Atlas (TCGA) database. LHX6 expression was significantly down-regulated in TCGA lung cancer as well as in lung adenocarcinoma and lung squamous carcinoma tissues compared with normal lung tissues (P<0.01) (Fig. 1B). Furthermore, LHX6 was down-regulated in lung cancer cell lines compared with a normal human bronchial epithelial (HBE) cell line both at mRNA and protein level (Fig. S1). These results suggested that LHX6 was down-regulated in lung cancer, indicating a role in lung cancer development.
LHX6 is down-regulated in lung tumor tissues. (A) Analysis of LHX6 expression levels in tumor and adjacent tissues of lung cancer patients using immunohistochemistry. LHX6 was down regulated in lung tumor tissues compared with adjacent tumor tissues. Scale bars represent 50 μm. (B) Down-regulation of LHX6 in lung cancer patients was further confirmed using public data sets in TCGA database. LHX6 mRNA expression in the TCGA lung cancer (LUNG) RNAseq (IlluminaHiSeq; n=1124), TCGA lung adenocarcinoma (LUAD) RNAseq (IlluminaHiSeq; n=571) and TCGA lung squamous carcinoma (LUSC) RNAseq (IlluminaHiSeq; n=553) data sets. Error bars indicate s.d. ** P<0.01.
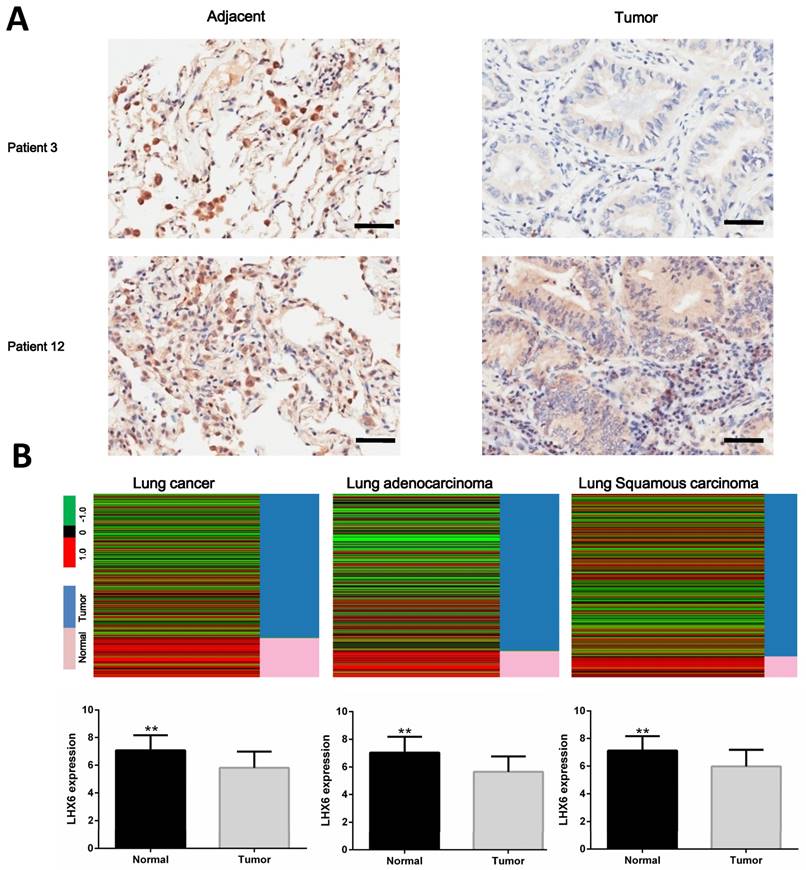
Relationship between LHX6 expression and overall survival (OS) of lung cancer patients retrieved from public datasets. (A) Kaplan-Meier survival analysis of LHX6 expression in lung cancer patients (n=1926). High expression of LHX6 predicted longer OS of lung cancer patients compared with low LHX6 expression. (B) Kaplan-Meier survival analysis of LHX6 expression in lung adenocarcinoma patients (n=720). High expression of LHX6 predicted longer OS of lung adenocarcinoma patients compared with low LHX6 expression. (C) Kaplan-Meier survival analysis of LHX6 expression in lung squamous carcinoma patients (n=525). The expression status of LHX6 was not correlated with the OS of lung squamous carcinoma patients.
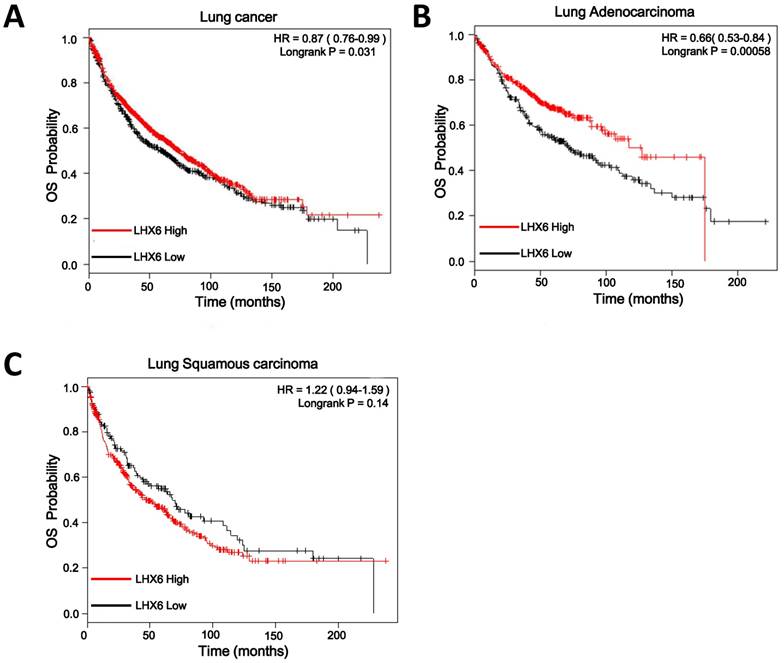
Low expression of LHX6 is significantly associated with poor clinical outcomes in lung adenocarcinoma patients
To investigate the clinical significance of LHX6, we first performed a meta-analysis to explore the association between LHX6 expression and prognostic outcomes in 1928 lung cancer patients using the Kaplan-Meier Plotter (KMplotter) software program (www. kmplot.com). The results showed that low expression of LHX6 was significantly associated with poor prognostic outcome in lung cancer patients (n=1926, Hazard ratio (HR)=0.87, 95% confidence interval (CI)=0.76-0.99, P=0.031) (Fig. 2A) by univariate analysis. A subgroup analysis based on histological types revealed that LHX6 was associated with overall survival (OS) of lung adenocarcinoma patients (n=720, HR=0.66, 95% CI =0.53-0.84, P<0.001) (Fig. 2B) but not of lung squamous carcinoma patients (n=525, HR=1.22, 95% CI=0.94-1.59, P=0.14) (Fig. 2C). These results indicated that LHX6 may be a favorable prognostic factor only in lung adenocarcinoma patients.
To confirm this observation, we conducted IHC on a TMA containing 88 lung adenocarcinoma tissues from patients with OS clinical statistics. After IHC, we used the scoring system to consolidate the results by the intensity and positive staining percentage of tumor cells. Based on the results, the staining was quantified and classified into two groups: high and low (Fig. S2). Survival analysis using the Kaplan-Meier method and log rank test showed that high expression of LHX6 presented a longer survival time than that of low LHX6 expression in total lung adenocarcinoma patients and patients with negative lymph nodes (Fig. 3 A and B). To correct the bias caused by univariate analysis, LHX6 expression as well as other parameters were examined in a multivariate Cox-regression analysis (after adjusting for gender, age, histological grade, clinical stage, tumor size and lymph node). In addition to clinical stage (HR=1.888, P=0.000), LHX6 expression was found to be an independent prognostic factor for OS of patients (HR=0.390, P=0.014) and the patients with negative lymph nodes (HR=0.212, P=0.014) in lung adenocarcinoma (Fig. 3 C and D). We further investigated the relationship between LHX6 expression and clinical parameters and found that LHX6 expression was evidently correlated with histological grade (n=88, P=0.006), tumor size (n=88, P=0.026) and lymph node status (n=84, P=0.039) of lung adenocarcinoma patients (Table S2). The incidence of LHX6 high expression was 31.57% (12/38) in lymph node-negative patients but 13.04% (6/46) in lymph node-positive patients and we found that LHX6 was down-regulated in two independent lung adenocarcinoma cohorts from TCGA data sets with metastatic lung cancer (Fig. S3 A and B), these results suggested that LHX6 may be involved in lung tumor metastasis. Furthermore, by analyzing a TCGA lung adenocarcinoma cohort with tumor stage data, LHX6 expression was found to be significantly related to the clinical stages (n=513, P=0.004) (Table S3).
High expression of LHX6 predicts a longer survival time in lung adenocarcinoma patients. (A) Kaplan-Meier survival analysis of LHX6 expression in lung adenocarcinoma patients. Long OS was observed in the high LHX6 expression group compared with the low LHX6 group. (B) Kaplan-Meier survival analysis of LHX6 expression in lung adenocarcinoma patients with metastasis-negative lymph nodes. Long OS was observed in the high LHX6 expression group compared with the low LHX6 group. (C) Multivariate Coxregression analysis of the relationship between LHX6 expression and OS of lung adenocarcinoma patients. LHX6 was determined to be an independent favorable prognostic factor. (D) Multivariate Coxregression analysis of the relationship between LHX6 expression and OS of lung adenocarcinoma patients with a metastasis-negative lymph node status. LHX6 was determined to be an independent favorable prognostic factor.
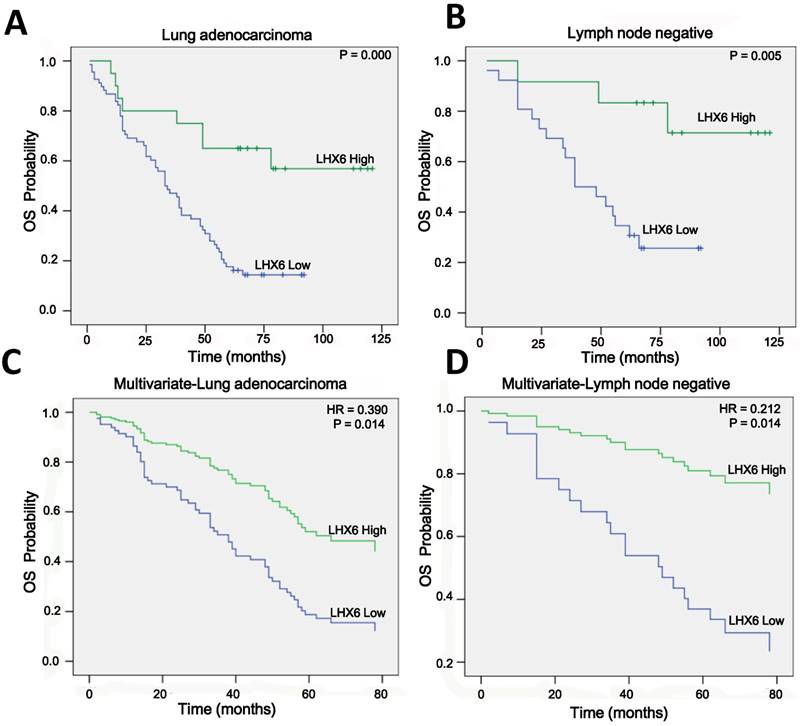
LHX6 suppresses tumor cell proliferation, migration and invasion. (A) Transfectants of the control vector and LHX6 were identified by WB in LTEP-a-2 and SPC-a-1 cells. (B) MTS assays were performed to examine the effect of LHX6 overexpression on the relative number of viable cells based on the absorbance. (C) Transwell assays were used to examine the effect of LHX6 on cell migration and invasion in LTEP-a-2 and SPC-a-1 cells. (D) Knockdown of LHX6 in LTEP-a-2-LHX6 stably cells was identified by WB. (E) MTS assays were used to examine the effect of LHX6 knockdown on the relative number of viable cells based on the absorbance. (F) Transwell assays were performed to examine the effect of LHX6 knockdown on cell migration and invasion in LTEP-a-2-LHX6 stably cells. Error bars indicate s.d. (n=3).* P<0.05; ** P<0.01.
LHX6 inhibits lung adenocarcinoma cell proliferation, migration and invasion in vitro
To investigate the role of LHX6 in lung adenocarcinoma progression, we established a gain-of-function cell model by transfecting an LHX6-expression vector into two lung adenocarcinoma cell lines, LTEP-a-2 and SPC-a-1, and the expression of exogenous LHX6 was confirmed by WB (Fig. 4A). We then examined the effects of LHX6 overexpression on cell proliferation, migration and invasion. Using a 4-days growth curve analysis, ectopic expression of LHX6 was found to significantly inhibit the proliferation of LTEP-a-2 and SPC-a-1 cells (Fig. 4B). The suppressive effect on lung adenocarcinoma cell growth was further confirmed by colony formation assays (Fig. S4A). We further investigated the effect of LHX6 on lung adenocarcinoma cell metastasis. Transwell assays with or without matrix gel showed that LHX6 significantly suppressed the migration and invasion ability of LTEP-a-2 and SPC-a-1 cells (Fig. 4C). To further confirm the role of LHX6 in lung adenocarcinoma cell growth and metastasis. We knocked down LHX6 expression in the LTEP-a-2-LHX6 stably cell line with LHX6-miRNA. LHX6 expression was significantly reduced in the cells transfected with LHX6-miRNA as shown by WB (Fig. 4D). Knockdown of LHX6 markedly enhanced cell viability and colony formation ability compared with the control group (Fig. 4E and Fig. S4B). Furthermore, knockdown of LHX6 reversed the inhibitory effect on cell metastasis of LTEP-a-2-LHX6 stably cells (Fig. 4F). Collectively these data suggested that LHX6 serves as a tumor suppressor by inhibiting tumor cell proliferation, migration and invasion in lung adenocarcinoma.
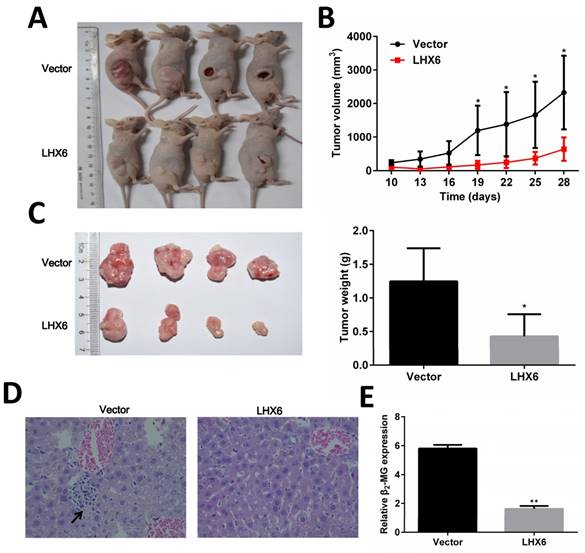
LHX6 inhibits tumor formation and metastasis in nude mice
To determine the anti-tumorigenic and anti-metastasis role of LHX6 in vivo, LTEP-a-2-LHX6 stably and vector control cells were subcutaneously injected into nude mice. The tumor volume was significantly smaller in the LHX6 over expression group compared with the control group (Fig. 5 A and B). The mean tumor weight was lower in nude mice injected with LHX6-overexpressing cells than in vector control mice (n=4, P=0.033) (Fig. 5C). Next, the liver metastasis was determined by both tissue observation using H&E staining and quantification of human-specific β2-MG (beta-2-microglobulin) [34] (Fig. 5 D and E). The results revealed that the nude mice of the LHX6 over expression group demonstrated a significantly reduced liver metastasis compared with the empty vector control mice. These data indicated that LHX6 had a significant effect on impeding tumor growth and metastasis, supporting LHX6 as a tumor suppressor in lung adenocarcinoma in vivo.
LHX6 inhibits tumor formation and metastasis in nude mice. (A) Control vector and LTEP-a-2-LHX6 cells stably expressing LHX6 (8 × 106) were subcutaneously injected into the right flank of nude mice. Pictures of BALB/c-nude mice and solid tumor tissues were taken after 4 weeks. (B) The tumor growth curve of LHX6-overexpressing cells was compared with vector control cells. (C) Tumor weights in the vector control and LHX6 groups were determined. Error bars indicate s.d. (n=4). (D) Liver metastases were observed by H&E staining in the control group but not in the LHX6 group. Arrows indicate the metastatic loci. (E) Liver metastasis was further quantified using RT-qPCR. Human-specific β2-MG levels were used to quantify metastatic human cancer cells with the mouse-specific β2-MG level as an internal control. The nude mice that were injected with LHX6-overexpressing cells demonstrated a significantly lower number of metastatic cancer cells in the liver compared with those that were injected with an empty control. Error bars indicate s.d. (n=4).* P<0.05; ** P<0.01.
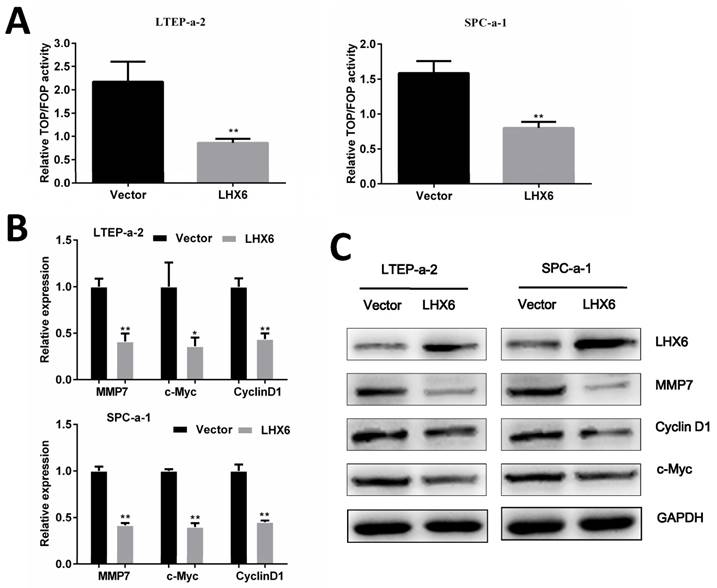
LHX6 represses the Wnt/β-catenin pathway by transcriptional silencing of β-catenin
We next explored the possible mechanism by which LHX6 suppresses lung adenocarcinoma progression. A TOP/FOP flash reporter assay showed that LHX6 significantly inhibited the transcriptional activity of β-catenin/T-cell factor (TCF) compared with the vector control transfectants in LTEP-a-2 and SPC-a-1 cell lines (Fig. 6A). We subsequently analyzed the expression of the downstream genes of the Wnt/β-catenin pathway and found that LHX6 decreased the mRNA and protein level of Cyclin D1, c-Myc and MMP7 (Fig. 6 B and C).
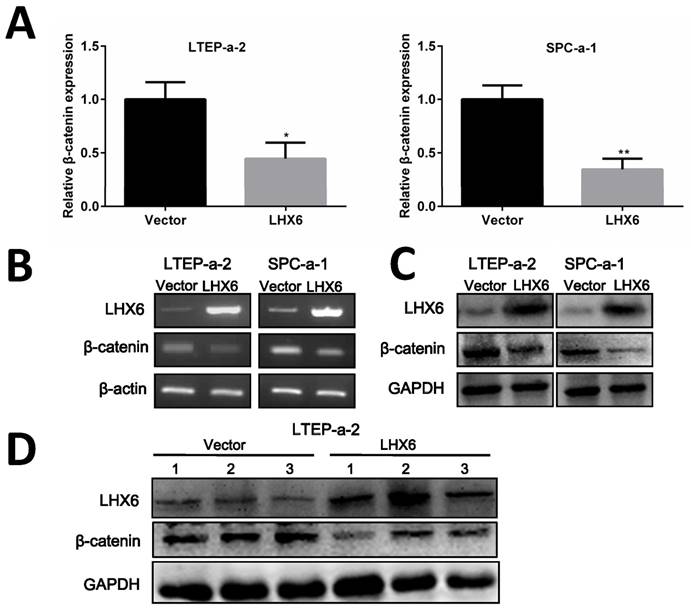
Next, we thought to determine the possible mechanism by which LHX6 could interrupt Wnt/β-catenin signaling. As an important signal transmitter involved in the Wnt/β-catenin pathway, the expression of β-catenin was first detected both at mRNA and protein level and the results showed that β-catenin was significantly down-regulated by LHX6 overexpression (Fig. 7A-C). This observation was further confirmed in the xenografted tumors derived from the nude mice (Fig. 7D). We further referred to an online data base to confirm whether LHX6 expression was reversely correlated with β-catenin expression. Interestingly, a reversed correlation was only found in the TCGA lung cancer patients (n=1124 P=0.038 R=-0.053) and lung adenocarcinoma patients (n=571 P=0.045 R=-0.071) (Fig. S5). However, we failed to observe this correlation in lung squamous carcinoma patients (n=553 P=0.272 R=0.026) (Fig. S5). These results indicated that LHX6 exerted its tumor suppressor function through down regulation of β-catenin in lung adenocarcinoma.
To further confirm that β-catenin was responsible for the suppressive effect of LHX6, a recovery assay was performed by re-introducing β-catenin into the LTEP-a-2-LHX6 cells. β-catenin re-expression reversed the inhibitory effect of LHX6 on cell proliferation and metastasis (Fig. S6). These data indicated that LHX6-mediated β-catenin down-regulation plays a crucial role in inhibiting lung adenocarcinoma cell proliferation and metastasis.
LHX6 suppresses the Wnt/β-catenin pathway. (A) LHX6 decreased the transcriptional activity of the Wnt/β-catenin pathway, as determined by a TOP flash/FOP flash reporter luciferase activity assay in LTEP-a-2 and SPC-a-1 cell lines. TOP and FOP flash plasmids were co-transfected with control vector or LHX6 and PRL-TK plasmids in LTEP-a-2 and SPC-a-1 cells. (B, C) LHX6 inhibited the expression of Wnt/β-catenin pathway target genes at both the mRNA and protein levels. Error bars indicate s.d. (n=3). * P<0.05; ** P<0.01.
LHX6 inhibits the expression of β-catenin. (A, B) Ectopic expression of LHX6 significantly reduced the expression of β-catenin on the mRNA level. The mRNA expression was examined by RT-qPCR and RT-PCR with β-actin as an internal control. Error bars indicate s.d. (n=3). * P<0.05; ** P<0.01. (C) Overexpression of LHX6 significantly decreased the expression of β-catenin on the protein level; GAPDH was used as an internal control. (D) LHX6 decreased the endogenous protein expression of β-catenin in xenografted tumors, as detected by WB.
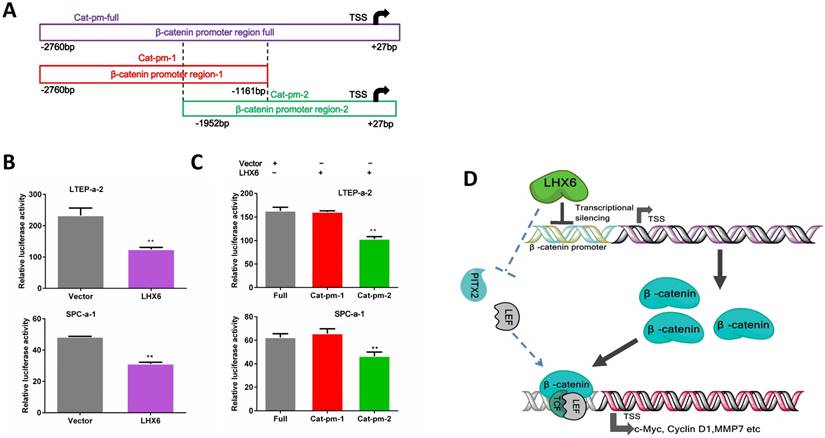
We next further investigated the possible mechanism by which LHX6 suppressed the expression of β-catenin. As a transcription factor, we first considered whether LHX6 could transcriptional silence the expression of β-catenin. An online database showed 42 predicted binding sites for LHX6 on the promoter region of β-catenin (Fig. S7). We next performed a luciferase assay. Over expression of LHX6 significantly suppressed the transcription of β-catenin (Fig. 8 A and B). To further confirm this observation, the full promoter region of β-catenin was divided into two overlapping regions, and LHX6 inhibited the luciferase reporter activity only when the catenin-promoter-2 was included, which indicated that the region ranging from -1161bp to +27bp was crucial for the inhibitory activity of LHX6 (Fig. 8C). Taken together, these results indicated that LHX6 suppresses lung adenocarcinoma cell proliferation and metastasis by transcriptional silencing β-catenin (Fig. 8D).
Discussion
In the present study, we investigated for the first time the clinical significance of LHX6 expression in lung cancer patients. Using IHC, we found that LHX6 was down-regulated in lung cancer patients, and these results were further confirmed by analyzing TCGA data base with larger patient samples. Moreover, LHX6 expression was an independent favorable prognostic factor in lung adenocarcinoma patients but not in lung squamous carcinoma patients, and its expression was tightly associated with clinical stages, histological type, tumor size and lymph node status of lung adenocarcinoma patients. In addition, our data indicated that LHX6 inhibited the proliferation and metastasis of lung adenocarcinoma cells in vitro and in vivo. Furthermore, we showed that LHX6 exerted anti-tumor activity via transcriptional silencing the expression of β-catenin.
LHX6 transcriptionally represses the expression of β-catenin. (A) The schema of different regions of the β-catenin promoter (Catenin-promoter-full, -1 and -2) were constructed into a luciferase reporter vector. The region in the dotted line indicates the overlap region of Catenin-promoter-1 and -2. TSS indicates the transcription start site. (B) LHX6 inhibited the luciferase activity in LTEP-a-2 and SPC-a-1 cells transfected with Catenin-promoter-full. (C) LHX6 inhibited the luciferase activity only when the Catenin-promoter-2 region was co-transfected into LTEP-a-2 and SPC-a-1 cells. This result indicated that the region ranging from -1161bp to +27bp is crucial for the inhibitory activity of LHX6. Error bars indicate s.d. (n=3). ** P<0.01. (D) Schematic diagram of the mechanisms of LHX6-mediated suppression of lung adenocarcinoma cell proliferation and metastasis based on our study and previous studies. LHX6 transcriptionally silences the expression of β-catenin, which subsequently inhibits the expression of Wnt/β-catenin pathway genes; the pathway shown with the dotted lines indicates the possible mechanism reported in mice odontogenesis, and this mechanism needs further investigation in a cancer model.
Previous studies have reported that different histological subtypes of lung cancer showed different molecular tumorigenesis events and prognostic outcomes [35-37], indicating the importance of characterizing new biomarkers separately in different subtypes of lung cancer. Our data showed that LHX6 expression was a favorable prognostic factor in lung adenocarcinoma but not in lung squamous carcinoma patients. Recent studies have reported that SOX30 was a favorable prognostic factor in lung adenocarcinoma but an unfavorable one in lung squamous carcinoma [8], and GPC5 was also found to be related to the clinical progression only in lung adenocarcinoma patients [9]. Furthermore, our clinical study suggested that LHX6 was an independent prognostic factor for lung adenocarcinoma patients and for patients with negative lymph node status. This observation demonstrated that LHX6 is a potential prognostic marker for lung adenocarcinoma patients.
Wnt/β-catenin signaling is a conserved molecular pathway involved in regulating a variety of physiological and pathological processes, including cell-fate determination, cell behavior and organ development [38-40]. Abnormal activation of Wnt/β-catenin signaling, an important oncogenic step in cancer progression [20, 27], is largely dependent on the fate of β-catenin. Previous studies have indicated different suppression mechanisms of β-catenin in the inhibition of the Wnt/β-catenin pathway in cancer progression, such as promotion of the degradation of β-catenin in breast cancer [41], by retention of β-catenin in the cytoplasm in lung cancer [25], physical interaction with β-catenin to impede Wnt/β-catenin signaling in colon carcinoma cells [42] and impede the translocation of β-catenin into the nucleus [9]. In that case, even though a recent study has found that LHX6 could inhibit breast cancer proliferation by interfering Wnt/β-catenin pathway, the exact regulation mechanism was not further explored [16]. In our study, we found that LHX6 inhibited the canonical Wnt/β-catenin signaling by transcriptional silencing the expression of β-catenin in lung adenocarcinoma. Our data indicated that the promoter region ranging from -1161bp to +27bp was crucial for the inhibition of β-catenin mediated by LHX6, and further, by bioinformatics analysis, we found three potential binding sites (CTCAATTAGG, CCTAATTGAC and AATAACTAAT) in this promoter region, and our future studies will try to determine the exact binding sequence. In addition to studies in cancer, a previous study has demonstrated that LHX6 could transcriptional silence the expression of PITX2 and LEF1 in the regulation of odontogenesis in mice [43], and studies have indicated that PITX2 and LEF1 function in the Wnt signaling pathway by recruiting and interacting with β-catenin to activate target genes [44, 45]. Together with our results, it can be concluded that LHX6 might be a conservative suppressor of the Wnt/β-catenin pathway not only in normal tissue development but also in tumor progression. Furthermore, because previous studies have indicated important roles of LHX6 in head and neck carcinomas and cervical cancer [11, 18], our present work may provide clues for the future mechanism investigation of these cancers.
Paradoxically, some studies have reported that over-expression of β-catenin predicted a prolonged survival time for NSCLC patients [46, 47], while others have reported completely adverse results [48, 49]. Interestingly, we found a reversed correlation between LHX6 and β-catenin expression in the lung adenocarcinoma patient data sets but not in lung squamous carcinoma patient data (Fig. S5), and low expression of β-catenin was found to predict a favorable OS in lung adenocarcinoma but an unfavorable OS in lung squamous carcinoma patients (Fig. S8). Accordingly, our data showed that LHX6 overexpression down regulated the expression of β-catenin and predicted a longer survival time in lung adenocarcinoma patients. These observations further consolidated our results that LHX6 suppressed lung adenocarcinoma progression via down-regulation of the expression of β-catenin. Furthermore, this observation could provide a possible explanation to the different prognostic results of LHX6 expression in lung adenocarcinoma and lung squamous carcinoma. Importantly, since the molecular tumorigenesis events vary in different histological subtypes of lung cancer [35, 37], the complex relationship between LHX6, β-catenin and their prognostic outcome in lung squamous carcinoma patients remains an interesting question. Our future studies may focus on this issue.
In summary, our study indicates for the first time that LHX6 expression is an independent favorable prognostic factor in lung adenocarcinoma patients and is tightly associated with tumor progression. This may provide a new prognostic biomarker for lung adenocarcinoma patients and a target for new therapies to treat lung adenocarcinoma. Furthermore, we showed that LHX6 inhibits the Wnt/β-catenin pathway by transcriptional silencing the expression of β-catenin. This not only adds to our current knowledge of the tumorigenesis process in lung adenocarcinoma but also provides clues for future studies of LHX6 in other types of cancer.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The authors thank the Institute of Cancer, Southwest Hospital for providing human lung cancer tissues. This work was supported by the National Natural Science Foundation of China [No. 81502551, 81172714 and 8157311].
Authors' contributions
JY, FH and JL conceived the study; JY, FH and WL designed the experiments. JY, YH, XH and MZ collected the patients' samples and conducted the experiments; XJ, LY and HC analyzed the online database; JY and FH interpreted the data and wrote the manuscript; HZ and JL supervised the study. JC and JL provided with valuable advices, and proofread the manuscript. All authors read and approved the final manuscript.
Email addresses
Juntang Yang: juntangyang@qq.com;
Fei Han: han16897723@163.com;
Wenbin Liu: wenbinliutmmu@sina.com; Mingqian Zhang: 2289462942@qq.com;
Yongsheng Huang: yongshengh@126.com; XianglinHao: 851479330@qq.com;
Xiao Jiang: 48644522@163.com;
Li Yin: yingli198609@163.com;
Hongqiang Chen: chenhongqiang1989@126.com; Jia Cao: caojia1962@126.com.
Ethics
This study was approved by the ethics committee of the Southwest Hospital, affiliated with Third Military Medical University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65:87-108
2. Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. Journal of the National Cancer Institute. 2014;106:djt356
3. Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nature reviews Cancer. 2010;10:760-74
4. Ridge CA, McErlean AM, Ginsberg MS. Epidemiology of lung cancer. Seminars in interventional radiology. 2013;30:93-8
5. Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clinics in chest medicine. 2011;32:605-44
6. Neri S, Yoshida J, Ishii G, Matsumura Y, Aokage K, Hishida T. et al. Prognostic impact of microscopic vessel invasion and visceral pleural invasion in non-small cell lung cancer: a retrospective analysis of 2657 patients. Annals of surgery. 2014;260:383-8
7. Wanders R, Steevens J, Botterweck A, Dingemans AM, Reymen B, Baardwijk A. et al. Treatment with curative intent of stage III non-small cell lung cancer patients of 75 years: a prospective population-based study. European journal of cancer. 2011;47:2691-7
8. Han F, Liu W, Xiao H, Dong Y, Sun L, Mao C. et al. High expression of SOX30 is associated with favorable survival in human lung adenocarcinoma. Scientific reports. 2015;5:13630
9. Yuan S, Yu Z, Liu Q, Zhang M, Xiang Y, Wu N. et al. GPC5, a novel epigenetically silenced tumor suppressor, inhibits tumor growth by suppressing Wnt/beta-catenin signaling in lung adenocarcinoma. Oncogene. 2016;35:6120-31
10. Lv S, Xue J, Wu C, Wang L, Wu J, Xu S. et al. Identification of A Panel of Serum microRNAs as Biomarkers for Early Detection of Lung Adenocarcinoma. Journal of Cancer. 2017;8:48-56
11. Jung S, Jeong D, Kim J, Yi L, Koo K, Lee J. et al. Epigenetic regulation of the potential tumor suppressor gene, hLHX6.1, in human cervical cancer. International journal of oncology. 2011;38:859-69
12. Cesario JM, Landin Malt A, Deacon LJ, Sandberg M, Vogt D, Tang Z. et al. Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2. Human molecular genetics. 2015;24:5024-39
13. Kim J, Yeon J, Choi SK, Huh YH, Fang Z, Park SJ. et al. The Evolutionarily Conserved LIM Homeodomain Protein LIM-4/LHX6 Specifies the Terminal Identity of a Cholinergic and Peptidergic C. elegans Sensory/Inter/Motor Neuron-Type. PLoS genetics. 2015;11:e1005480
14. Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T. et al. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron. 2014;82:350-64
15. Zhou C, Yang G, Chen M, He L, Xiang L, Ricupero C. et al. Lhx6 and Lhx8: cell fate regulators and beyond. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2015;29:4083-91
16. Hu Z, Xie L. LHX6 inhibits breast cancer cell proliferation and invasion via repression of the Wnt/beta-catenin signaling pathway. Molecular medicine reports. 2015;12:4634-9
17. Liu WB, Jiang X, Han F, Li YH, Chen HQ, Liu Y. et al. LHX6 acts as a novel potential tumour suppressor with epigenetic inactivation in lung cancer. Cell death & disease. 2013;4:e882
18. Estecio MR, Youssef EM, Rahal P, Fukuyama EE, Gois-Filho JF, Maniglia JV. et al. LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene. 2006;25:5018-26
19. Takahashi-Yanaga F, Kahn M. Targeting Wnt Signaling: Can We Safely Eradicate Cancer Stem Cells? Clinical Cancer Research. 2010;16:3153-62
20. Teng Y, Wang XW, Wang YW, Ma DX. Wnt/beta-catenin signaling regulates cancer stem cells in lung cancer A549 cells. Biochem Bioph Res Co. 2010;392:373-9
21. Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789-99
22. Ma L, Wang X, Jia T, Wei W, Chua MS, So S. Tankyrase inhibitors attenuate WNT/beta-catenin signaling and inhibit growth of hepatocellular carcinoma cells. Oncotarget. 2015;6:25390-401
23. Crawford HC, Fingleton BM, Rudolph-Owen LA, Goss KJ, Rubinfeld B, Polakis P. et al. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883-91
24. Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422-6
25. Chen X, Meng J, Yue W, Yu J, Yang J, Yao Z. et al. Fibulin-3 suppresses Wnt/beta-catenin signaling and lung cancer invasion. Carcinogenesis. 2014;35:1707-16
26. Chen X, Song X, Yue W, Chen D, Yu J, Yao Z. et al. Fibulin-5 inhibits Wnt/beta-catenin signaling in lung cancer. Oncotarget. 2015;6:15022-34
27. Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M. et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51-62
28. Han F, Liu W, Jiang X, Shi X, Yin L, Ao L. et al. SOX30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene. 2015;34:4391-402
29. Han F, Dong Y, Liu WB, Ma XX, Shi RH, Chen HQ. et al. Epigenetic Regulation of Sox30 Is Associated with Testis Development in Mice. PloS one. 2014:9
30. Kobayashi M, Aida M, Nagaoka H, Begum NA, Kitawaki Y, Nakata M. et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22375-80
31. Mhyre AJ, Marcondes AM, Spaulding EY, Deeg HJ. Stroma-dependent apoptosis in clonal hematopoietic precursors correlates with expression of PYCARD. Blood. 2009;113:649-58
32. Li Q, Dashwood WM, Zhong X, Al-Fageeh M, Dashwood RH. Cloning of the rat beta-catenin gene (Ctnnb1) promoter and its functional analysis compared with the Catnb and CTNNB1 promoters. Genomics. 2004;83:231-42
33. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer (vol 8, e82241, 2013). PloS one. 2014:9
34. Lupberger J, Kreuzer KA, Baskaynak G, Peters UR, le Coutre P, Schmidt CA. Quantitative analysis of beta-actin, beta-2-microglobulin and porphobilinogen deaminase mRNA and their comparison as control transcripts for RT-PCR. Molecular and cellular probes. 2002;16:25-30
35. McDoniels-Silvers AL, Nimri CF, Stoner GD, Lubet RA, You M. Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8:1127-38
36. McDoniels-Silvers AL, Stoner GD, Lubet RA, You M. Differential expression of critical cellular genes in human lung adenocarcinomas and squamous cell carcinomas in comparison to normal lung tissues. Neoplasia. 2002;4:141-50
37. Pao W, Girard N. New driver mutations in non-small-cell lung cancer. The Lancet Oncology. 2011;12:175-80
38. Ille F, Sommer L. Wnt signaling: multiple functions in neural development. Cellular and molecular life sciences: CMLS. 2005;62:1100-8
39. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781-810
40. Wang Y, Li YP, Paulson C, Shao JZ, Zhang X, Wu M. et al. Wnt and the Wnt signaling pathway in bone development and disease. Frontiers in bioscience. 2014;19:379-407
41. Tan ZY, Zheng HQ, Liu XX, Zhang WH, Zhu JR, Wu GY. et al. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/beta-catenin signaling in breast cancer. Oncotarget. 2016;7:24076-87
42. Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SCJ. et al. Sox17 and Sox4 Differentially Regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802-15
43. Zhang Z, Gutierrez D, Li X, Bidlack F, Cao H, Wang J. et al. The LIM homeodomain transcription factor LHX6: a transcriptional repressor that interacts with pituitary homeobox 2 (PITX2) to regulate odontogenesis. The Journal of biological chemistry. 2013;288:2485-500
44. Amen M, Liu X, Vadlamudi U, Elizondo G, Diamond E, Engelhardt JF. et al. PITX2 and beta-catenin interactions regulate lef-1 isoform expression. Mol Cell Biol. 2007;27:7560-73
45. Amendt BA, Liu XM, Amen M, Engelhardt JF. Wnt signaling regulates lymphoid enhancer factor (Lef-1) isoform expression through functional interactions between PITX2, beta-catenin and Lef-1. Faseb Journal. 2007;21:A656-A
46. Hommura F, Furuuchi K, Yamazaki K, Ogura S, Kinoshita I, Shimizu M. et al. Increased expression of beta-catenin predicts better prognosis in nonsmall cell lung carcinomas. Cancer. 2002;94:752-8
47. Kren L, Hermanova M, Goncharuk VN, Kaur P, Ross JS, Pavlovsky Z. et al. Downregulation of plasma membrane expression/cytoplasmic accumulation of beta-catenin predicts shortened survival in non-small cell lung cancer. A clinicopathologic study of 100 cases. Ceskoslovenska patologie. 2003;39:17-20
48. Xu C, Xie D, Yu SC, Yang XJ, He LR, Yang J. et al. beta-Catenin/POU5F1/SOX2 transcription factor complex mediates IGF-I receptor signaling and predicts poor prognosis in lung adenocarcinoma. Cancer Res. 2013;73:3181-9
49. Xu X, Sun PL, Li JZ, Jheon S, Lee CT, Chung JH. Aberrant Wnt1/beta-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2011;6:716-24
Author contact
![]() Corresponding authors: Professor Huidong Zhang or Professor Jinyi Liu. 30 Gaotanyan Street, Shapingba District, Chongqing 400038, PR China E-mail: huidong.zhangcom or jinyiliutmmucom
Corresponding authors: Professor Huidong Zhang or Professor Jinyi Liu. 30 Gaotanyan Street, Shapingba District, Chongqing 400038, PR China E-mail: huidong.zhangcom or jinyiliutmmucom

 Global reach, higher impact
Global reach, higher impact