3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(13):2626-2635. doi:10.7150/jca.19381 This issue Cite
Research Paper
CDC27 Induces Metastasis and Invasion in Colorectal Cancer via the Promotion of Epithelial-To-Mesenchymal Transition
1. Sun Yat-sen University Cancer Center; State Key Laboratory of Oncology in South China; Collaborative Innovation Center for Cancer Medicine, Guangzhou;
2. Department of Hematology/Oncology, Guangzhou Women and Children's Medical center, Guangzhou Medical University, Guangzhou, Guangdong;
3. Guangdong Lung Cancer Institute, Guangdong General Hospital and Guangdong Academy of Medical Sciences, Guangzhou.
* These authors contributed equally to this work
Received 2017-1-28; Accepted 2017-5-22; Published 2017-8-21
Abstract
Distant metastasis is the primary cause of cancer-related death among patients with colorectal cancer (CRC), and the discovery of novel therapeutic targets by further exploring the molecular mechanisms of CRC metastasis is therefore urgently needed. We previously illustrated that CDC27 overexpression promoted proliferation in CRC, but no studies have emphasized the role of CDC27 in cancer metastasis thus far. Our previous data indicated that the expression of CDC27 was significantly associated with distant metastasis in patient tissues, and therefore, in this study, we focused on the investigation of the potential mechanisms of CDC27 in CRC metastasis. The results revealed that CDC27 promoted the metastasis, invasion and sphere-formation capacity of DLD1 cells, but that the inhibition of CDC27 in HCT116 cells suppressed metastasis both in vitro and in vivo. Mechanistic analyses revealed that CDC27 promoted epithelial-to-mesenchymal transition (EMT), as demonstrated by the reduced expression of the epithelial markers ZO-1 and E-cadherin and the enhanced expression of the mesenchymal markers ZEB1 and Snail in HCT116 and DLD1 cells. Further mechanistic investigation indicated that CDC27 promoted metastasis and sphere-formation capacity in an ID1-dependent manner. In conclusion, we first demonstrated the role of CDC27 in cancer metastasis and showed that CDC27 may serve as a promising therapeutic target for CRC.
Keywords: CDC27, colorectal cancer, metastasis, EMT, ID1
Introduction
Distant metastasis is the leading cause of cancer-related death among colorectal cancer (CRC) patients [1].The majority of CRC patients with hepatic metastasis cannot be cured with surgical therapy, and the 5-year survival rate following pathological diagnosis is below 10% [2]. In contrast, the 5-year survival rate of patients with early-stage CRC is higher than 90% [3-5]. Understanding the molecular mechanisms that lead to the metastatic phenotype of CRC, along with the subsequent discovery of novel therapeutic targets, is therefore urgently needed to develop efficient medical approaches that improve the prognosis of CRC patients with metastasis.
Cancer metastasis is a complex multistep biological process [6]. Compelling evidence indicates that epithelial-to-mesenchymal transition (EMT) plays crucial roles in promoting cancer progression through various mechanisms [7,8]. EMT endows cells with migratory and invasive properties, contributes to the acquisition of stem cell properties, and prevents cell apoptosis and senescence [9]. Therefore, the mesenchymal state is associated with the capacity of cells to migrate to distant tissues, facilitating their subsequent differentiation into multiple cell types during the multiple steps of metastasis.
Over the past few years, accumulating evidence has revealed that CDC27 mutates in various cancer types, such as prostate cancer, osteosarcoma, and CRC [10-13]. There is new evidence that CDC27 is upregulated in breast cancer and that CDC27 expression may be a significant predictor of breast cancer recurrence [14]. In our previous study, we reported that downregulation of CDC27 inhibits the proliferation of CRC cells via the accumulation of p21, and that CDC27 expression is significantly correlated with tumor progression and poor survival among CRC patients. Moreover, we also performed immunohistochemistry (IHC) to evaluate endogenous CDC27 expression in paraffin-embedded tissue sections (n=166) derived from cases of histopathologically confirmed CRC. The results implied that CDC27 expression is significantly correlated with distant metastasis in patients with CRC (p=0.03) [15]. However, no studies have reported the role of CDC27 in cancer metastasis thus far.
ID1 is a helix-loop-helix (HLH) protein that heterodimerizes with the basic HLH transcription factors and inhibits them from binding to DNA, thereby regulating gene transcription [16,17]. ID1 is involved in regulating a variety of cellular processes including the cell cycle, apoptosis, senescence, and differentiation [18,19]. Previous studies have confirmed that ID1 promotes metastasis and EMT in various cancer types, such as lung cancer and esophageal cancer [20-22]. In addition, ID1 contributes to the self-renewal capacity of murine cortical neural stem cells, and a murine model of hematopoiesis indicated that Id1-/- whole-bone marrow displayed decreased self-renewal capacity compared with that of the wild-type control [23-24].
In the present study, we sought to determine whether CDC27 play a crucial role in CRC metastasis. The results suggested that the CDC27-ID1 axis may serve as a promising therapeutic target for CRC patients with metastasis.
Results
CDC27 is a positive regulator of CRC metastasis
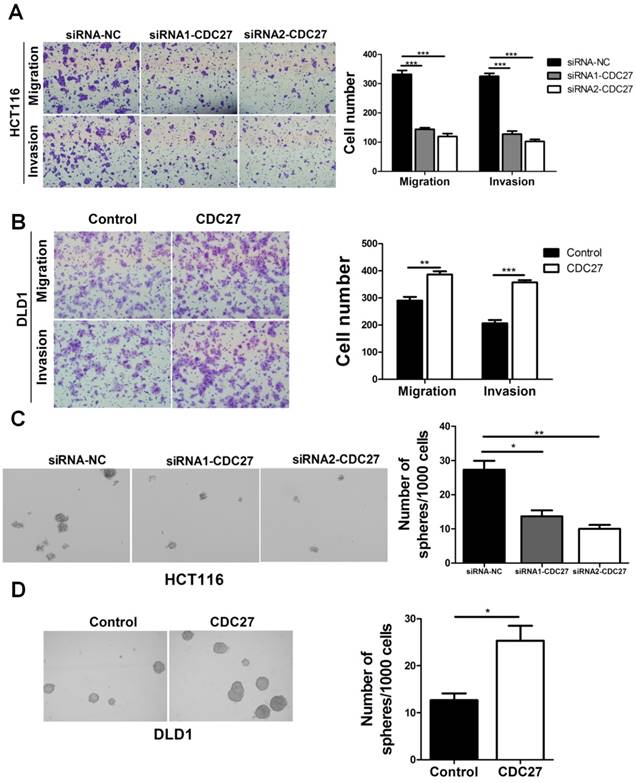
To determine the function of CDC27 in the metastasis and invasiveness of CRC, we performed transwell and Matrigel invasion assays. SiRNA targeting CDC27 (siRNA1-CDC27 or siRNA2-CDC27) or negative control siRNA (siRNA-NC) was transiently transfected into HCT116 cells, while DLD1 cells were transiently transfected with either a CDC27-carrying plasmid or an empty plasmid. The expression levels of CDC27 were determined via western blotting (Supplemental Figure S1). The results showed that knockdown of CDC27 significantly suppressed both the migratory and invasive abilities of HCT116 cells compared with the control treatment (p<0.001, Figure 1A). Conversely, ectopic expression of CDC27 promoted DLD1 cell migration and invasion (p<0.01, Figure 1B). In addition, colony sphere-formation assays were performed to assess whether CDC27 affects the sphere-formation capacity of CRC cells. The data indicated that the sphere-formation capacity of HCT116 cells was significantly weakened upon downregulation of CDC27 (p<0.05, Figure 1C). In contrast, CDC27 overexpression remarkably enhanced the sphere-formation capacity of DLD1 cells (p<0.05, Figure 1D). Our results provided evidence that CDC27 is a positive regulator of CRC metastasis.
Downregulation of CDC27 suppresses EMT in CRC
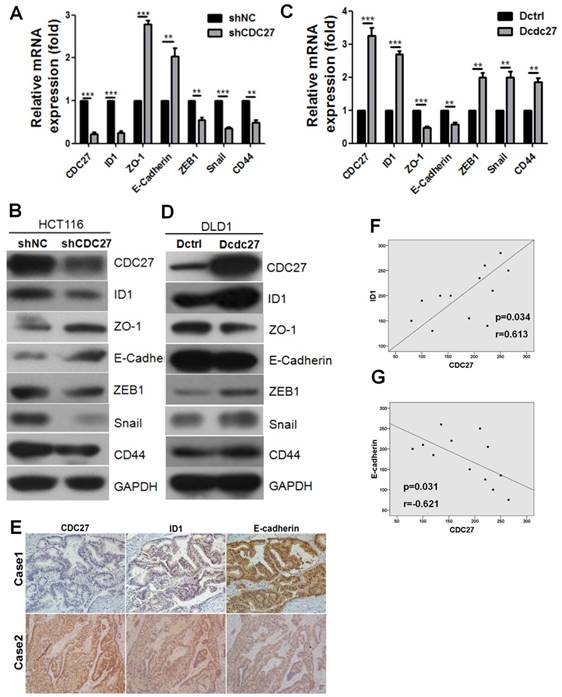
In our previous study, we demonstrated that CDC27 promotes the expression of ID1, which plays a key role in tumorigenesis and promotes EMT in various tumors. Given our above findings, we examined whether CDC27 has the potential to induce EMT in CRC cell lines. For this purpose, we constructed a DLD1 cell line stably overexpressing CDC27 (Dcdc27) and a control DLD1 cell line (Dctrl). Considering that siRNA2-CDC27 was more efficient than siRNA1-CDC27 in silencing CDC27 expression, we selected the siRNA2-CDC27 sequence to generate shCDC27 and established HCT116 cell lines in which CDC27 was stably knocked down; alternatively, a control shRNA was used (below, these cells lines are referred to as shCDC27 and shNC, respectively). As shown in Figure 2A and 2B, the mRNA and protein expression levels of the epithelial markers ZO-1 and E-cadherin were remarkably increased in stable shCDC27 cell lines, and the mRNA and protein expression levels of the mesenchymal markers ZEB1 and Snail were decreased. Similar results were observed in the stable Dcdc27 and Dctrl cell lines (Figure 2C, 2D). Furthermore, we observed that the expression of the epithelial stemness marker CD44 was significantly decreased in shCDC27 cell lines (Figure 2A, 2B), but that it was increased in Dcdc27 cell lines compared with expression in the control cell line (Figure 2C, 2D). These findings supported the above results that CDC27 promoted the sphere-formation capacity of CRC cells (Figure 1C, 1D). Therefore, the above results suggested that CDC27 has the potential to induce EMT in CRC cells, which at least partially accounts for the activities of CDC27 in promoting the metastasis and invasiveness of CRC cells.
Given that CDC27 modulated the ID1 and the EMT program in HCT116 and DLD1 cells, we sought to determine whether the expression of ID1 or E-cadherin was correlated with CDC27 expression in tumor tissues derived from CRC patients. IHC staining was performed to analyze the relationship between the expression of CDC27 and E-cadherin or ID1 in tumor tissues from 12 CRC patients. As shown in Figure 2E, 2F, and 2G, ID1 expression was significantly and positively correlated with CDC27 expression (R=0.613, p=0.034), and E-cadherin expression was negatively correlated with CDC27 expression (R=-0.621, p=0.031).
CDC27 promotes the metastasis, invasion and sphere-formation capacity of CRC cells. (A and B) Representative images show the migration and invasion abilities of HCT116 cells and DLD1 cells with transient CDC27 knockdown or overexpression. The number of cells was quantified. *, p<0.05, **, p<0.01, ***, p<0.001, based on Student's t-test. (C and D) Representative images show the sphere-formation capability of HCT116 cells and DLD1 cells with transient CDC27 knockdown or overexpression. *, p<0.05, **, p<0.01 based on Student's t-test. These experiments were repeated at least three times. Error bars, mean ± SD.
CDC27 induces the EMT process in CRC. (A and B) Real-time PCR and western blotting were performed to detect the expression of EMT markers in HCT116 stable cell lines. The means ± SD of triplicate samples are shown. **, p<0.01, ***, p<0.001 based on Student's t-test. (C and D) Real-time PCR and Western blotting were performed to detect the expression of EMT markers in DLD1 stable cell lines. The means ± SD of triplicate samples are shown. **, p<0.01, ***, p<0.001 based on Student's t-test. (E) Representative IHC images of CDC27, ID1, and E-cadherin expression in CRC tumor tissues (×100 magnification). (F and G) Pearson correlation coefficients between CDC27 and either ID1 or E-cadherin expression in 12 CRC patient tissues were analyzed.
CDC27 mediates EMT via the modulation of ID1 expression
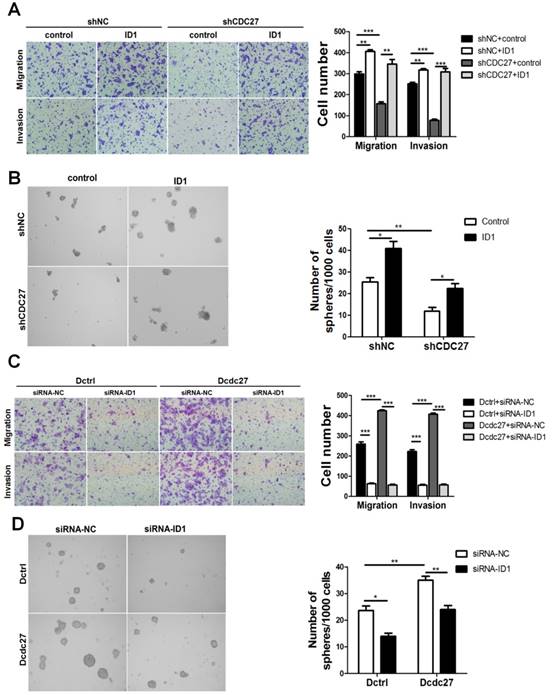
Considering that ID1 participates in EMT, contributes to CSC properties in multiple cancer cell lines and is positively regulated by CDC27, we next sought to determine whether ID1 is involved in metastasis and sphere-formation capacity of CRC cells caused by CDC27 alterations. To examine this issue, an exogenous ID1 expression plasmid (pReceiver-M02-ID1) or an empty vector (pReceiver-M02) was transfected into stable shCDC27 or shNC HCT116 cells. Consistent with our expectation, the findings suggested that ID1 impaired the inhibition of metastasis, invasion, and sphere-formation capability induced by CDC27 downregulation in HCT116 stable cell lines (Figure 3A, 3B). Moreover, we observed a similar phenomenon when we transiently knocked down ID1 expression. Knockdown of ID1 dramatically suppressed metastasis, invasion and sphere-formation capability of DLD1 stable cell lines (Figure 3C, 3D).
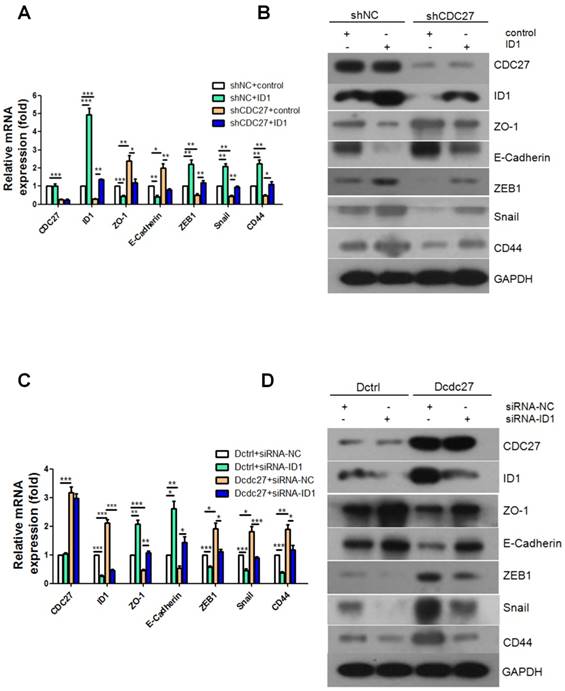
Given the results obtained thus far, we next explored whether CDC27 also regulates EMT progression by regulating ID1. Alterations in the expression of the EMT markers noted above were examined via real-time PCR and western blotting. Intriguingly, the results revealed that the expression levels of the epithelial markers ZO-1 and E-cadherin were dramatically down-regulated and that the expression levels of the mesenchymal markers ZEB1, Snail and CD44 were restored upon exogenous overexpression of ID1 (Figure 4A, 4B). Furthermore, siRNA targeting ID1 (siRNA1-ID1, siRNA2-ID1) or negative control siRNA (siRNA-NC) was transfected into stable Dcdc27 or Dctrl cells. Similarly, the results showed that transient knockdown of ID1 remarkably reversed the decrease in ZO-1 and E-cadherin expression caused by CDC27 upregulation and dramatically inhibited the accumulation of ZEB1, Snail and CD44 in stable Dcdc27 cells (Figure 4C, 4D). Therefore, we concluded that CDC27 affect EMT progression in an ID1-dependent manner.
CDC27 promotes metastasis in a xenograft mouse model
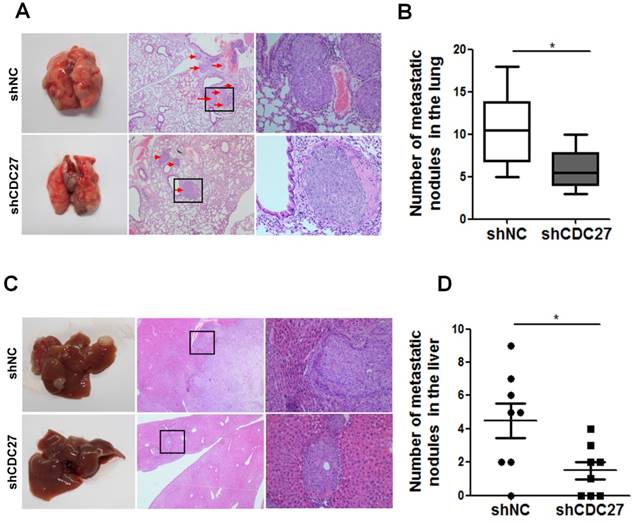
The above findings prompted us to examine whether CDC27 affect the metastatic process in CRC cells in vivo. Therefore, we established mouse models of lung and hepatic metastasis using the stable shCDC27 and shNC HCT116 cell lines. The results indicated that knockdown of CDC27 led to a significant decrease in the number of lung metastatic tumor nodules (n=8, Figure 5A, 5B). Moreover, metastatic nodules were observed in the liver of most of the mice from the shNC group, and the number of metastatic nodules was distinctly greater in the shNC group than that in the shCDC27 group (n=8, Figure 5C, 5D).
CDC27 promotes metastasis in CRC cell lines in an ID1-dependent manner. (A and B) ID1 was transiently overexpressed in HCT116 stable cell lines. Representative images of migration, invasion transwell, and sphere-formation rescue assays are shown. Quantification of the results is presented. The means ± SD of triplicate samples are shown. *, p<0.05, **, p<0.01, ***, p<0.001 based on Student's t-test. (C and D) ID1 was transiently knocked down in DLD1 stable cell lines. Representative images of migration, invasion transwell, and sphere-formation rescue assays are shown. Quantification of the results is also presented. The means ± SD of triplicate samples are shown. *, p<0.05, **, p<0.01, ***, p<0.001 based on Student's t-test.
Discussion
In our previous study, we reported that the downregulation of CDC27 suppresses the proliferation of CRC cells and that CDC27 expression is a negative prognostic factor for survival among CRC patients [15]. However, no previous study has focused on the role of CDC27 in cancer metastasis. Moreover, our previous IHC analysis results indicated that CDC27 expression is correlated with distant metastasis in CRC patients. All of these previous findings drove our interest in investigating whether CDC27 participates in the regulation of CRC metastasis and invasion. In this study, we focused on determining the function of CDC27 in CRC metastasis and elucidating its underlying mechanisms to identify potential targets for the treatment of CRC patients with metastasis.
It has been well described that EMT is closely linked to tumor metastasis and invasion [25]. The hallmarks of the EMT process are loss of E-cadherin-mediated cell-cell adhesion, acquisition of mesenchymal marker expression and thus increased cancer cell motility and invasiveness [7]. Given that CDC27 enhances the migration ability of CRC cells (Figure 1A, 1B), we first examined whether CDC27 affects the EMT process. Our findings showed that the expression levels of E-cadherin and ZO-1 were dramatically increased and that the expression levels of ZEB1, CD44, Snail were correspondingly significantly decreased in stable HCT116 shCDC27 cells (Figure 2A, 2B). Similar findings were obtained using stable Dctrl and Dcdc27 cells (Figure 2C, 2D).
CDC27 induces the EMT process via the modulation of ID1 expression in CRC. (A and B) Real-time PCR and western blotting were performed to determine the expression of EMT markers in the HCT116 stable cell lines with transient ID1 overexpression. (C and D) Real-time PCR and western blotting were performed to analyze the expression of EMT markers in the DLD1 stable cell lines with transient ID1 knockdown. The means ± SD of triplicate samples are shown. *, p<0.05, **, p<0.01, ***, p<0.001 based on Student's t-test.
CDC27 promotes the metastasis of CRC cells in nude mouse models. (A and B) For the lung metastasis model, lung sections were subjected to H&E staining. Representative macroscopic images of metastatic lung nodules are shown, and the arrows indicate the metastatic nodules. The number of metastatic nodules in the lung specimens was analyzed. *, p<0.05. (C and D) For the hepatic metastasis model, liver sections were subjected to H&E staining. Representative macroscopic images of metastatic liver tumors are shown, and the arrows indicate the metastatic nodules. The number of metastatic nodules in the liver specimens was analyzed. *, p<0.05.
However, many topics remain to be further elucidated. In our previous and current studies, we demonstrated that downregulation of CDC27 inhibits the proliferation and metastasis of CRC cell lines by modulating ID1 expression. Considering that ID1 plays an oncogenic role in various cancer types and that high expression of CDC27 was reported to be a significant predictor of breast cancer recurrence [14], we highly suspect that the CDC27-ID1 axis may also play a crucial role in other forms of cancer. Therefore, whether the CDC27-ID1 axis exerts its oncogenic effect specifically in CRC or universally in a variety of cancer types warrants further exploration.
In addition, the molecular mechanisms upstream of CDC27 remain obscure. Recent studies revealed that knockdown of poly(rC)-binding protein 1 (PCBP1) abrogates the silencing of CDC27 mRNA translation, resulting in constitutive expression of CDC27, which leads to chromosomal instability and aneuploidy [14]. More importantly, PCBP1 is involved in several critical biological processes. Previous studies have demonstrated that PCBP1 is a suppressor of metastasis, that it is involved in the EMT process and that PCBP1 downregulation predicts poor prognosis of various cancer types [31-33], such as non-small cell lung cancer and gastric cancer [34-36]. Intriguingly, in addition to its function as an EMT suppressor, PCBP1 has been reported to contribute to the maintenance of CSC properties and to the inhibition of CD44 expression [36,37]. Based on our results that CDC27 promoted cell mobility and enhanced the expression of CD44, we strongly speculate that PCBP1 might be involved in the upstream regulation of CDC27 in CRC, which plausibly explains our findings in this study.
Moreover, considering the mounting evidence from sequencing studies in recent years that CDC27 is mutated in several tumor types [10-12,38,39], there is a possibility that a CDC27 mutation itself might lead to aberrant, constitutive activation of CDC27 in CRC, and this possibility needs to be investigated further in our future studies.
In conclusion, the present study demonstrates that CDC27 promotes metastasis in CRC. Therefore, CDC27 may emerge as a novel marker of metastasis, and CDC27 may serve as a potential therapeutic target for the prevention and treatment of CRC.
Materials and Methods
Cell culture
DLD-1 cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS, PAA), and HCT116 cells were cultured in DMEM containing 10% FBS. All cell lines were tested for mycoplasma contamination (New MycoProbe Mycoplasma Detection Kit, R&D Systems) at the indicated time points.
Patient tissue specimens and clinicopathological characteristics
The patients for whom follow-up information was not available were excluded from this study. The tissue specimens used conformed to the following criteria: contained matched tumor tissue (percentage of tumor cells > 70%) and corresponding normal mucosal tissue (>5 cm laterally from the edge of the tumor region). The patients underwent follow-up once every 3 months during the first 2 years after surgery, once every 6 months during the third and fourth years, and once a year thereafter. Patients who had a single primary lesion and no neoadjuvant chemotherapy were included. Patients provided informed consent, and the use of clinical specimens for research purposes was approved by the Institutional Research Ethics Committee.
Immunohistochemistry
These procedures were performed as previously described [15]. A primary antibody targeting CDC27 (Santa, sc-9972, 1:50) was used. CDC27 immunostaining intensity was scored as follows: negative = 0, weak = 1, moderate = 2, or strong = 3. The final H score was calculated by multiplying the CDC27 immunostaining intensity score by the percentage of CDC27-immunostained area. Receiver operating characteristic (ROC) curve analysis was used to determine a cutoff value for defining high or low CDC27 expression. A ROC curve was generated to plot the sensitivity and specificity for the outcome, and the H score that was closest to the point of maximal sensitivity and specificity was selected as the cutoff value.
RNA extraction and qRT-PCR
These procedures were performed as previously described [15]. Briefly, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and cDNA was synthesized using the GoScript Reverse Transcription System (Promega). The primers used are shown in Supplementary Table S1.
Western blotting analysis
This procedure was performed as described previously [15]. Briefly, cells were collected and lysed in RIPA buffer (Beyotime, P0013B). Total protein samples were loaded and separated on sodium dodecyl sulfate-polyacrylamide gradient gels, followed by transfer to PVDF membranes. Then, the membranes were blocked with 5% non-fat milk for 2 h at room temperature. Next, the membranes were incubated with the primary antibody followed by a horseradish peroxidase-conjugated secondary antibody. Then, the immunoreactive protein bands were detected using an electrochemiluminescence system (KeyGENBioTECH). Primary antibodies against the following antigens were used: CDC27 (Santa Cruz, sc-9972; 1:200), ID1 (Santa Cruz, sc-488; 1:200), ZO-1 (Cell Signaling Technology, #8193; 1:1000), E-cadherin (Cell Signaling Technology, #3195; 1:1000), ZEB1 (Cell Signaling Technology, #3396; 1:1000), Snail (Cell Signaling Technology, #3879; 1:1000), CD44 (Cell Signaling Technology, #3570; 1:1000), and GAPDH (Proteintech, 60004-1-lg, 1:10000).
Plasmid generation and stable cell line construction
Stable cell lines were constructed as described previously [15].
Migration and invasion assays
Transfected cells (1×105) in 200 μL of serum-free DMEM or RPMI 1640 medium were plated on uncoated or Matrigel-coated cell culture inserts (8-μm pores; BD Falcon, San Jose, CA). DMEM or RPMI 1640 medium containing 10% FBS was added to the bottom chamber. After 18-24 hours, the cells on the lower surface of the filter were fixed, stained, and examined using a microscope. The number of migrated cells in three randomly selected optical fields (×100 magnification) from triplicate filters was averaged.
Sphere-formation assay
For the sphere-formation assays, cells were resuspended and seeded in DMEM/F12 medium supplemented with 20 ng/mL epidermal growth factor (Invitrogen) and 1% penicillin/streptomycin. The size and number of cell spheres were evaluated via microscopy after 12 days.
Animal experiments
Animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-Sen University Cancer Center. Male BALB/c nude mice (4-5 weeks old, 15-18 g) were purchased from the Slaccas Experimental Animal Center (Shanghai, China). Mice were injected with stable HCT116 cells to produce the lung and hepatic metastasis models.
Hepatic metastasis model
Mice were randomly assigned to two groups (n=8), both of which were anesthetized with isoflurane and subjected to laparotomy. In all, 1×106 cells (shCDC27 or shNC) were resuspended in 100 µL of PBS and were injected into the distal tip of the spleen using an insulin syringe. Eight weeks after injection, the mice were sacrificed, and the spleens and livers were collected. The sections were stained with hematoxylin and eosin (H&E). Metastatic nodules were counted under a microscope.
Lung metastasis model
Mice were randomly assigned to two groups (n=8). A total of 2 × 106 cells in 100 μL of PBS were injected into the tail vein of mice. Ten weeks after injection, the animals were euthanized, the lungs were harvested and the sections were stained with H&E. Metastatic nodules were counted under a microscope [40].
Statistical analysis
All statistical analyses were completed using SPSS version 16.0 software. Normally distributed data are expressed as the means ± SD. Student's t-test and the two-tailed chi-square test were used. Pearson correlation coefficients were calculated for correlation analysis. A p-value of less than 0.05 was considered statistically significant.
Supplementary Material
Supplementary Figure S1 and Table S1.
Acknowledgements
We thank Jiemin Chen and Chen Yao for their helpful assistance with the manuscript. This work was supported by the National Natural Science Foundation of China (No. 81272513, No. 81272638) and the National High Technology Research and Development Program of China (863 Program, No. 2012AA02A204). The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (www.researchdata.org.cn), with the approval RDD number as RDDB2017000140.
Author Contributions
JXW and WLH designed the experiments; LQ conducted experiments; RYL and SC provided research materials and methods; XT, JXL, and RG analyzed the data; LQ wrote the manuscript. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298-306
2. Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814-23
3. Chang GJ, Rodriguez-Bigas MA, Skibber JM. et al. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. J Natl Cancer Inst. 2007;99:433-41
4. Chen W, Zheng R, Zeng H. et al. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35:73
5. Yamagishi H, Kuroda H, Imai Y. et al. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer. 2016;35:4
6. Gumireddy K, Li A, Gimotty PA. et al. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297-304
7. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818-29
8. Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679-95
9. Acloque H, Adams MS, Fishwick K. et al. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438-49
10. Litchfield K, Summersgill B, Yost S. et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun. 2015;6:5973
11. Juhlin CC, Goh G, Healy JM. et al. Whole-exome sequencing characterizes the landscape of somatic mutations and copy number alterations in adrenocortical carcinoma. J Clin Endocrinol Metab. 2015;100:E493-E502
12. Reimann E, Kõks S, Ho XD. et al. Whole exome sequencing of a single osteosarcoma case-integrative analysis with whole transcriptome RNA-seq data. Hum Genomics. 2014;8:20
13. Lawrence MS, Stojanov P, Mermel CH. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495-501
14. Link LA, Howley BV, Hussey GS. et al. PCBP1/HNRNP E1 protects chromosomal integrity by translational regulation of CDC27. Mol Cancer Res. 2016;14:634-46
15. Qiu L, Wu J, Pan C. et al. Downregulation of CDC27 inhibits the proliferation of colorectal cancer cells via the accumulation of p21Cip1/Waf1. Cell Death Dis. 2016;7:e2074
16. Benezra R, Davis RL, Lockshon D. et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49-59
17. Meteoglu I, Meydan N, Erkus M. Id-1: regulator of EGFR and VEGF and potential target for colorectal cancer therapy. J Exp Clin Cancer Res. 2008;27:69
18. Yuen HF, Chan YP, Chan KK. et al. Id-1 and id-2 are markers for metastasis and prognosis in oesophageal squamous cell carcinoma. Br J Cancer. 2007;97:1409-15
19. Jankovic V, Ciarrocchi A, Boccuni P. et al. Id1 Restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc Natl Acad Sci U S A. 2007;104:1260-5
20. Fong S, Itahana Y, Sumida T. et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100:13543-8
21. Gupta GP, Perk J, Acharyya S. et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506-11
22. Zhao ZR, Zhang ZY, Zhang H. et al. Overexpression of id-1 protein is a marker in colorectal cancer progression. Oncol Rep. 2008;19:419-24
23. Perry SS, Zhao Y, Nie L. et al. Id1, But not Id3, directs long-term repopulating hematopoietic stem-cell maintenance1. Blood. 2007;110:2351-60
24. Ciarrocchi A, Jankovic V, Shaked Y. et al. Id1 restrains p21 expression to control endothelial progenitor cell formation. PLOS ONE. 2007;2:e1338
25. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-8
26. Ling MT, Lau TC, Zhou C. et al. Overexpression of id-1 in prostate cancer cells promotes angiogenesis through the activation of vascular endothelial growth factor (VEGF). Carcinogenesis. 2005;26:1668-76
27. Sikder HA, Devlin MK, Dunlap S. et al. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525-30
28. Alani RM, Young AZ, Shifflett CB. Id1 Regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci U S A. 2001;98:7812-6
29. Nieborowska-Skorska M, Hoser G, Rink L. et al. Id1 transcription inhibitor-matrix metalloproteinase 9 axis enhances invasiveness of the breakpoint cluster region/abelson tyrosine kinase-transformed leukemia cells. Cancer Res. 2006;66:4108-16
30. Tobin NP, Sims AH, Lundgren KL. et al. Cyclin D1, Id1 and EMT in breast cancer. BMC Cancer. 2011;11:417
31. Chaudhury A, Hussey GS, Ray PS. et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via TranScript-selective translational induction of Dab2 and ILEI. Nat Cell Biol. 2010;12:286-93
32. Wang H, Vardy LA, Tan CP. et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18:52-62
33. Zhou M, Tong X. Downregulated poly-C binding protein-1 is a novel predictor associated with poor prognosis in acute myeloid leukemia. Diagn Pathol. 2015;10:147
34. Liu Y, Gai L, Liu J. et al. Expression of poly(C)-binding protein 1 (PCBP1) in NSCLC as a negative regulator of EMT and its clinical value. Int J Clin Exp Pathol. 2015;8:7165-72
35. Zhang ZZ, Shen ZY, Shen YY. et al. HOTAIR Long noncoding RNA promotes gastric cancer metastasis through suppression of Poly r(C)-Binding protein (PCBP) 1. Mol Cancer Ther. 2015;14:1162-70
36. Zhang T, Huang XH, Dong L. et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer. 2010;9:72
37. Chen Q, Cai ZK, Chen YB. et al. Poly r(C) binding protein-1 is central to maintenance of cancer stem cells in prostate cancer cells. Cell Physiol Biochem. 2015;35:1052-61
38. Lindberg J, Mills IG, Klevebring D. et al. The mitochondrial and autosomal mutation landscapes of prostate cancer. Eur Urol. 2013;63:702-8
39. Ahn JW, Kim HS, Yoon JK. et al. Identification of somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers. Genome Med. 2014;6:18
40. Zhou Y, Wu J, Fu X. et al. OTUB1 promotes metastasis and serves as a marker of poor prognosis in colorectal cancer. MOL CANCER. 2014;13:258
Author contact
![]() Corresponding authors: Wenlin Huang, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, No. 651 Dongfeng East Road, Guangzhou 510060, PR China. Phone: 86-20-8734-3146; Fax: 862087343146; E-mail: hwenlsysu.edu.cn; Jiangxue Wu, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, No. 651 Dongfeng East Road, Guangzhou 510060, PR China. Phone: 86-20-8734-3146; Fax: 862087343146; E-mail: gladyswcom
Corresponding authors: Wenlin Huang, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, No. 651 Dongfeng East Road, Guangzhou 510060, PR China. Phone: 86-20-8734-3146; Fax: 862087343146; E-mail: hwenlsysu.edu.cn; Jiangxue Wu, State Key Laboratory of Oncology in South China, Sun Yat-sen University Cancer Center, No. 651 Dongfeng East Road, Guangzhou 510060, PR China. Phone: 86-20-8734-3146; Fax: 862087343146; E-mail: gladyswcom

 Global reach, higher impact
Global reach, higher impact