3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(14):2699-2703. doi:10.7150/jca.19947 This issue Cite
Short Research Communication
Evaluation Analysis of miRNAs Overexpression in Liquid-Based Cytology Endometrial Samples
1. Department of Cytopathology, University General Hospital “ATTIKON”, School of Medicine, National and Kapodistrian University of Athens, Athens 12464, Greece
2. Department of Obstetrics and Gynaecology, “Saint Savvas” Anticancer Hospital, Athens 11522, Greece
3. 3 rd Department of Gynaecology and Obstetrics, University General Hospital “ATTIKON”, School of Medicine, National and Kapodistrian University of Athens, Athens 12464, Greece
4. Department of Obstetrics and Gynaecology, IASO Hospital, 37-39 Kifisias Av., Maroussi, 15123, Athens, Greece
5. Colposcopy Unit, 1 st Department Department of Gynaecology and Obstetrics, General Hospital “Elena Venizelou”, Athens, 11521, Greece
6. 2 nd Department of Pathology, University General Hospital “ATTIKON”, School of Medicine, National and Kapodistrian University of Athens, Athens 12464, Greece
*Joint first authors
† In loving memory of a dedicated doctor, an outstanding scientist, and a beloved teacher.
Received 2017-3-6; Accepted 2017-6-7; Published 2017-8-22
Abstract
Background: miRNAs have an important role as their deregulation is linked to endometrial cancer.
Methods: A custom miScript® miRNA PCR Array was used to investigate for the first time the expression of eight miRNAs in forty-nine histologically confirmed Liquid Based cytology endometrial samples. The expression profile of the same miRNAs was also examined in sixty formalin-fixed tissue samples.
Results: Expression of seven miRNAs was significantly higher in malignant samples with three of them (mir-182, mir-141 and mir-205) performing optimally.
Conclusion: These results suggest the potential use of this non-invasive method of sampling for miRNA expression studies. Furthermore miRNA overexpression could serve as an ancillary or reflex test for optimal identification of malignant samples especially in morphologically inadequate samples.
Keywords: miRNAs, endometrial cancer, Liquid Based Cytology
Introduction
Endometrial cancer (EC) is the seventh most common cancer among women worldwide and the most common gynaecologic malignancy in Western countries [1]. Despite its high prevalence, the molecular mechanisms with a key role in the development of this cancer are still not fully elucidated. Apart from hormonal association in the progression of EC, several genetic implications have so far been described, including gene mutations, oncogene activation or tumour suppression gene inactivation [2, 3]. The role of miRNAs has been clearly considered important, since their deregulation is linked to cancer, a number of studies having so far elucidated this relationship [4, 5]. In endometrial cancer studies, a differential expression of miRNAs between endometrioid samples and normal tissue has been identified and their role in diagnosis, treatment and prevention has been described to be of utmost importance [6-8].
Evaluation of abnormal uterine bleeding is usually performed via dilatation and curettage. However, collection of endometrial cells via direct endometrial sampling and cytological evaluation through Liquid Based Cytology (LBC) has been used as an alternative, less interventional diagnostic procedure [9]. This study aims to explore the expression profile of miRNAs, whose important role in endometrial carcinogenesis has been described [10], in histologically confirmed LBC endometrial samples and compare the results to formalin-fixed paraffin embedded (FFPE) samples.
Materials and methods
A hundred and nine (109) samples were enrolled from women (28-85 years of age, mean 57.1). Forty-nine samples were direct endometrial cytological samples stored in PreservCyt solution from women attending the outpatient clinics of both the 3rd Department of Obstetrics and Gynaecology of the University General Hospital “ATTIKON” and the Department of Obstetrics and Gynaecology of “Saint Savvas” Anticancer Hospital between May 2012 and May 2015. These included samples from women referred due to abnormal uterine bleeding or due to increased endometrial thickness in ultrasound examination. Sixty FFPE samples were used to establish miRNA expression levels. As controls, in both LBCs and FFPEs groups, histological confirmed benign samples from women who underwent hysterectomy for non-EC related reasons or endometrial curettage were used. Ethical approval was obtained from the Bioethics Committee of the hospitals and all women gave a written consent.
RNA from FFPE and LBC samples was extracted with the miRNeasy FFPE Kit (Qiagen) and miRNeasy mini kit (Qiagen). RNA's quantity and quality was assessed using QIAxpert (Qiagen). Reverse transcription and amplification was performed in one step with the use of a custom miScript® miRNA PCR Array (Qiagen). The array plate was designed for Applied Biosystems 7500 Fast Real-Time PCR (Applied Biosystems, Foster City, CA, USA) to profile the expression of mir-9-5p, mir-125-5p, mir-141-3p, mir-182-5p, mir-200b-3p, mir-200c-3p, mir-205-5p and mir-222-3p. As normalization controls of the array data two small nucleolar RNAs (SNORD61, SNORD68) were used as endogenous controls. Replicate positive PCR controls as well as reverse transcription controls were included and all samples were run in duplicates.
Normalization of the miRNAs was performed using the geometric mean of all the histological benign samples and the relative expression was calculated with the ΔΔCt method. Stability of normalization was verified using the geNorm macro, which displayed the lowest variation when using the two SNORDs (M=0.56). One polyp and an endometrial carcinoma, from the FFPE samples, and two benign as well as one carcinoma, from the LBC samples displayed Ct values>32 for the SNORDs and were excluded from the analysis. All statistical analysis was performed in SPSS using non-parametric tests for difference of means (Mann-Whitney) and correlations (Spearman).
Results
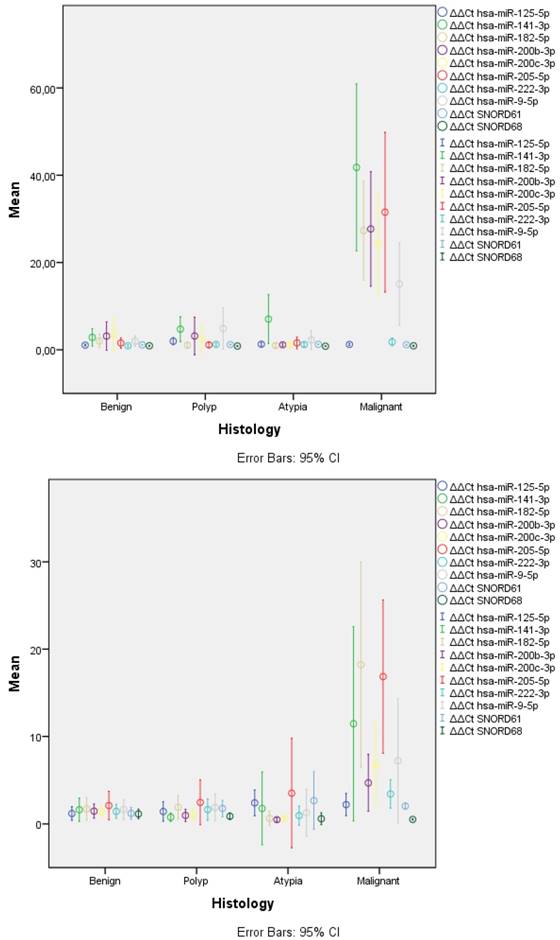
The only miRNA that remained stable across all samples was mir-125-5p (Table 1, Figure 1). Most of the remaining were significantly overexpressed in malignant samples when compared to either the benign samples or any of the non-malignant categories. mir-222-3p and mir-9-5p were significantly overexpressed in malignant samples compared only to benign for both types of samples. However, they displayed variable results when compared to other categories. The only miRNA that showed a gradual increase as histology progressed from benign to polyp, atypical and malignancy was mir-141-3p reaching significance across all categories. The rest of the overexpressed miRNAs had similar levels of expression to benign samples and a steep overexpression was observed in malignancy.
In general, LBC samples displayed higher variance in expression of miRNAs and this was evident across all categories (Table 1, Figure 1). This was caused, most probably, due to lower concentration of isolated miRNAs as mirrored by the significant higher Ct values for SNORDs (28.75 vs. 19.95, p<0.001) and most miRNAs across histology categories.
Several significant correlations were identified. For FFPE samples, the gradual increase of expression of mir-141-3p was also mirrored by the highest correlation with worse histology with a spearman correlation co-efficiency (CC) of 0.729 (P<0.001). Lower CCs were identified for mir-205-5p (0.705), mir-182-5p (0.621), mir-200b-3p (0.614), mir-200c-3p (0.605) and mir-9-5p (0.505). Among the miRNAs, the most significant correlation was identified among mir-200b-3p with mir-200c-3p (0.989), mir-182-5p (0.964) and mir-205-5p (0.904). Similar results were found for LBC samples with lower CCs; i.e. mir-141-3p (p=0.015, CC 0.446) and mir-205-5p (0.002, 0.560).
Concerning malignancy identification, ROC analysis identified mir-182-5q as having the highest area under the curve (0.979) and a 4-fold overexpression cut-off displayed a sensitivity of 93.8% with a specificity of over 95%. Keeping the same lower limit for specificity, similar results were identified for a 13-fold increase of mir-141-3p (AUC 0.964, Sensitivity 81.3) and an 8-fold increase of mir-205-5p (0.964, 81.3). Less optimal results were identified for a 13-fold increase of mir-200b-3p (0.954, 68.8), a 13-fold increase of mir-200c-3p (0.949, 68.8). The remaining miRs had lower AUC and sensitivity under 65%. Similar results were found for LBC samples with lower AUCs; i.e. mir-141-3p (0.955), mir-182-5p (0.942) with a sensitivity of 85.7% and a specificity of 100% for a 5-fold increase (table 1).
Expression levels of miRNAs and SNORDs across histological categories. FFPE samples (left) displayed significantly lower variance in all categories than LBC samples (right). Similar overexpression of most miRNAs was observed, with LBC samples having lower overexpression ratios, especially for hsa-miR-200b-3p and hsa-mir-200c-3p; LBC: Liquid Based Cytology, FFPE: Formalin-Fixed Paraffin Embedded.
List of miRNAs tested in LBC endometrial samples and FFPE samples. The overexpression fold change, the p-value and the AUC of the malignant samples ROC analysis are presented; LBC: Liquid Based Cytology, FFPE: Formalin-Fixed Paraffin Embedded, AUC: Area Under Curve, ROC: Receiver Operating Characteristic.
| FFPE Histology (N=58) | Benign (N=24) Fold change (95CI) | p-value | Polyp (N=10) Fold change (95CI) | p-value | Hyperplasia with atypia (N=8) Fold change (95CI) | p-value | Malignant (N=16) Fold change (95CI) | AUC for malignant (95CI) |
| miR-9-5p | 1.96 (0.73-0.32) | <0.001 | 4.86 (0.08-9.6) | 0.060 | 2.23 (0.05-4.49) | 0.005 | 15.05 (5.69-24.42) | 0,82(0,70-0.94) |
| miR-125-5p | 1.06 (0.84-1.28) | 0.318 | 1.96 (1.14-2.79) | 0.027 | 1.28 (0.82-1.73) | 0.742 | 1.22 (0.97-1.47) | 0.48 (0.32-0.64) |
| miR-141-3p | 2.83 (0.82-4.84) | <0.001 | 4.27 (1.86-7.58) | <0.001 | 7.02 (1.41-12,62) | 0.001 | 41.79 (22.66-60.92) | 0.96 (0.92-1) |
| miR-182-5p | 2.01 (0.50-3.53) | <0.001 | 1.06 (0.39-1.74) | <0.001 | 0.95 (0.47-1.43) | <0.001 | 27.30 (15.94-38.65) | 0.97 (0.94-1) |
| miR-200b-3p | 3.13 (0.00-6.39) | <0.001 | 3.16 (0.00-7.47) | <0.001 | 1.14 (0.77-1.51) | <0.001 | 27.68 (14.56-40.81) | 0.95 (0.90-1) |
| miR-200c-3p | 3.43 (0.00-7.19) | <0.001 | 2.51 (0.00-5.91) | <0.001 | 1.12 (0.74-1.51) | <0.001 | 24.40 (12.68-36.11) | 0.94 (0.89-1) |
| miR-205-5p | 1.55 (0.33-2.76) | <0.001 | 1.12 (0.52-1.71) | <0.001 | 1.58 (0.21-2.96) | <0.001 | 31.52 (13.20-49.84) | 0.96 (0.91-1) |
| miR-222-3p | 0.91 (0.60-1.21) | 0.018 | 1.23 (0.84-1.62) | 0.737 | 1.20 (0.77-1.63) | 0.806 | 1.80 (0.87-2.73) | 0.64 (0.49-0.79) |
| LBC Histology (N=46) | Benign (N=18) Fold change (95CI) | p-value | Polyp (N=10) Fold change (95CI) | p-value | Hyperplasia with atypia (N=6) Fold change (95CI) | p-value | Malignant (N=12) Fold change (95CI) | AUC for malignant (95CI) |
| miR-9-5p | 1.65 (0.51-2.78) | 0.004 | 1.89 (0.34-3.44) | 0.010 | 1.28 (-1.3-3.96) | 0.073 | 7.21 (0.12-14.30) | 0.89 (0.77-1) |
| miR-125-5p | 1.19 (0.40-1.97) | 0.189 | 1.42 (0.30-2.52) | 0.109 | 2.40 (0.92-3.89) | 0.788 | 2.21 (0.92-3.49) | 0.67 (0.44-0.90) |
| miR-141-3p | 1.63 (0.29-2.96) | 0.004 | 0.76 (0.28-1.24) | <0.001 | 1.77 (-2.39-5.93) | 0.042 | 11.45 (0.33-22.57) | 0.95 (0.88-1) |
| miR-182-5p | 1.71 (0.39-3.03) | 0.004 | 1.89 (0.54-3.25) | 0.002 | 0.64 (0.17-1.46) | 0.006 | 18.21 (6.41-30.00) | 0.94 (0.82-1) |
| miR-200b-3p | 1.46 (0.65-2.27) | 0.029 | 0.97 (0.26-1.68) | 0.003 | 0.49 (0.27-0.70) | 0.006 | 4.69 (1.44-7.94) | 0.90 (0.77-1) |
| miR-200c-3p | 1.41 (0.71-2.11) | 0.006 | 1.06 (0.35-1.78) | 0.001 | 0.62 (0.44-0.80) | 0.006 | 6.27 (2.00-11.52) | 0.94 (0.83-1) |
| miR-205-5p | 2.10 (0.47-3.73) | <0.001 | 2.4 (0.09-5.02) | <0.001 | 3.52 (-2.4-9.78) | 0.012 | 16.85 (8.07-25.63) | 0.96 (0.93-1) |
| miR-222-3p | 1.41 (0.64-2.23) | 0.021 | 1.64 (0.42-2.82) | 0.033 | 0.95 (0.14-2.05) | 0.024 | 3.43 (1.81-5.05) | 0.85 (0.67-1) |
Discussion
Our data represents, for the first time to our best knowledge, analysis of miRNAs in histologically confirmed cytological material collected via direct endometrial sampling and processed by Liquid Based Cytology. According to the statistical calculations, the most robust overexpression was recorded for mir-182, mir-141 and mir-205 for both LBC and FFPE samples.
miRNA-182 targets gene transcription of the tumour suppressor Transcription Elongation Factor A-like 7 (TCEAL7) gene in Endometrial Carcinoma [1, 11]. As shown mir-182 functions as an oncogenic miRNA in EC [1] and its overexpression is related to cell proliferation promotion by targeting the tumour suppressor gene TCEAL7 while it also modulates the activity of its downstream effectors c-Myc, cyclin D1 and NFκB [12, 13]. Also, up-regulation of mir-182 suppresses FOXO1 gene which in endometrial cancer is down-regulated compared to normal tissue playing an important role in endometrial tumorigenesis [7].
According to Dong et al., [6], the up-regulation of mir-205 and mir-141, which is a member of mir-200 family, might be related to hormone receptor status and prognosis in endometrial carcinomas. The importance of mir-205 and mir-141 miRNAs in endometrial cancer is also reflected to in their ability to regulate the expression of factors important for epithelial mesenchymal transition (EMT) and tumour metastasis [14].
In the present study, our effort was to elucidate whether miRNA analysis overexpression in an endometrial sample type, which is easier accessible as far the patient convenience is concerned, has robust and valuable results. The results of this study, suggest that the expression profiles of mir-182, mir-141 and mir-205, which are proven to have a strong relationship with tumour progression of this gynaecologic malignancy, could potentially serve as biomarkers to differentiate among non-malignant and malignant samples when an endometrial LBC sample is used. More significantly, four histologically verified carcinomas that were cytologically evaluated as inadequate (n=3) or benign (n=1) would all be identified by using those three miRNAs, since in all four cases, two of these three miRNAs were overexpressed by a value higher than the cut-off set to still have 100% specificity. Certainly, a larger cohort of samples is needed to be examined, so that the trend that is described here would be reproduced. However, these preliminary results are promising, as they may have a high prognostic impact without unnecessary interventional procedures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Guo Y, Liao Y, Jia C, Ren J, Wang J, Li T. MicroRNA-182 promotes tumor cell growth by targeting transcription elongation factor A-like 7 in endometrial carcinoma. Cell Physiol Biochem. 2013;32:581-90
2. Rosa-Rosa JM, Leskela S, Cristobal-Lana E, Santon A, Lopez-Garcia MA, Munoz G. et al. Molecular genetic heterogeneity in undifferentiated endometrial carcinomas. Mod Pathol. 2016;29:1594
3. Chang YS, Huang HD, Yeh KT, Chang JG. Genetic alterations in endometrial cancer by targeted next-generation sequencing. Experimental and molecular pathology. 2016;100:8-12
4. Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer: the impact of mutant p53 on Drosha complex activity. Journal of experimental & clinical cancer research: CR. 2016;35:45
5. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6:235-46
6. Dong Y, Si JW, Li WT, Liang L, Zhao J, Zhou M. et al. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int J Clin Exp Pathol. 2015;8:2864-75
7. Lee TS, Jeon HW, Kim YB, Kim YA, Kim MA, Kang SB. Aberrant microRNA expression in endometrial carcinoma using formalin-fixed paraffin-embedded (FFPE) tissues. PLoS One. 2013;8:e81421
8. Sun H, Yan L, Tu R, Zhang Y, Ma L, Tang W. et al. Expression Profiles of Endometrial Carcinoma by Integrative Analysis of TCGA Data. Gynecologic and obstetric investigation. 2017;82:30-8
9. Margari N, Pouliakis A, Anoinos D, Terzakis E, Koureas N, Chrelias C. et al. A reporting system for endometrial cytology: Cytomorphologic criteria-Implied risk of malignancy. Diagn Cytopathol. 2016;44:888-901
10. Sianou A, Galyfos G, Moragianni D, Andromidas P, Kaparos G, Baka S. et al. The role of microRNAs in the pathogenesis of endometrial cancer: a systematic review. Arch Gynecol Obstet. 2015;292:271-82
11. Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW, Yim SF. et al. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358-65
12. Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J. et al. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008;27:7223-34
13. Lafferty-Whyte K, Bilsland A, Hoare SF, Burns S, Zaffaroni N, Cairney CJ. et al. TCEAL7 inhibition of c-Myc activity in alternative lengthening of telomeres regulates hTERT expression. Neoplasia. 2010;12:405-14
14. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-5601
Author contact
![]() Corresponding author: Correspondence: Christine Kottaridi, MSc, MPH, PhD; phone: +30-210-5831959; E-mail: ckottaridicom
Corresponding author: Correspondence: Christine Kottaridi, MSc, MPH, PhD; phone: +30-210-5831959; E-mail: ckottaridicom

 Global reach, higher impact
Global reach, higher impact