Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(15):2992-3000. doi:10.7150/jca.21038 This issue Cite
Research Paper
Overexpression of LAMC1 predicts poor prognosis and enhances tumor cell invasion and migration in hepatocellular carcinoma
1. Department of Hepatobiliary Surgery, Sun Yat-Sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, China
2. Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, China
3. Department of Pathology, Sun Yat-Sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, China
4. Department of Surgery, Qianfoshan Hospital, Shandong Province, Jinan, 250014, China
5. Department of Hepatobiliary Surgery, The Affiliated Hospital of Shandong University, Shandong Province, Jinan, 250000, China
* These two authors contribute to this work equally
Received 2017-5-16; Accepted 2017-7-3; Published 2017-8-25
Abstract
LAMC1 encodes an extracellular matrix protein, laminin γ1 chain, which is involved in several biological and pathological processes including tissue development, tumor cell invasion and metastasis. In present study, we demonstrated that both LAMC1 protein and mRNA levels were elevated in HCC tissue samples compared with non-cancerous tissue samples according to western blot analyses, immunohistochemistry (IHC) and microarray. Moreover, high LAMC1 expression was positively correlated with incomplete encapsulation (p=0.014), poor overall (OS, p=0.02) and disease-free survival (DFS, p=0.014). Using cell lines, we demonstrated that the levels of LAMC1 mRNA and protein were significantly higher in HCC cell lines than that in LO2 cell line. After the expression of LAMC1 was depressed by siRNA technique, the cell proliferation, migration and invasion were depressed significantly. Taken together, these data suggest that LAMC1 is enriched in HCC; overexpression of LAMC1 predicts poor prognosis, and enhances tumor cell invasion and migration. LAMC1 might be a new biomarker predictive of HCC prognosis and might also be a useful treatment target.
Keywords: LAMC1, hepatocellular carcinoma, prognosis, invasion, metastasis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer mortality worldwide. Approximately 700,000 people die of HCC each year. More importantly, the global incidence is still rising [1, 2]. The prognosis of HCC is poor because the disease is often at a fairly advanced stage at the time of diagnosis. Liver resection remains the main curative option for HCC with a 5-year overall survival rate of 50-70% after curative hepatectomy [2, 3]. However, the long term prognosis is still unsatisfactory due to the high recurrence rate. Although more and more molecular markers with high sensitivity and specificity have been proposed, none has justified its routine use in clinical practice till now [3, 4]. Hence, a better understanding of more molecular markers and pathways that contribute to the progression and recurrence of HCC is critical to develop more effective, targeted therapies [5, 6].
LAMC1 encodes laminin γ1 chain, a protein essential for assembly of basement membrane. Laminins have been described in a wide variety of biological and pathological processes including tissue development, tumor cell invasion and metastasis [7-9]. Moreover, laminin proteins are present in the extracellular matrix and cell membrane, serving as potential biomarkers for detection [7]. The laminin γ1 chain has been reported to be involved in carcinogenesis of the liver through Sp1-mediated transactivation at the LAMC1 promoter [10]. Moreover, C-16, a peptide derived from laminin γ1 chain, had been shown to significantly promote cell adhesion, enhance pulmonary metastasis and migration of murine melanoma B-16 cells, possibly by inducing the secretion of matrix metalloproteasis-9 (MMP-9) [11]. More recently, it was demonstrated that LAMC1 is upregulated in uterine carcinoma, and its expression was significantly associated with FIGO stage, myometrial invasion, cervical/adnexal involvement, angiolymphatic invasion and lymph node metastasis [12,13]. However, there is lack of the systematical investigation focusing on the expression features of LAMC1, clinical relevance, and biological function in HCC.
In present study, we first assessed the LAMC1 expression in HCC and its associations with clinical characters, tumor recurrence and patient survivals. Our results suggest that LAMC1 might be a new biomarker predictive of HCC prognosis and might also be a useful treatment target.
Materials and Methods
Patients and tissue specimens
The fresh frozen HCC tissues (n=59) and paraffin-embedded HCC specimens (n = 196) from cases that were histologically confirmed and did not undergo neo-adjuvant treatment were obtained from Sun Yat-Sen University Cancer Center in Guangzhou. Written informed consent was obtained from each patient, and the study was approved by the Institute Research Ethics Committee at the Cancer Center. The including criteria for present study were: 1) no previous treatment for HCC before surgery; 2) histologic confirmation of HCC; 3) R0 resection; 4) no lymph node or extrahepatic metastasis; 5) HCV negative, 6) the follow-up period ≥6.0months. After surgery, patients were followed up 3 monthly for the first 2 years, then 6 monthly thereafter with physical examination, blood tests for AFP and liver function, and contrast abdominal computed tomography (CT). The last follow-up date for patients still alive was in December 2016.
Cell culture
Normal primary NPECs (NPECw and NPEC01) were established as described previously, and cultured in keratinocyte/serum-free medium (Invitrogen, Carlsbad, CA, USA). The HCC cell lines (LO2, MHCC-97H, MHCC-97L, SNU-449, SK-Hep1, HCCLM3, HLE, Mahlavu, PLC/PRF/5, Hep3B, Huh7, and HepG2) were cultured in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), and in a humidified 5% CO2 incubator at 37°C.
Quantitative RT-PCR analysis
Total RNA was extracted from fresh tissue specimens and HCC cell lines by the TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions. After measuring the concentration and quality of RNA by NanoDrop spectrophotometer (ND-1000, Thermo Scientific, Massachusetts, USA), 2 ug of the total RNA was used to synthesized the complementary DNA (cDNA) using a reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA). Quantitative Real-time RT-PCR (qRT-PCR) using the Power SYBR Green qPCR SuperMix-UDG (Invitrogen, Grand Island, NY, USA) was used to determine the mRNA level of the target genes on a Light Cycler 480 SYBR Green I Master (Roche, Indianapolis, IN, USA). β-actin was used as an internal control. Three independent experiments were performed to make sure the quantitative determination.
Western blotting analysis
The proteins extracted from tissues and HCC cells were stored in 1x SDS sample buffer with protein inhibitor, cocktail (F. Hoffmann-La Roche Ltd, Basel, Switzerland). The concentration of proteins was measured by the BCA method using Spectramax M5 (Molecular Devices, Sunnyvale, CA, USA). After heat denaturation at 98°C for 10 minutes, equal amounts of protein (20ug for tissues, 10ug for cells) were electrophoretically separated in 9% SDS polyacrylamide gels, and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). 5% skimmed milk or 5% bovine serum albumin dissolved by PBS-T was used for blocking the membrane for 1 hour. Then the membrane was incubated overnight at 4°C with indicated antibodies. Then the membrane was incubated at normal temperature for 40 minutes with the secondary antibody (AB) (Pierce, Thermo Fisher Scientific Inc., Rockford, IL, USA). The signals were detected using enhanced chemiluminescence (ECL) (Amersham Pharmacia Biotech, Piscataway, NJ, USA). The membranes were stripped and normalized with a mouse monoclonal antibody against human β-tubulin (diluted 1:3000; Santa Cruz Biotechnology, Texas, USA) to confirm equal loading of the samples. Then the membrane treated with an HRP-conjugated secondary Ab (Pierce, Thermo Fisher Scientific Inc., Rockford, IL, USA). Equal amounts of whole tissue or cell lysates were electrophoretically separated in a 9% SDS polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane (Pall, Port Washington, USA). 5% skimmed milk was used to block the membrane for 1 hour (h) at room temperature. Then the membrane was incubated with the indicated antibodies overnight at 4°C. The signals were detected using an enhanced chemiluminescence (GE Healthcare, New Jersey, USA). The membranes were stripped and probed with a rabbit monoclonal antibody against human β-tubulin (diluted 1:3000; Santa Cruz Biotechnology, Texas, USA) to confirm equal loading of the samples.
Immunohistochemical analysis
The paraffin-embedded HCC samples were cut into 4-µm sections. Then the sections were baked for 2 hour at 65ºC, deparaffinized in xylenes and rehydrated with alcohol to distilled water. Three percent hydrogen peroxide was used to block endogenous peroxidase activity at room temperature for 10 min, and then the sections were boiled in Citrate Antigen Retrieval Solution (PH=6.0) for 5 min in an electric pressure cooker antigen retrieval. After that, the sections were incubated with the rabbit polyclonal anti-LAMC1 antibody (diluted 1:300, Abcam, Cambridge, MA, USA) overnight at 4 ºC in a moist chamber and a secondary antibody for 30 min at 37 ºC on the next day. Subsequently, the sections were stained for protein detection in 3, 3-diaminobenzidine for 2 min and counterstained with Mayer's hematoxylin to stain nucleus. Finally, the sections were dehydrated and mounted. As a negative control, the primary antibody was replaced by normal rabbit serum.
Each section was independently evaluated by two pathologists who were blinded to the clinical status of the patients. The biomarker was evaluated according to staining intensity scored as (0, negative; 1, weak; 2, moderate; 3, strong) and the extent of staining scored as the percentage of positive cells. The final immunoreactivity score (IRS) is the product of intensity score multiply the extent score.
SiRNA transfection
The siRNA oligoribonucleotides were purchased from Sigma (ID: NM_001042422). The scramble RNA duplex for the siRNA was nonhomologous to any human genome sequences. Hep3B and HepG2 cells (2 × 105) were seeded on 6-well plates for 24 h, and then transfected with 12.5 nM of the RNA duplex and 5 μL of Lipofecta-mine RNAiMAX (Invitrogen, Grand Island, NY, USA), according to the manufacturer's instructions. After 48h or 72 h, cells were harvested for further experiments.
MTS assays [3-(4,5-dimethyl-2-thiazolyl)-2,5- diphenyl-2-H-tetrazolium bromide assay)
MTS assays were used to measure cell viability. 48 hours after transfection with siScramble, siLAMC1 (3000cells/well) were plated in six plicate in 96-well cultured with DMEM containing 10% FBS. By the indicated time point, each well was added 20 μ L MTS, then cells were incubated for 3 h at 37°C. The absorbance was measured at 450 nm using the Spectramax M5 (Molecular Devices, Sunnyvale, CA, USA).
Cell invasion and migration assay
After being subjected to serum-starvation for 24 hours, 2×104 Hep3B cells were plated into the upper chambers (Corning Costar Corp., Cambridge, MA, USA) of transwell plates with or without a Matrigel coating (BD Biosciences, Bedford, MA, USA). Each of the lower chambers was filled with 500 μl DMEM supplemented with 10% fetal bovine serum. After incubation for 12 (migration) or 24 (invasion) hours, the cells that migrated to the reverse sides of the inserts were fixed in methanol for 10 min, stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 10 min, photographed, and counted (4 random 100× fields per well). Migrated cells were plotted as the average number of cells per field of view from 3 different experiments.
Statistical analysis
Two-tailed Mann-Whitney test was used to analysis the differences between groups. ROC analysis was used to determine the cut-off value for LAMC1. The chi-square test was used to analyze the relationship between LAMC1 expression and the clinicopathological characteristics. Kaplan-Meier method and compared by log-rank test were used to calculated the OS, DFS and sub-analysis by TNM staging. The prognostic variables in predicting OS and DFS were assessed by multivariate Cox proportional hazards regression analysis. Variables that proved to be significant in the univariate analysis were tested subsequently with the multivariate Cox proportional hazard model. The forward selection method was used for multivariate Cox proportional analysis. All statistical tests were two-sided, and p< 0.05 was considered significant difference. All the statistical were performed using the SPSS 16.0 statistical software (SPSS Company, Chicago, Illinois, USA).
Results
Expression of LAMC1 was up-regulated within HCC tumor tissues
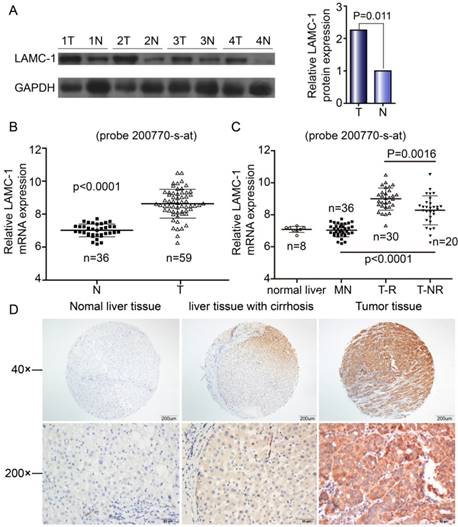
Western blotting and qRT-PCR were performed to explore the expression of LAMC1 in 59 HCC patients. The difference in both western blot and qRT-PCR was obvious between tumor tissues and normal samples (Fig 1A, Fig 1B). By microarray, the expression of LAMC1 mRNA was significantly higher in HCC tumor than that in match normal tissues (p<0.001) and significantly higher in tumor with early recurrence than that without recurrence (p<0.001) (Fig 1C). As shown in Fig 1D, the LAMC1 expression is higher in HCC tumor than that in normal liver tissue and matched liver with cirrhosis. All in all, expression of LAMC1 was up-regulated in HCC tumor.
High level of LAMC1 correlates with poorer progress in HCC patients
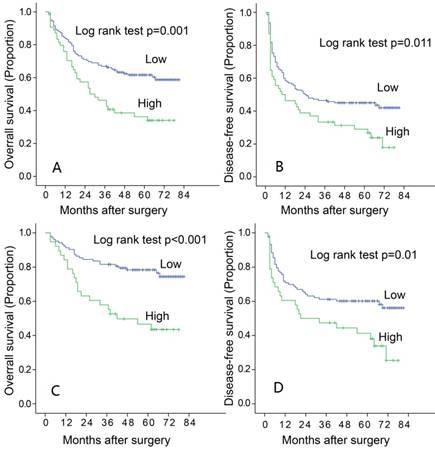
To find out the relationship between the expression of LAMC1 and the progression of HCC, and the association between LAMC1 and clinicopathology variables, 196 HCC tumors were subjected IHC staining with a human LAMC1 antibody. The median follow-up period was 49 months, and the median overall survival (OS) was 51.9±17.9 months. The 1, 3, 5-year OS and disease-free survivals (DFS) for whole group were 81.6%, 61.2% and 53.8% respectively; and 57.1%, 42.9% and 39.8% respectively. According to the ROC analysis, we determined the cut-off value of LAMC1 as 0.4. Tumors exceeding 0.4 were defined as high expression. Of the 196 tumors, 54 (27.5%) were high expression of LAMC1. There were relationship between LAMC1 and tumor capsule (Table 1, p=0.014). According to the Kaplan-Meier analysis, patients with higher LAMC1 expression had shorter OS and DFS (p=0.001, Figure 2A, 2B). Stratification analysis based on TNM staging I and II showed that higher LAMC1 suggested poorer prognosis in early-median stages in both OS and DFS (p<0.001, p=0.001 respectively Figure 2C, 2D).
Correlations between LAMC1 expression and clinicopathalogic characteristics in 196 patients with HCC
| Clinicopathalogic characteristics | All | High LAMC1 | Low LAMC1 | P value |
|---|---|---|---|---|
| (n=196) | (n=54) | (n=142) | ||
| Age(year) | 48.46±11.64 | 47.74±12.59 | 48.72±11.29 | 0.595 |
| ≤50 | 107 | 32 | 75 | 0.418 |
| >50 | 89 | 22 | 67 | |
| Gender | ||||
| male | 166 | 47 | 119 | 0.574 |
| female | 30 | 7 | 23 | |
| HBsAg | ||||
| negative | 31 | 10 | 21 | 0.523 |
| positive | 165 | 44 | 121 | |
| AFP | ||||
| ≤400ng/ml | 107 | 28 | 79 | 0.635 |
| >400ng/ml | 89 | 26 | 63 | |
| Macro-vascular invasion | ||||
| negative | 160 | 47 | 113 | 0.228 |
| positive | 36 | 7 | 29 | |
| Tumor size(cm) | 7.1±4.0 | 7.3±3.2 | 7±4.3 | 0.23 |
| ≤5cm | 75 | 19 | 56 | 0.584 |
| >5cm | 121 | 35 | 86 | |
| Tumor Number | ||||
| single | 153 | 43 | 110 | 0.744 |
| multiple | 43 | 11 | 32 | |
| Tumor capsule | ||||
| incomplete | 94 | 36 | 58 | 0.014 |
| complete | 102 | 18 | 84 | |
| TNM staging | ||||
| I-II | 141 | 38 | 103 | 0.736 |
| IIIa-IIIb | 55 | 16 | 39 | |
| PLT(109) | 190.9±82.3 | 215.4±111.3 | 181.6±66.3 | 0.058 |
| ALT(U/ml) | 55.0±80.8 | 49.35±41.1 | 57.2±91.5 | 0.888 |
| AST(U/ml) | 48.1±42.6 | 47.1±32.3 | 48.4±50.0 | 0.559 |
| ALB(g/L) | 42.4±4.0 | 42.1±4.1 | 42.5±3.9 | 0.503 |
| TBIL(umol/L) | 19.1±21.7 | 16.7±7.1 | 20.0±25.1 | 0.278 |
| CRE(umol/L) | 77.1±15.2 | 76.2±15.8 | 77.5±15.1 | 0.603 |
LAMC 1 expression is increased in HCC patients. A. LAMC 1 and GAPDH expression in tumor tissues (T) compared with adjacent matched normal tissues (N), as assessed by western blotting. (n = 4 paired, p=0.011, paired Wilcoxon signed-rank test). B. Expression of LAMC1 mRNA in tumor and match normal tissues by microarray (P<0.0001). C. Expression of LAMC1 mRNA in HCC with recurrence (T-R), HCC without recurrence (T-NR), match normal liver (MN) and normal liver (N) tissues by microarray. (TR and T-NR vs. MN, T-R vs. T-NR P<0.0001, P=0.0016 respectively) D. Expression of LAMC1 in normal liver tissue, hepatocellular carcinoma tumor tissue and matched liver with cirrhosis.
Increased LAMC 1 expression is correlated with poor prognosis of HCC. A. LAMC 1 expression was associated with OS according to Kaplan-Meier analysis. (n=196, p=0.001). B. LAMC 1 expression was associated with DFS according to Kaplan-Meier analysis. (n=196, p=0.011). C. Subgroup analysis for OS based on TNM staging I-II (n=141 P<0.001). D. Subgroup analysis for DFS based on TNM staging I-II (n=141, P=0.001).
Cox proportional hazards model was used to find out the relationship between LAMC1 expression and prognosis. After univariate Cox regression analysis, all variables with significance were put into analyzed by multivariate analysis. The multivariate analysis showed that LAMC1 expression (HR 1.662, p< 0.001), pTNM staging (HR 1.972, p=0.02) were independent predictors in OS (Table 2) and LAMC1 expression (HR 2.732, p< 0.001), pTNM staging (HR 1.619, p=0.014) and macro-vascular invasion (HR 1.662, p=0.001) were independent predictors in DFS (Table 3).
Analyses of prognostic factors for overall survival
| Variables | univariate analysis | multivariate analysis | |
|---|---|---|---|
| p-value | Hazard ratio (95% CI) | p-value | |
| Gender(male/female) | 0.115 | ||
| Age (≤50/>50) | 0.155 | ||
| TNM staging (I II/III) | <0.001 | 3.393(1.804-6.382) | <0.001 |
| Number(single/multiple) | <0.001 | ||
| Diameter(≤5cm/>5cm) | <0.001 | ||
| Macro-vascular invasion(negative/positive) | <0.001 | ||
| Capsule(incomplete/complete) | 0.021 | ||
| AFP (≤400/>400) | 0.01 | ||
| HBsAg (-/+) | 0.823 | ||
| ALB | 0.917 | ||
| PLT | 0.229 | ||
| ALT | 0.557 | ||
| AST | 0.002 | ||
| TBIL | 0.81 | ||
| LAMC1(low/high) | 0.002 | 1.972(1.27-3.062) | 0.02 |
Analyses of prognostic factors for disease-free survival
| Variables | univariate analysis | multivariate analysis | |
|---|---|---|---|
| p-value | Hazard ratio (95% CI) | p-value | |
| Gender (male/female) | 0.227 | ||
| Age(≤50/>50) | 0.122 | ||
| TNM staging(I II/III) | <0.001 | 2.732(1.558-4.790) | < 0.001 |
| Number(single/multiple) | <0.001 | ||
| Diameter(≤5cm/>5cm) | <0.001 | ||
| Macro-vascular invasion(negative/positive) | <0.001 | 1.662(1.132-2.440) | 0.01 |
| Capsule(incomplete/complete) | 0.002 | ||
| AFP(≤400/>400) | 0.016 | ||
| HBsAg(-/+) | 0.433 | ||
| ALB | 0.936 | ||
| PLT | 0.349 | ||
| ALT | 0.447 | ||
| AST | <0.001 | ||
| TBIL | 0.885 | ||
| LAMC1(low/high) | 0.015 | 1.619(1.103-2.378) | 0.014 |
Expression level of LAMC1 in normal and HCC cell lines
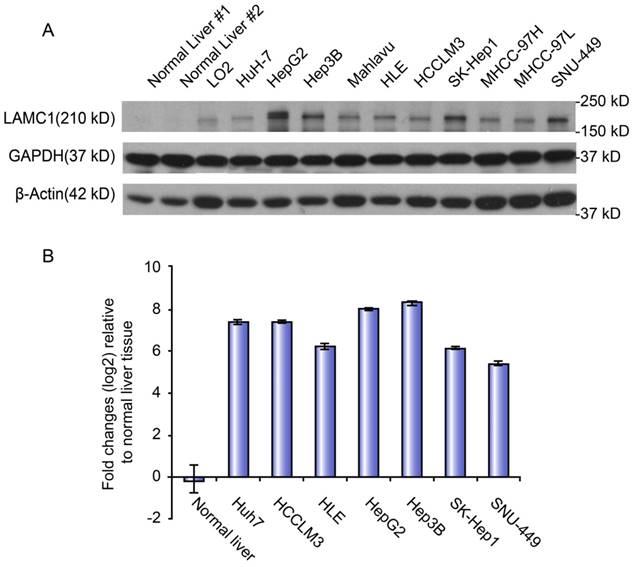
The mRNA and protein levels of LAMC1 were used to determine the expression level of LAMC1 in normal liver, immortalized liver cell line (LO2) and panel cell lines (HuH-7, HepG2, Hep3B, Mahlavu, HLE, HCCLM3, SK, MHCC-97H, -97L, and SNU-449). Both qRT-PCR and Western blot showed that the level of LAMC1 in panel cell lines, especially HepG2 and Hep3B, was obviously higher than the normal liver and immortalized liver cell line (LO2) (Figure 3).
Knockdown of LAMC1 suppresses the proliferation, migration and invasion of HCC cells
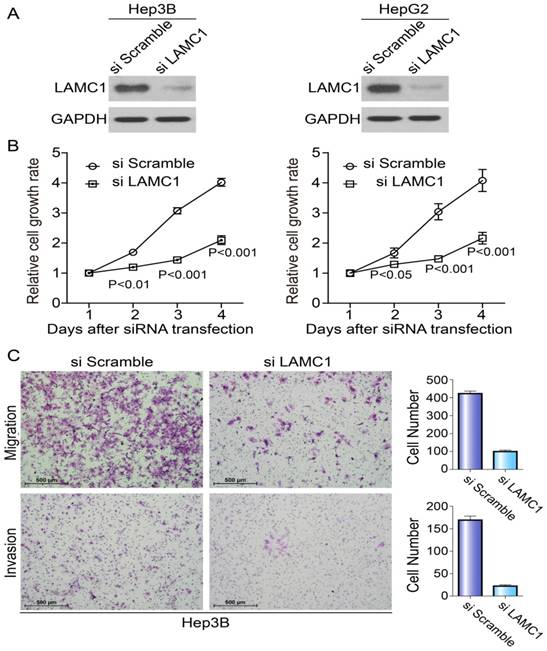
To explore the potential roles of LAMC1 in HCC tumorigenesis, we knocked down the LAMC1 in HepG2 and Hep3B with distinct siRNA duplexes. The efficiency of downregulation was validated by Western blotting. As Fig. 4A showed, the level of LAMC1 was nearly abolished in siLAMC1 transfectants. At the indicated times, we performed MTS to determine the viability. Compared to the control, siLAMC1 transfectants displayed significantly lower proliferation (Fig. 4B). As shown in Figure 4C, the ability of migration and invasion in Hep3B siLAMC1cells was decreased.
Discussion
In present study, we demonstrated that the expression of LAMC1 was significantly higher in HCC tissue compared to that in normal liver tissue, high expression of LAMC1 was significantly associated with incomplete tumor encapsulation and tumor recurrence, and for the first time, we revealed that high expression of LAMC1 predicted poorer OS and DFS for patients with HCC after hepatectomy, especially for patients with early-mediate stage HCC. We also demonstrated that when the expression of LAMC1 was depressed, the cell proliferation, invasion and migration were depressed significantly.
Extracellular matrix proteins constitute an integral part of the tumor microenvironment and play critical roles in regulating tumor cell proliferation, survival, autophagy, migration, and invasion [14]. Laminins are heterotrimeric extracellular matrix glycoproteins that contain an α-chain, β-chain, and γ-chain, which are encoded by five α (α1-α5), four β (β1-β4), and three γ (γ1-γ3) different laminin (LAM) genes[7-9,15,16]. These chains account for the 15 laminin trimeric assemblies (laminin 1-15) to date, and 15 members of this family are present in different basement membranes, which serve different functions. Among these different polypeptides, laminin-γ1 is present in all laminin isoforms, except in laminin-5, and it has a widespread distribution in most basement membranes [7-9].
Our results demonstrated that the expression of LAMC1 was up-regulated in HCC tumor tissues and cell lines, compared to normal liver tissue and cell lines. Similarly, Lietard J et al had demonstrated that LAMC1 mRNA was 2.8-fold increase in human HCCs compared with control livers, and the laminin γ1 chain was involved in carcinogenesis of the liver through Sp1-mediated transactivation at the LAMC1 promoter [10]. Wewer and Albrechtsen had shown an increased deposition of laminins in the sinusoids, and an active process of basement membrane remodeling was suggested in human livers during the development of both primary and secondary cancers [17]. These results hypothesized that LAMC1 might contribute to the tumor development. However, little is known about the underlying mechanisms.
Tumor cell adhesion to substratum is a first step for invasion, and laminin plays an important role in this process followed by degradation of this matrix and invasion of infiltrating cell into new intracellular space [18]. It was also reported that in mouse melanoma cells, lamininγ1 chain peptide C-16 promotes migration, matrix metalloproteinase (MMP)-9 secretion, and pulmonary metastasis. The interaction of the tumor cells with laminin initiates the protease cascade leading to the production of active MMP-2 and to the degradation of the basement membrane [11]. In uterine carcinoma, Kashima et al. also revealed that LAMC1 expression was significantly associated with myometrial invasion, cervical/adnexal involvement, angiolymphatic invasion and lymph node metastasis [13]. In agreement with previous studies, our results demonstrated that high LAMC1 expression was positively correlated with incomplete encapsulation and tumor recurrence after surgery. It indicated that overexpression of LAMC1 was associated with tumor growth and invasion.
LAMC 1 expression in different HCC cell lines. A. LAMC 1 protein expression in different HCC cell lines, as assessed by western blotting. B. LAMC 1 mRNA levels in different HCC cell lines, as assessed by qRT-PCR.
Knockdown of LAMC1 suppresses the proliferation, migration and invasion of HCC cells. A. The effect of LAMC 1 knockdown with siRNA and si Scramble was verified by western blotting 48 hours after transfection. B. Knockdown of LAMC1 by siRNA significantly attenuated the proliferation of HCC cells (Hep3B and HepG2) (P<0.001). C. Knockdown of LAMC1 by siRNA significantly decreased the migration and invasion ability of Hep3B cell.
In present study, for the first time, we demonstrated that LAMC1 was an independent predictor of prognosis, according to univariate and multivariate analyses, and high expression of LAMC1 predicted poorer overall and disease-free survival for patients with HCC after hepatectomy. The subgroup analysis based on TNM staging revealed that LAMC1 might accurately predicate the prognosis in early-mediate stage HCC after curative resection. Only few studies reported the association between LAMC1 expression and tumor patient prognosis [19-21]. In glioblastoma, Ishibashi M et al. reported that high expression of LAMC1 predicted shorter survival [19]. Ke HL et al. also revealed that the LAMC1 expression is associated with grade, recurrence, and progression-free survival in meningioma [20]. Furthermore, lamin A/C was also described as a marker for colorectal cancer and for colorectal cancer displaying a more aggressive phenotype [21,22]. Our results highlighted the potential use of LAMC1 as a new biomarker predictive of HCC prognosis.
Little is known about the underlying mechanisms of LAMC1 in HCC tumorigenesis. As a transmembrane protein, LAMC1 may mediate cell adhere and migration. So the relationship between the LAMC1 expression and tumor cell invasion and migration was investigated in the present study. Our results demonstrated that the ability of proliferation, invasion and migration was significantly decreased in siLAMC1cells, compared to the normal tumor cell lines. Adhesion of cancer cells to the extracellular matrix play a pivotal role in regulating a variety of cellular processes during tumor development, such as motility, invasion and metastasis. Recent studies revealed that laminin regulates neurite growth and neuronal migration via integrin signaling through the AKT/GSK-3β pathway, loss of laminin expression impaired phosphorylation of FAK [23-25]. In the prostate cancer, Kong L et al. revealed that knockdown or overexpression of lamin A/C resulted in inhibition or stimulation, respectively, of cell growth, colony formation, migration and invasion, and knockdown or overexpression of lamin A/C decreased or increased, respectively, protein levels of phosphor-AKT in cancer cells, suggesting lamin A/C proteins are positively involved in malignant behavior through the PI3K/AKT pathway [26].
In summary, we investigated the expression and potential role, as well as the underlying molecular mechanisms, of LAMC1 in HCC. Our data revealed that LAMC1 up-regulation was correlated with tumor recurrence and poor prognoses, and suggested that LAMC1 might be a new biomarker predictive of HCC prognosis and might also be a useful treatment target.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (NSFC 81572387 and 81602143), the Scientific Project of Guangdong Province (2017A020215034).
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent for publication
Publication consent was obtained from all authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24(1):1-17
2. Zhao C, Nguyen MH. Hepatocellular carcinoma screening and surveillance: practice guidelines and real-life practice. J Clin Gastroenterol. 2016;50(2):120-133
3. Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835-853
4. Dhir M, Melin AA, Douaiher J, Lin C, Zhen WK, Hussain SM, Geschwind JF, Doyle MB, Abou-Alfa GK, Are C. A review and update of treatment options and controversies in the management of hepatocellular carcinoma. Ann Surg. 2016;263(6):1112-1125
5. European Association for the Study of the Liver, European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908-943
6. Zhou DS, Wang HB, Zhou ZG, Zhang YJ, Zhong Q, Xu L, Huang YH, Yeung SC, Chen MS, Zeng MS. TACC3 promotes stemness and is a potential therapeutic target in hepatocellular carcinoma. Oncotarget. 2015;6(27):24163-24177
7. Simon-Assmann P. The laminin family: Founding members of the basement membrane. Cell Adhes Migr. 2013;7(1):44-47
8. Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells. Cancer Lett. 2005;223(1):1-10
9. Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12(3):197-207
10. Liétard J, Musso O, Théret N, L'Helgoualc'h A, Campion JP, Yamada Y, Clément B. Sp1-mediated transactivation of LamC1 promoter and coordinated expression of laminin-gamma1 and Sp1 in human hepatocellular carcinomas. Am J Pathol. 1997;151(6):1663-1672
11. Kuratomi Y, Nomizu M, Tanaka K, Ponce ML, Komiyama S, Kleinman HK, Yamada Y. Laminin gamma 1 chain peptide, C-16 (KAFDITYVRLKF), promotes migration, MMP-9 secretion, and pulmonary metastasis of B16-F10 mouse melanoma cells. Br J Cancer. 2002;86(7):1169-1173
12. Kuhn E, Kurman RJ, Soslow RA, Han G, Sehdev AS, Morin PJ, Wang TL, Shih I. The diagnostic and biological implications of laminin expression in serous tubal intraepithelial carcinoma. Am J Surg Pathol. 2012;36(12):1826-1834
13. Kashima H, Wu R, Wang Y, Sinno AK, Miyamoto T, Shiozawa T, Wang T, Fader AN, Shih I. Laminin C1 expression by uterine carcinoma cells is associated with tumor progression. Gynecol Oncol. 2015;139(2):338-344
14. van der Zee JA, van Eijck CH, Hop WC, Biermann K, Dicheva BM, Seynhaeve AL, Koning GA, Eggermont AM, Ten Hagen TL. Tumour basement membrane laminin expression predicts outcome following curative resection of pancreatic head cancer. Br J Cancer. 2012;107(7):1153-1158
15. Aumailley M. The laminin family. Cell Adhes Migr. 2013;7(1):48-55
16. Kikkawa Y, Hozumi K, Katagiri F, Nomizu M, Kleinman HK, Koblinski JE. Laminin-111-derived peptides and cancer. Cell Adhes Migr. 2013;7(1):150-159
17. Albrechtsen R, Wewer UM, Thorgeirsson SS. De novo deposition of laminin-positive basement membrane in vitro by normal hepatocytes and during hepatocarcinogenesis. Hepatology. 1988;8(3):538-46
18. Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A. Tumor exosome-mediated promotion of adhesion to mesothelial cells in gastric cancer cells. Oncotarget. 2016;7(35):56855-56863
19. Ishibashi M, Bottone FG, Taniura S, Kamitani H, Watanabe T, Eling TE. The cyclooxygenase inhibitor indomethacin modulates gene expression and represses the extracellular matrix protein laminin γ1 in human glioblastoma cells. Exp Cell Res. 2005;302(2):244-252
20. Ke HL, Ke RH, Li B, Wang XH, Wang YN, Wang XQ. Association between laminin gamma1 expression and meningioma grade, recurrence, and progression-free survival. Acta Neurochir (Wien). 2013;155(1):165-171
21. Willis ND, Cox TR, Rahman-Casañs SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruïne A, Hutchison CJ. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One. 2008;3(8):e2988
22. Akhavan A, Griffith OL, Soroceanu L, Leonoudakis D, Luciani-Torres MG, Daemen A, Gray JW, Muschler JL. Loss of cell-surface laminin anchoring promotes tumor growth and is associated with poor clinical outcomes. Cancer Res. 2012;72(10):2578-2588
23. Vinayagam A, Stelzl U, Foulle R, Plassmann S, Zenkner M, Timm J, Assmus HE, Andrade-Navarro MA, Wanker EE. A directed protein interaction network for investigating intracellular signal transduction. Sci Signal. 2011;4(189):s8
24. Chen Z, Haegeli V, Yu H, Strickland S. Cortical deficiency of laminin γ1 impairs the AKT/GSK-3β signaling pathway and leads to defects in neurite outgrowth and neuronal migration. Dev Biol. 2009;327(1):158-168
25. Sun L, Liu L, Liu X, Wang Y, Li M. MGr1-Ag/37LRP induces cell adhesion-mediated drug resistance through FAK/PI3K and MAPK pathway in gastric cancer. Cancer Sci. 2014;105(6):651-659
26. Kong L, Schafer G, Bu H, Zhang Y, Zhang Y, Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33(4):751-759
Author contact
![]() Corresponding author: Minshan Chen, M.D. Ph.D., Department of Hepatobiliary Surgery, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, China. Telephone: (8620) 8734 3117; Fax: (8620) 8734 3117; Email: Chminshsysu.edu.cn
Corresponding author: Minshan Chen, M.D. Ph.D., Department of Hepatobiliary Surgery, Sun Yat-sen University Cancer Center, 651 Dongfeng Road East, Guangzhou 510060, China. Telephone: (8620) 8734 3117; Fax: (8620) 8734 3117; Email: Chminshsysu.edu.cn

 Global reach, higher impact
Global reach, higher impact