Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(13):2643-2652. doi:10.7150/jca.20234 This issue Cite
Research Paper
A BAP1 Mutation-specific MicroRNA Signature Predicts Clinical Outcomes in Clear Cell Renal Cell Carcinoma Patients with Wild-type BAP1
1. Department of Urology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing 210006, China.
2. Department of Urology, Nantong Hospital of Traditional Chinese Medicine, 41 Jianshe Road, Nantong 226006, China.
3. Department of Pathology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing 210006, China.
4. Department of Radiation Oncology, JiangSu Armed Police General Hospital, 8 Jiangdu South Road, Yangzhou 225003, China
5. Department of Urology, Huaian First People's Hospital, Nanjing Medical University, 6 Beijing West Road, Huaian 223300, China.
*These three authors (Yu-Zheng Ge, Lu-Wei Xu, and Chang-Cheng Zhou) contributed equally to this work.
Received 2017-3-24; Accepted 2017-6-25; Published 2017-8-21
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is the most prevalent histologic subtype of kidney cancers in adults, which could be divided into two distinct subgroups according to the BRCA1 associated protein-1 (BAP1) mutation status. In the current study, we comprehensively analyzed the genome-wide microRNA (miRNA) expression profiles in ccRCC, with the aim to identify the differentially expressed miRNAs between BAP1 mutant and wild-type tumors, and generate a BAP1 mutation-specific miRNA signature for ccRCC patients with wild-type BAP1.
Methods: The BAP1 mutation status and miRNA profiles in BAP1 mutant and wild-type tumors were analyzed. Subsequently, the association of the differentially expressed miRNAs with patient survival was examined, and a BAP1 mutation-specific miRNA signature was generated and examined with Kaplan-Meier survival, univariate and multivariate Cox regression analyses. Finally, the bioinformatics methods were adopted for the target prediction of selected miRNAs and functional annotation analyses.
Results: A total of 350 treatment-naïve primary ccRCC patients were selected from The Cancer Genome Atlas project, among which 35 (10.0%) subjects carried mutant BAP1 and had a shorter overall survival (OS) time. Furthermore, 33 miRNAs were found to be differentially expressed between BAP1 mutant and wild-type tumors, among which 11 (miR-149, miR-29b-2, miR-182, miR-183, miR-21, miR-365-2, miR-671, miR-365-1, miR-10b, miR-139, and miR-181a-2) were significantly associated with OS in ccRCC patients with wild-type BAP1. Finally, a BAP1 mutation-specific miRNA signature consisting of 11 miRNAs was generated and validated as an independent prognostic parameter.
Conclusions: In summary, our study identified a total of 33 miRNAs differentially expressed between BAP1 mutant and wild-type tumors, and generated a BAP1 mutation-specific miRNA signature including eleven miRNAs, which could serve as a novel prognostic biomarker for ccRCC patients with wild-type BAP1.
Keywords: clear cell renal cell carcinoma, BAP1, mutation, microRNA, prognosis
Introduction
Renal cell carcinoma (RCC) comprises 2-3% of all human malignancies and represents the ninth leading cancer in European Union with approximately 84,400 new RCC cases reported in 2012 [1]. In the United Sates, a total of 63,990 newly diagnosed RCC cases and 14,400 RCC-related deaths are projected to occur in 2017 [2]. Histologically, RCC encompasses a broad spectrum of entities and clear cell RCC (ccRCC) accounts for 70-80% of all RCC cases [3, 4]. The last few decades have witnessed the substantial developments and advances in both ccRCC understanding and treatment. To date, several histopathological variables, such as tumor-node-metastasis (TNM) stage, Fuhrman nuclear grade, microvascular invasion, and tumor necrosis, have been identified as the independent prognostic factors of ccRCC, and recommended to be applied in routine clinical practice [5]. However, the predictive accuracy of the currently-established prognostic systems needs to be improved, as the patients with similar clinical and pathological features could have various outcomes, which highlights the need for novel molecular biomarkers [6-8].
BRCA1 associated protein-1 (BAP1) is a nuclear-localized deubiquitinating enzyme, with ubiquitin carboxyl hydrolase activity and two nuclear localization signal motifs [9]. Functionally, BAP1 is a potent tumor suppressor, which could regulate various fundamental cellular processes including proliferation, death, differentiation, gluconeogenesis, and the DNA damage response [10, 11]. The BAP1 gene is located on chromosome 3p21.1, a genomic region frequently mutated in a variety of human malignancies including approximately 6-15% ccRCC [12-17]. The BAP1 mutant ccRCC patients have a worse prognosis than those with wild-type BAP1 [14-16], and the BAP1 mutant tumors exhibit a specific gene expression signature [13, 14], which overall suggests that BAP1 mutation status could serve as a novel molecular classification parameter of ccRCC.
MicroRNAs (miRNAs) are small non-coding and single-stranded RNAs with about 19-25 nucleotides in length, which regulate a wide array of pathophysiological processes by modulating gene expression through base pairing with the 3' untranslated region of target mRNAs [18, 19]. Increased data suggests that miRNAs exert important functions in the development and progression of human malignancies, and could serve as potential biomarkers for the early diagnosis, prognosis prediction, and therapy decision [20-24]. Aberrantly expressed miRNAs between ccRCC tumors and normal kidney tissues have been investigated by various researchers, and different miRNA expression profiles with clinical significance have been identified in ccRCC [25-27]. Recently, a tumor-specific miRNA signature consisting of 22 miRNAs was identified within a cohort of 411 ccRCC patients from The Cancer Genome Atlas (TCGA) project, which could function as an independent prognostic factor of ccRCC [28]. However, to date, no study has been conducted to compare the miRNA expression profiles between BAP1 mutant and wild-type tumors, and no specific miRNA signature was establsished for ccRCC patients with wild-type BAP1.
Hence, we stringently designed a stepwise study using the data from TCGA project to: 1) ascertain the differential miRNA expression profiles between wild-type and mutant BAP1 tumors; 2) identify the miRNAs with prognostic potential from the differential expression profiles; 3) generate a BAP1 mutation-specific miRNA signature for ccRCC patients with wild-type BAP1; and 4) predict target genes and potential involved biological pathways for selected miRNAs.
Materials and Methods
Patients and samples
All subjects in the current study were identified from those who underwent partial nephrectomy or radical nephrectomy for sporadic ccRCC at multiple centers of TCGA consortium, between 1998 and 2010 [16]. The full clinical and pathological data were downloaded from the TCGA data portal (January 10th, 2017), and re-evaluated according to the predesigned selection criteria: 1) pathologically diagnosed ccRCC; 2) with no history of other malignancies; 3) no chemotherapy or radiation therapy prior to surgery; 4) determined BAP1 mutation status; and 5) genome-wide miRNA expression profiles analyzed. Furthermore, two experienced urological pathologist (YCZ and XBY) double-checked the pathological TNM stage and Fuhrman nuclear grade according to the 2010 TNM classification of American Joint Committee on Cancer (AJCC) and 2004 World Health Organization criteria, respectively [29, 30]. Overall, a total of 350 ccRCC patients were included in the present study, with full annotation of the corresponding clinical and pathological parameters such as age, gender, race, AJCC TNM information, Fuhrman grade, surgical types, and detailed follow-up information (Table 1). The study protocol was approved by the institutional review board of Nanjing First Hospital, Nanjing Medical University, informed consent was obtained from all individual participants, and the data collection and procession were performed in accordance with the human subject protection and data access policies of TCGA project.
Clinical characteristics of patients with clear cell renal cell carcinoma
| Category | Total (n = 350) | BAP1 Status | P Valuea | ||
|---|---|---|---|---|---|
| WT (n = 315) | MUT (n=35) | ||||
| Age, Median (IQR) | 60 (41, 79) | 60 (41, 79) | 60 (41,79) | 0.464 | |
| Ethnicity | 0.450 | ||||
| Caucasian | 332 (94.9%) | 299 (94.9%) | 33 (94.3%) | ||
| African | 11 (3.1%) | 9 (2.9%) | 2 (5.7%) | ||
| Asian | 7 (2.0%) | 7 (2.2%) | 0 (0.0%) | ||
| Gender | <0.001b | ||||
| Male | 225 (64.3%) | 213 (67.6%) | 12 (34.3%) | ||
| Female | 125 (35.7%) | 102 (32.4%) | 23 (65.7%) | ||
| AJCC Stage | 0.006b | ||||
| Stage Ⅰ | 152 (43.4%) | 146 (46.3%) | 6(17.1%) | ||
| Stage Ⅱ | 36 (10.3%) | 31 (9.8%) | 5 (14.3%) | ||
| Stage Ⅲ | 103 (29.4%) | 90 (28.6%) | 13 (37.1%) | ||
| Stage Ⅳ | 59 (16.9%) | 48 (15.2%) | 11 (31.4%) | ||
| Tumor Size | 0.009b | ||||
| T1 | 157 (44.9%) | 150 (47.6%) | 7 (20.0%) | ||
| T2 | 45 (12.9%) | 37 (11.7%) | 8 (22.9%) | ||
| T3 | 143 (40.9%) | 123 (39.0%) | 20 (57.1%) | ||
| T4 | 5 (1.4%) | 5 (1.6%) | 0 (0.0%) | ||
| Lymph Node | 0.088 | ||||
| N0 | 159 (45.4%) | 138 (43.8%) | 21 (60.0%) | ||
| N1 | 11 (3.1%) | 9 (2.9%) | 2 (5.7%) | ||
| NX | 180 (51.4%) | 168 (53.3%) | 12 (34.3%) | ||
| Metastasis Status | 0.029b | ||||
| M0 | 291 (83.1%) | 267 (84.8%) | 24(68.6%) | ||
| M1 | 59 (16.9%) | 48 (15.2%) | 11 (31.4%) | ||
| Fuhrman Grade | 0.003b | ||||
| G1 | 5 (1.4%) | 5 (1.6%) | 0 (0.0%) | ||
| G2 | 136 (38.9%) | 126 (40.0%) | 10 (28.6%) | ||
| G3 | 150 (42.9%) | 137 (43.5%) | 13 (37.1%) | ||
| G4 | 58 (16.6%) | 47 (14.9%) | 11(31.4%) | ||
| GX | 1 (0.3%) | 0 (0.0%) | 1 (2.9%) | ||
| Surgical Type | 0.648 | ||||
| Partial nephrectomy | 64 (18.3%) | 59 (18.7%) | 5 (14.3%) | ||
| Radical nephrectomy | 286 (81.7%) | 256 (81.3%) | 30 (85.7%) | ||
BAP1, BRCA1 associated protein-1; WT, wild-type; MUT, mutant; IQR, Inter-Quartile Range; AJCC, American Joint Committee on Cancer; NX, regional lymph node unknown; GX, Fuhrman Grade unknown.
a P values derived from the comparisons between BAP1 mutant and wild-type groups;
b statistical significant results (in bold).
BAP1 mutation analysis
The whole-exome sequencing data was downloaded from TCGA data portal (January 20th, 2017) for the analysis of BAP1 mutation status, which was acquired on the following two platforms: Illumina HiSeq (Illumina Inc, San Diego, CA, USA) and SOLiD (Applied Biosystems Inc., Foster City, CA, USA), and partially validated on another two sequencing platforms: 454 (Roche, Basel, Switzerland) and Ion Torrent (Life Technologies, Grand Island, NY, USA) [16]. Furthermore, the BAP1 mutation status for each individual was double-checked by applying the online analytical tool: cBioPortal for Cancer Genomics as previously described [31].
Microarray data procession
The genome-wide miRNA expression profiling was performed with the Illumina Genome Analyzer and HiSeq platforms (Illumina Inc, San Diego, CA, USA), and presented as reads per million counts (RPM). After downloaded from TCGA data portal (February 2nd, 2017), the summary miRNA expression data was processed with BRB-Array tools (version 4.5.1; National Cancer Institute, Bethesda, MD, USA) which were developed by the BRB-Array Tools Development Team [32]. In brief, the miRNAs meet the following criteria were retained and log2 transformed for further analysis: 1) more than 1 RPM in at least 10% of all samples, and 2) had changes of more than 1.5 fold from the median value in at least 20% of samples.
Bioinformatics analysis
The miRNA target prediction was conducted using three computational algorithms including TargetScan (version 7.1; http://www.targetscan.org) [33], miRDB (http://www.mirdb.org/) [34], and the Good mirSVR score collection within miRanda (http://www.microrna.org) [35]. The genes predicted by at least two independent tools were uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) program (version 6.8; https://david.ncifcrf.gov/) [36, 37] for further annotation analyses including Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, as we previously reported [21].
Statistical analysis
The continuous variables were presented as mean ± standard deviation or median and Inter-Quartile Range (IQR), and compared using Student's t-test or Mann-Whitney U test. The categorical variables were presented as counts and percentages, and compared with Chi-square statistics (or Fisher's exact test when appropriate).
The miRNA expression levels between BAP1 mutant and wild-type tumors were ascertained with Student's t-test embedded in BRB-Array tools (significance level set as 0.01), and the unsupervised hierarchical cluster analysis was performed by Euclidian distance and average linkage methods. To identify the prognosis-associated miRNAs, the univariate Cox proportional hazards regression analysis was applied, and a risk score formula was developed by combining the expression level (Expr) weighted by the regression coefficient (B), which was calculated as follows: Risk Score=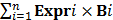 [23, 28]. Survival curves were generated by the Kaplan-Meier method and compared using the log-rank test. The variables identified by the univariate Cox analysis with a threshold P value <0.10 were included in the multivariate Cox proportional hazards regression analysis. The results generated by Cox regression analyses were reported as hazard ratio (HR) and the corresponding 95% confidential interval (CI). The statistical analyses were performed with the use of BRB-Array Tools and SPSS (version 21.0; SPSS Institute Inc, Chicago, IL, USA), and the statistical significance was taken as a two-sided P value <0.05 unless specifically indicated.
[23, 28]. Survival curves were generated by the Kaplan-Meier method and compared using the log-rank test. The variables identified by the univariate Cox analysis with a threshold P value <0.10 were included in the multivariate Cox proportional hazards regression analysis. The results generated by Cox regression analyses were reported as hazard ratio (HR) and the corresponding 95% confidential interval (CI). The statistical analyses were performed with the use of BRB-Array Tools and SPSS (version 21.0; SPSS Institute Inc, Chicago, IL, USA), and the statistical significance was taken as a two-sided P value <0.05 unless specifically indicated.
Results
Baseline clinical and pathological characteristics
All 350 participants enrolled in this study were clinically and pathologically diagnosed with ccRCC with no history of other malignancies or neoadjuvant therapy. The median age for all these subjects was 60 years with an IQR of 41-79 years, and the median follow-up time was 33.9 months (IQR: 11.3-55.1 months). Overall, a total of 110 patients (31.4%) died after a median follow-up time of 19.8 months (IQR: 10.3-42.1 months), and the detailed demographic and clinicopathologic characteristics were summarized in Table 1.
BAP1 mutation status and clinical outcomes
Among all 350 ccRCC patients, a summary of 35 (10.0%) mutations were detected, which included 9 (2.6%) nonsense, 15 (4.3%) missense, 3 (0.8%) splice site, and 8 (2.3%) frameshift mutations (Figure 1 and Supplementary Table S1). The ccRCC patients with mutated BAP1 showed a trend towards higher AJCC stage (P=0.006), pathologic T stage (P=0.009), Fuhrman grade (P=0.003) and distant metastasis (P=0.029) compared with those with wild-type BAP1 (Table 1). As depicted in Figure 1, the patients with BAP1 mutant tumors had a worse prognosis in terms of median overall survival (OS; BAP1 mutant vs. wild type: 31.7 vs. 86.7 months; P=0.008). Furthermore, the univariate Cox regression analysis confirmed that patients with mutant BAP1 had a significantly higher probability of death (HR =1.909, 95% CI: 1.175-3.102; P=0.009); however, unfortunately, it did not pass the multivariate analysis after adjusting for the other significant prognostic variables (Supplementary Table S2).
Differentially expressed miRNAs between BAP1 mutant and wild-type tumors
The miRNA expression in BAP1 mutated (n=35) and wild-type (n=315) tumors was profiled, and a total of 33 miRNAs were found to be expressed differentially after adjustment for multiple testing (Supplementary Table S3). Among all the 33 miRNAs, 15 miRNAs (45.5%) were up-regulated and the remaining 18 (54.5%) were down-regulated in tumors with mutated BAP1 compared with those with wild-type BAP1. In addition, the unsupervised hierarchical clustering with the 33 miRNAs expression data could clearly separate the tumors with mutated and wild-type BAP1 (Figure 2).
BAP1 mutation status correlates with overall survival. A. Pie chart representing the distribution of different BAP1 mutation types in 350 ccRCC patients; B. Kaplan-Meier curve analysis of the association between BAP1 mutation status and overall survival in 350 ccRCC patients.
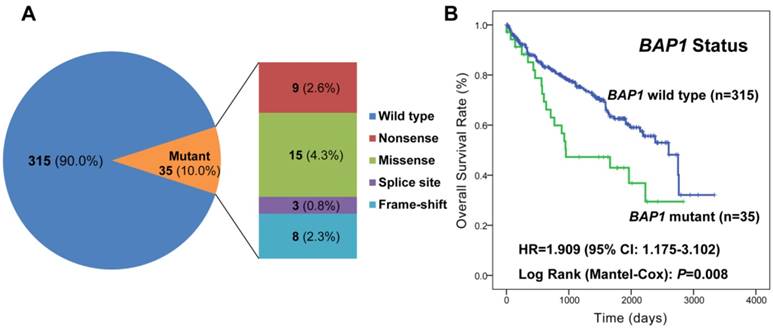
Unsupervised cluster analysis of the 33 differentially expressed miRNAs between BAP1 mutant and wild-type tumors. The transformed log2 intensities of significant miRNAs were mean centered across 350 ccRCC samples and a hierarchical clustering with average linkage was conducted.
Establishment of BAP1 mutation-specific miRNA signature
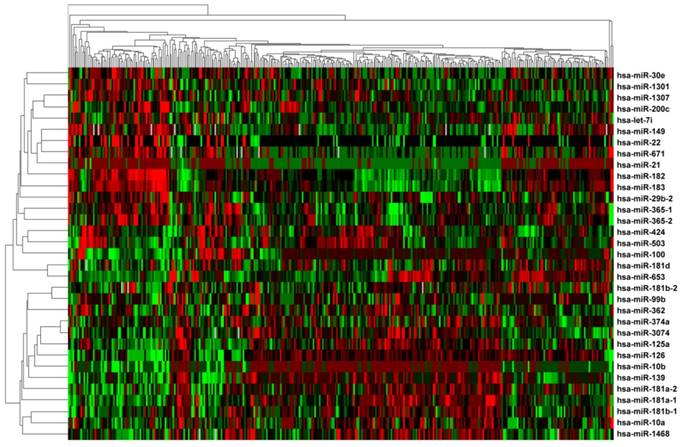
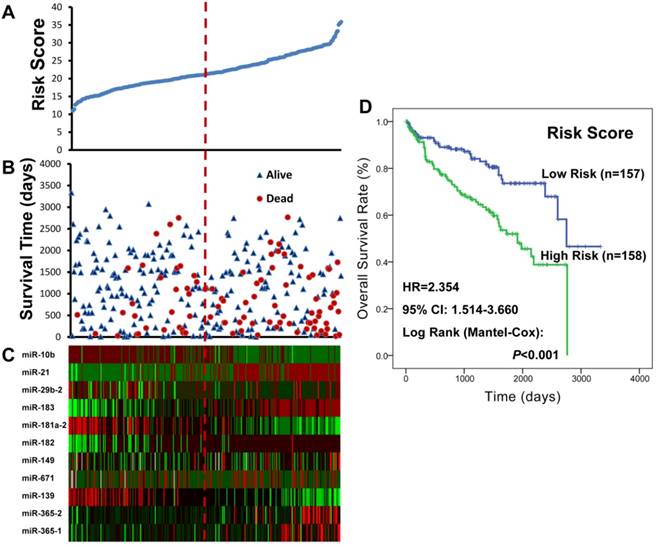
To identify miRNAs of prognostic values for ccRCC patients with wild-type BAP1, the 33 differentially expressed miRNAs were subjected to univariate Cox proportional hazard regression analysis. A total of eleven miRNAs (miR-149, miR-29b-2, miR-182, miR-183, miR-21, miR-365-2, miR-671, miR-365-1, miR-10b, miR-139, and miR-181a-2) were found to be significantly related with patient survival (Supplementary Table S4), and confirmed by Kaplan-Meier survival curves (Figure 3). Among the 11 significant miRNAs, eight miRNAs (miR-149, miR-29b-2, miR-182, miR-183, miR-21, miR-365-2, miR-671, and miR-365-1) were negatively associated with patients' OS and the other three (miR-10b, miR-139, and miR-181a-2) were positively associated. Finally, an eleven-miRNA signature risk score was calculated, and the 315 ccRCC patients with wild-type BAP1 were divided into high-risk (n= 158) and low-risk (n=157) groups (Figure 4).
Validation of the eleven-miRNA signature as an independent prognostic factor for ccRCC patients with wild-type BAP1
As presented in the Kaplan-Meier survival curves (Figure 4), the high-risk patients had a shorter median OS compared with the low-risk ones (high- vs. low-risk: 63.7 vs. 91.7 months; P<0.001). The differences in patients' OS corresponded to a HR of 2.354 (95% CI: 1.514-3.660; P<0.001) derived from the univariate Cox regression analysis. After adjusting for the other significant variables including age, AJCC stage, pathologic T, metastasis status, Fuhrman grade, and surgical type (Table 2), the eleven-miRNA signature was proven as an independent prognostic factor for BAP1 wild-type ccRCC patients (HR=1.602, 95% CI: 1.012-2.536; P=0.044).
Univariate and multivariate analysis of parameters associated with overall survival in ccRCC patients with wild-type BAP1
| Variables | HR | 95% CI | P value |
|---|---|---|---|
| Univariate Cox regression analysis | |||
| Age (≥ 60 vs. <60) | 2.849 | 1.772-4.581 | <0.001 |
| Gender (male vs. female) | 0.949 | 0.613-1.469 | 0.815 |
| Ethnicity (Caucasians vs. non-Caucasians) | 1.147 | 0.362-3.635 | 0.816 |
| AJCC stage (Ⅲ+Ⅳ vs. Ⅰ+Ⅱ) | 4.874 | 3.023-7.857 | <0.001 |
| Tumor size (T3+T4 vs. T1+T2) | 3.791 | 2.427-5.922 | <0.001 |
| Metastasis status (M1 vs. M0) | 4.720 | 3.074-7.247 | <0.001 |
| Fuhrman grade (G3+G4 vs. G1+G2) | 2.645 | 1.619-4.319 | <0.001 |
| Surgical type (RN vs. PN) | 5.131 | 1.882-13.99 | 0.001 |
| Risk Score (High Risk vs. Low Risk) | 2.354 | 1.514-3.660 | <0.001 |
| Multivariate Cox regression analysis a | |||
| Age (≥ 60 vs. <60) | 2.341 | 1.452-3.775 | <0.001 |
| AJCC stage (Ⅲ+Ⅳ vs. Ⅰ+Ⅱ) | 2.664 | 1.529-4.642 | 0.001 |
| Metastasis status (M1 vs. M0) | 2.069 | 1.251-3.420 | 0.005 |
| Risk Score (High Risk vs. Low Risk) | 1.602 | 1.012-2.536 | 0.044 |
ccRCC, clear cell renal cell carcinoma; BAP1, BRCA1 associated protein-1; HR, hazard ratio; 95% CI, 95% confidential interval; AJCC, AJCC, American Joint Committee on Cancer; RN, radical nephrectomy; PN, partial nephrectomy.
a The final results after stepwise analysis with Backward (LR) method
Kaplan-Meier curve analysis of miRNAs for the overall survival in ccRCC patients with wild-type BAP1. A total of 11 miRNAs were presented, including miR-149, miR-29b-2, miR-182, miR-183, miR-21, miR-365-2, miR-671, miR-365-1, miR-10b, miR-139, and miR-181a-2.
The BAP1 mutation-specific miRNA risk score analysis of ccRCC patients with wild-type BAP1. A. miRNA risk score distribution; B. Patients' survival status and time; C. Heatmap of the miRNA expression profiles; rows represented miRNAs, and columns indicated patients. The red dotted line represented the miRNA signature cutoff value (median), which divided patients into low-risk and high-risk groups. D. Kaplan-Meier curve analysis of the miRNA signature for the overall survival in 315 ccRCC patients with wild-type BAP1.
Target gene prediction and functional annotation analysis
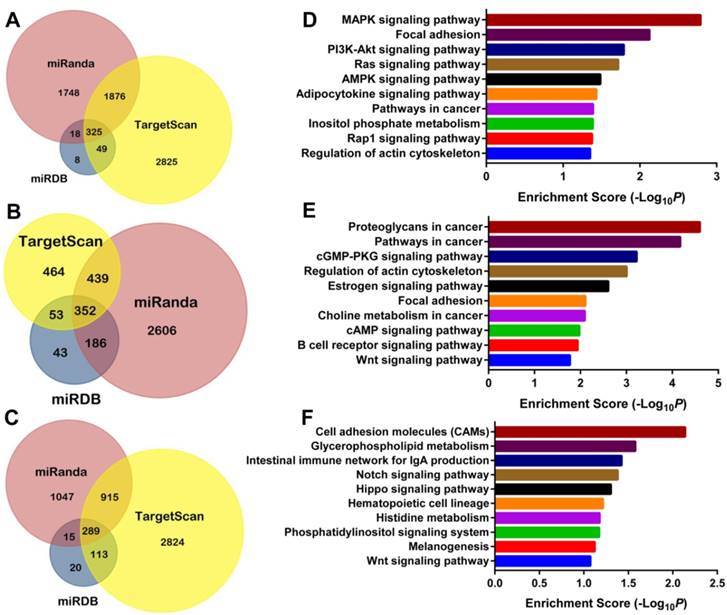
Three computational algorithms (TargentScan, miRDB, and miRanda) were applied to predict miRNA targets, and a total of 2268 target genes were selected for miR-29b-2, 1030 for miR-182, and 1332 for miR-671 by at least two independent tools (Figure 5 and Supplementary Table S5). The GO enrichment analysis of miRNA target genes identified a summary of 281 terms for miR-29b-2, 337 for miR-182, and 278 for miR-671, which indicated a wide range of biological function during the development and progression of ccRCC (Supplementary Table S5). Furthermore, the KEGG pathway enrichment analysis demonstrated 32 miR-29b-2 associated, 62 miR-182 related, and 25 miR-671 linked pathways (Supplementary Table S5). As shown in Figure 5, the top 10 related pathways for each miRNA suggested that they were widely involved in the cancer-related pathways. Of note, the results provided direct evidence that miR-182 was involved in the pathway “renal cell carcinoma” through modulating eight target genes.
Discussion
The identification and validation of novel biomarkers for ccRCC comprise a significant area of practical cancer research [38, 39]. The major findings of the present study are as follows: 1) BAP1 was mutated in about 10.0% of ccRCC patients, and those with mutated BAP1 had a worse prognosis; 2) thirty-three miRNAs expressed differentially between wild-type and mutated BAP1 tumors; 3) a BAP1 mutation-specific miRNA signature consisting of eleven miRNAs could serve as an independent prognostic parameter for ccRCC patients with wild-type BAP1. To the best of our knowledge, this is the first study to explore the genome-wide miRNA expression in BAP1 mutant and wild-type ccRCC tumors, which could help predict patient prognosis in the era of precision medicine.
Predicted miRNA target genes and enriched KEGG pathways. The Venn Diagrams of predicted targets for miR-29b-2 (A), miR-182 (B), and miR-671 (C); the top 10 related KEGG pathway enriched by target genes for miR-29b-2 (D), miR-182 (E), and miR-671 (F).
BAP1 is a classical two-hit tumor suppressor gene with important functions in cell proliferation and growth inhibition [40]. The immunohistochemistry assays of RCC tumor tissues have demonstrated that loss of BAP1 protein expression was specific for ccRCC compared with other histological subtypes of RCC [41, 42], and could function as a potent prognostic factor for ccRCC [43-45]. In the current study, we observed that BAP1 was mutated in 10.0% of the 350 ccRCC patients, with the similar frequency as the previous immunohistochemistry [41, 43, 44] and mutational analyses studies [12-17]. The ccRCC patients with mutated BAP1 were more likely to present high AJCC stage, pathologic T stage, Fuhrman grade and distant metastasis, and had a shorter median OS (BAP1 mutant vs. wild type: 31.7 vs. 86.7 months; P=0.008). In the Cox regression analysis, a HR of 1.909 (95% CI: 1.175-3.102; P=0.009) was obtained from the univariate analysis; however, no significant result for the prognostic value of BAP1 mutation was found after including the other variables for multivariate analysis, which could be explained by the limited sample size. It is conceivable that BAP1 mutation could serve as a potent prognostic parameter, as previous studies and the current study have indicated that ccRCC patients with mutated BAP1 had a shorter median survival time [14, 15, 17].
Previous studies have demonstrated that BAP1 mutant tumors exhibit a highly specific gene expression signature [13, 14]; however, to date, the miRNA expression profiles between BAP1 mutant and wild-type tumor have never been compared. As a class of small, endogenous, non-coding RNAs, miRNAs have been suggested to regulate a wide range of pathophysiological processes including carcinogenesis and metastasis [19]. In the current study, we explored the genome-wide miRNA expression profiles in BAP1 mutant and wild-type ccRCC tumors, and found a total of 33 differentially expressed miRNAs. Subsequently, eleven miRNAs (miR-149, miR-29b-2, miR-182, miR-183, miR-21, miR-365-2, miR-671, miR-365-1, miR-10b, miR-139, and miR-181a-2) were confirmed to be of prognostic values for ccRCC patients with wild-type BAP1, and a BAP1 mutation-specific miRNA signature combining all the 11 miRNAs was generated. Among the 11 miRNAs, eight except miR-29b-2, miR-182, and miR-671 were overlapped with the tumor-specific miRNA signature of 22 miRNAs reported by our group [28], partially due to the fact that BAP1 wild-type ccRCC patients accounted for 90.0% of all subjects.
All the three miRNAs (miR-29b-2, miR-182, and miR-671) were risky miRNAs, as the higher expression levels were negatively associated with patient survival. miR-29b-2 is transcribed from a locus at chromosome 1q32.2 with 2268 predicted target genes, and functions as a double-edged sword in the development and progression of human malignancies [46, 47]. Even though miR-29b-2 was well reputed as a tumor suppressor, the emerging evidence has demonstrated that it could exert oncogenic functions under certain conditions [48, 49], which was in line with the current study. miR-182 is a member of the miR-183 family, which could function as an tumor promoter in the urologic neoplasms including prostate cancer and bladder cancer [50, 51]. In this study, a total of 1030 target gene of miR-182 were predicted, and 62 pathways were enriched including “proteoglycans in cancer”, “pathway in cancer”, “Wnt signaling pathway”, “renal cell carcinoma”, and others. Contrary to the current study, Xu and colleagues found the down regulation of miR-182 in RCC tissues compared with the matched normal tissues, and demonstrated that miR-182 could suppress the proliferation and carcinogenesis of RCC cell lines by in vitro and in vivo studies [52]. As for miR-671, the biological functions in the development and progression of human cancers remains not yet understood. The bioinformatics analysis identified a total of 1332 target genes and enriched 278 GO terms and 25 pathways including “cell adhesion molecules”, ''Notch signaling pathway,'' ''Hippo signaling pathway'', et al for miR-671.
Some limitations of the current study should be acknowledged. First, due to the limited follow-up information, the endpoint of the present study was only OS [14], which could hinder the yielding of more interesting analyses. Second, in the multivariate Cox regression analysis, BAP1 mutation was not proved as an independent prognostic factor, which could be partially attributed to the limited sample size. Third, even though we strictly selected the subjects to control the potential heterogeneity and applied the leave-one-out cross validation method, the false positive results do potentially exist, which need an external validation cohort and functional studies to validate the results.
Conclusions
In summary, by employing a large independent ccRCC patient cohort, our study identified a total of 33 miRNAs differentially expressed between BAP1 mutant and wild-type tumors, and generated a BAP1 mutation-specific miRNA signature consisting of eleven miRNAs, which could serve as a novel prognostic biomarker for ccRCC patients with wild-type BAP1. Further studies focusing on the verifications of our findings in prospectively designed clinical trials, and the functional elucidation of these miRNAs are warranted.
Supplementary Material
Additional File 1Supplementary Tables S1-S4.
Supplementary Table S5.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (81370853 and 81570613) to RPJ, and Research and Innovation Program for Graduates of Jiangsu Province (CXZZ13_0583), Jiangsu Provincial Medical Youth Talent (QNRC2016073), Key Program of Science and Technology Foundation from Nanjing Medical University (2016NJMUZD042), and the National Natural Science Foundation of Jiangsu Province (BK20170135) to YZG. We would like to express our sincere thanks to The Cancer Genome Atlas (TCGA) project for generating, curating and providing high-quality biological and clinical data about clear cell renal cell carcinoma.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European journal of cancer. 2013;49:1374-403
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: a cancer journal for clinicians. 2017;67:7-30
3. Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387:894-906
4. Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. European urology. 2016;70:93-105
5. Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M. et al. EAU guidelines on renal cell carcinoma: 2014 update. European urology. 2015;67:913-24
6. Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A. et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. European urology. 2015;67:85-97
7. Wang H, Wu J, Gu W, Wang B, Wan F, Dai B. et al. Serum Adiponectin Level May be an Independent Predictor of Clear Cell Renal Cell Carcinoma. Journal of Cancer. 2016;7:1340-6
8. He X, Guo S, Chen D, Yang G, Chen X, Zhang Y. et al. Preoperative Albumin to Globulin Ratio (AGR) as Prognostic Factor in Renal Cell Carcinoma. Journal of Cancer. 2017;8:258-65
9. Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG. et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer research. 2008;68:6953-62
10. Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nature reviews Cancer. 2013;13:153-9
11. Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G. et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nature communications. 2015;6:8471
12. Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y. et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics. 2012;44:17-9
13. Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44:751-9
14. Kapur P, Pena-Llopis S, Christie A, Zhrebker L, Pavia-Jimenez A, Rathmell WK. et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. The lancet oncology. 2013;14:159-67
15. Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M. et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:3259-67
16. Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43-9
17. Gossage L, Murtaza M, Slatter AF, Lichtenstein CP, Warren A, Haynes B. et al. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes, chromosomes & cancer. 2014;53:38-51
18. Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516-9
19. Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nature reviews Genetics. 2016;17:719-32
20. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. The lancet oncology. 2012;13:e249-58
21. Ge YZ, Xu LW, Xu Z, Wu R, Xin H, Zhu M. et al. Expression Profiles and Clinical Significance of MicroRNAs in Papillary Renal Cell Carcinoma: A STROBE-Compliant Observational Study. Medicine. 2015;94:e767
22. Ge YZ, Xin H, Lu TZ, Xu Z, Yu P, Zhao YC. et al. MicroRNA expression profiles predict clinical phenotypes and prognosis in chromophobe renal cell carcinoma. Scientific reports. 2015;5:10328
23. Xu Z, Yu YQ, Ge YZ, Zhu JG, Zhu M, Zhao YC. et al. MicroRNA expression profiles in muscle-invasive bladder cancer: identification of a four-microRNA signature associated with patient survival. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:8159-66
24. Das F, Dey N, Bera A, Kasinath BS, Ghosh-Choudhury N, Choudhury GG. MicroRNA-214 Reduces Insulin-like Growth Factor-1 (IGF-1) Receptor Expression and Downstream mTORC1 Signaling in Renal Carcinoma Cells. The Journal of biological chemistry. 2016;291:14662-76
25. Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C. et al. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835-41
26. Wu X, Weng L, Li X, Guo C, Pal SK, Jin JM. et al. Identification of a 4-microRNA signature for clear cell renal cell carcinoma metastasis and prognosis. PloS one. 2012;7:e35661
27. Osanto S, Qin Y, Buermans HP, Berkers J, Lerut E, Goeman JJ. et al. Genome-wide microRNA expression analysis of clear cell renal cell carcinoma by next generation deep sequencing. PloS one. 2012;7:e38298
28. Ge YZ, Wu R, Xin H, Zhu M, Lu TZ, Liu H. et al. A tumor-specific microRNA signature predicts survival in clear cell renal cell carcinoma. Journal of cancer research and clinical oncology. 2015;141:1291-9
29. Kim SP, Alt AL, Weight CJ, Costello BA, Cheville JC, Lohse C. et al. Independent validation of the 2010 American Joint Committee on Cancer TNM classification for renal cell carcinoma: results from a large, single institution cohort. The Journal of urology. 2011;185:2035-9
30. Sun M, Lughezzani G, Jeldres C, Isbarn H, Shariat SF, Arjane P. et al. A proposal for reclassification of the Fuhrman grading system in patients with clear cell renal cell carcinoma. European urology. 2009;56:775-81
31. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401-4
32. Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer informatics. 2008;6:9-15
33. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015:4
34. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research. 2015;43:D146-52
35. Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome biology. 2010;11:R90
36. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44-57
37. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1-13
38. Ricketts CJ, Crooks DR, Sourbier C, Schmidt LS, Srinivasan R, Linehan WM. SnapShot: Renal Cell Carcinoma. Cancer cell. 2016;29:610-e1
39. Barbieri CE, Chinnaiyan AM, Lerner SP, Swanton C, Rubin MA. The Emergence of Precision Urologic Oncology: A Collaborative Review on Biomarker-driven Therapeutics. European urology. 2017;71:237-46
40. Brugarolas J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer journal. 2013;19:324-32
41. Ho TH, Kapur P, Joseph RW, Serie DJ, Eckel-Passow JE, Parasramka M. et al. Loss of PBRM1 and BAP1 expression is less common in non-clear cell renal cell carcinoma than in clear cell renal cell carcinoma. Urologic oncology. 2015;33:23 e9-14
42. Joseph RW, Kapur P, Serie DJ, Parasramka M, Ho TH, Cheville JC. et al. Clear Cell Renal Cell Carcinoma Subtypes Identified by BAP1 and PBRM1 Expression. The Journal of urology. 2016;195:180-7
43. Joseph RW, Kapur P, Serie DJ, Eckel-Passow JE, Parasramka M, Ho T. et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer. 2014;120:1059-67
44. Kapur P, Christie A, Raman JD, Then MT, Nuhn P, Buchner A. et al. BAP1 immunohistochemistry predicts outcomes in a multi-institutional cohort with clear cell renal cell carcinoma. The Journal of urology. 2014;191:603-10
45. Eckel-Passow JE, Serie DJ, Cheville JC, Ho TH, Kapur P, Brugarolas J. et al. BAP1 and PBRM1 in metastatic clear cell renal cell carcinoma: tumor heterogeneity and concordance with paired primary tumor. BMC urology. 2017;17:19
46. Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. European journal of cell biology. 2013;92:123-8
47. Yan B, Guo Q, Fu FJ, Wang Z, Yin Z, Wei YB. et al. The role of miR-29b in cancer: regulation, function, and signaling. OncoTargets and therapy. 2015;8:539-48
48. Shaker O, Maher M, Nassar Y, Morcos G, Gad Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene. 2015;560:77-82
49. Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y. et al. miR-29a/b Enhances Cell Migration and Invasion in Nasopharyngeal Carcinoma Progression by Regulating SPARC and COL3A1 Gene Expression. PloS one. 2015;10:e0120969
50. Casanova-Salas I, Rubio-Briones J, Calatrava A, Mancarella C, Masia E, Casanova J. et al. Identification of miR-187 and miR-182 as biomarkers of early diagnosis and prognosis in patients with prostate cancer treated with radical prostatectomy. The Journal of urology. 2014;192:252-9
51. Pignot G, Cizeron-Clairac G, Vacher S, Susini A, Tozlu S, Vieillefond A. et al. microRNA expression profile in a large series of bladder tumors: identification of a 3-miRNA signature associated with aggressiveness of muscle-invasive bladder cancer. International journal of cancer Journal international du cancer. 2013;132:2479-91
52. Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y. et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Molecular cancer. 2014;13:109
Author contact
![]() Corresponding author: Rui-Peng Jia, M.D., Ph.D., Professor of Urology, Chief of Department of Urology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing 210006, China. E-mail: urojiarpcom OR urojiarpedu.cn; Tel: +86-25-52271061; Fax: +86-25-52271061
Corresponding author: Rui-Peng Jia, M.D., Ph.D., Professor of Urology, Chief of Department of Urology, Nanjing First Hospital, Nanjing Medical University, 68 Changle Road, Nanjing 210006, China. E-mail: urojiarpcom OR urojiarpedu.cn; Tel: +86-25-52271061; Fax: +86-25-52271061

 Global reach, higher impact
Global reach, higher impact