3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(16):3212-3225. doi:10.7150/jca.19794 This issue Cite
Research Paper
Long non-coding RNAs in Colorectal Cancer: Progression and Future Directions
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai, 200032, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 200032, China.
Received 2017-2-24; Accepted 2017-8-29; Published 2017-9-15
Abstract
Identification of the colorectal adenoma-carcinoma sequence with its corresponding genetic and epigenetic alterations has significantly increased our knowledge of the etiopathogenesis of colorectal cancer (CRC). However, the molecular mechanisms of colorectal carcinogenesis and metastasis haven't been clearly elucidated. Long non-coding ribonucleic acids (lncRNAs) are key participants of gene regulations rather than “noises”. Accumulative studies have implicated that the aberrant expressions of lncRNAs are tightly corelated to CRC screening, diagnosis, prognosis and therapeutic outcomes. Our review focuses on recent findings on the involvement of lncRNAs in CRC oncogenesis and the lncRNA-based clinical implications in patients with CRC.
Keywords: Long noncoding RNA (lncRNA), colorectal cancer (CRC), tumorigenesis, biomarker
Introduction
Genomic studies over the past several decades have confirmed that only approximately 2% of human gene transcripts are stably translated into proteins, generating numerous non-protein-coding ribonucleic acids (ncRNAs)[1]. Mounting evidence suggests that these non-coding transcriptional “noises” encompass a wide variety of subclasses, including not only “housekeeping” ncRNAs [(such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs, also known as u-RNAs), small nucleolar RNAs (snoRNAs), and Ribonuclease P (RNase P)] but also “regulatory” ncRNAs[2]. The growing numbers of “regulatory” ncRNAs contain two main groups: the short ncRNAs [such as microRNAs (miRNAs), short interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs)] and long noncoding RNAs (lncRNAs)[3]. All of these ncRNAs form a network-like RNA infrastructure[4].
Over recent years, the different subclasses of short regulatory ncRNAs, such as miRNAs, have been relatively well recognized and studied based on their structural features and biologic functions. For example, we previously reported that the elevation of oncofetal miR-17-5p targets the nucleolar and coiled-body phosphoprotein 1 (NOLC1, also known as P130) gene, and subsequently activates the Wnt/β-catenin pathway during oncogenesis and CRC progression[5]. Moreover, the weak miR-150 expression status of CRC patients links with a poorer response to adjuvant chemotherapy and is considered a potential prognostic biomarker in CRC[6].
Most recently, our knowledge has been advanced by the emergence of next-generation sequencing (NGS) technology and the development of bioinformatics methods to investigate lncRNAs[7]. lncRNAs are typically defined as endogenous noncoding RNA molecules (> 200 nucleotides) that govern many genes expressions at the epigenetic, transcriptional, or post-transcriptional levels[8]. Building upon their genomic proximity to protein-coding genes, lncRNAs may be crudely divided into different groups: intergenic transcripts (lincRNAs), intronic transcripts, overlapping transcripts either in sense or in antisense, and processing-type transcripts that cannot be placed in any of the other categories[9, 10]. The past decade has witnessed a steep rise in interest in lncRNA research. Referring to the latest database, NONCODE v4.0 (http://www.bioinfo.org/noncode/), more than 54,000 and 46,400 lncRNA genes have been identified in humans and in mice, respectively[11]. However, a majority of lncRNAs remain to be characterized. Advances in understanding the basic mechanisms of the involvement of lncRNAs in cancer have raised the field of cancer research well beyond original expectations. Changes of lncRNAs expression profiles have been observed in many human CRC, and such discovery may open a new avenue for the lncRNA-based clinical applications in CRC[12].
As the fourth leading reason of cancer-related mortality, CRC is associated with tremendous social and economic burden worldwide[13]. The pathogenetic mechanisms underlying CRC development are complex. The “adenoma-carcinoma sequence” multi-step model proposed by Fearon and Vogelstein suggests that accumulating genetic and epigenetic mutations in the colorectum drive epithelial dysplasia and proliferation, ultimately resulting in CRC[14].
Both intrinsic (such as age, diabetes, obesity, adenomatous polyposis, and inflammatory bowel disease) and extrinsic (such as smoking, inadequate fiber, fish and vitamin D intake, hard-drinking , and high-fat diet) factors have been shown to be associated with CRC risk. Regarding intrinsic factors, We previously conducted a systematic review in approximately 9,000,000 participants from 41 prospective studies[15]. We confirmed that both general obesity and central obesity are positively associated with CRC risk[15]. Regarding extrinsic factors, we previously summarized 9 prospective studies with approximately one million participants for meta-analysis, and found inverse correlation between vitamin D intake/ blood 25-hydroxyvitamin D levels and CRC risk[16]. Moreover, although previous studies have documented the dysregulation of many oncogenes [including the Kirsten rat sarcoma viral oncogene homolog (KRAS) and v-myc avian myelocytomatosis viral oncogene homolog (MYC, also known as c-Myc)] and oncosuppressive genes (including the adenomatous polyposis coli (APC), β‑catenin (CTNNB1), deleted in colorectal cancer (DCC) and P53) in CRC, the molecular and genetic basis of colorectal carcinogenesis and metastasis remain largely unknown[17]. Therefore, there is still an urgent need to improve our understanding of the CRC etiopathogenesis and to develop additional tumor markers and therapeutic targets for this disease.
In this review, we will first provide an overview of the biology and function of lncRNAs. We will then focus on the involvement of lncRNA dysregulation in CRC etiopathogenesis. Finally, we will discuss the latest advance of the clinical implications of lncRNAs and their association with the diagnosis, prognosis and potential treatment of CRC.
Origins and functions of lncRNAs
Understanding the origins and molecular mechanisms of lncRNAs is critical for characterizing lncRNA function. The emergence of functional noncoding RNAs can be divided into the following non-mutually exclusive hypotheses[18]: (1) frame disruptions of a protein-coding gene generates a lncRNA; (2) two untranscribed and divided sequences are juxtaposed following chromosomal rearrangement and are transformed into a multi-exon lncRNA; (3) duplication of a noncoding gene following retrotransposition gives rise to lncRNAs; (4) tandem duplication events of neighboring repeats give rise to a lncRNA; and (5) a transposable element (TE) inserted into a gene emerges as a new lncRNA. However, the origins of many lncRNAs remain elusive except for several typical transcripts, such as X-inactive specific transcript (Xist). Recent studies reported by Derrien at al[10] and Ulitsky at al[19] suggest that a novel lncRNA gene can occur de novo either from untranscribed sequence or from TE, which is in contrast to protein coding genes.
While the origin and evolution of lncRNAs are still obscure, even less is known regarding their precise molecular functions. Although previously regarded as “junk DNA” because of their overwhelming burden of transposons, pseudogenes, and simple repeats, there is mounting evidence that lncRNAs exert a diverse spectrum of regulatory functions, including X chromosome inactivation (XCI), genomic imprinting, tissue regeneration, cell cycle control, apoptosis, aging and carcinogenesis[1, 20].
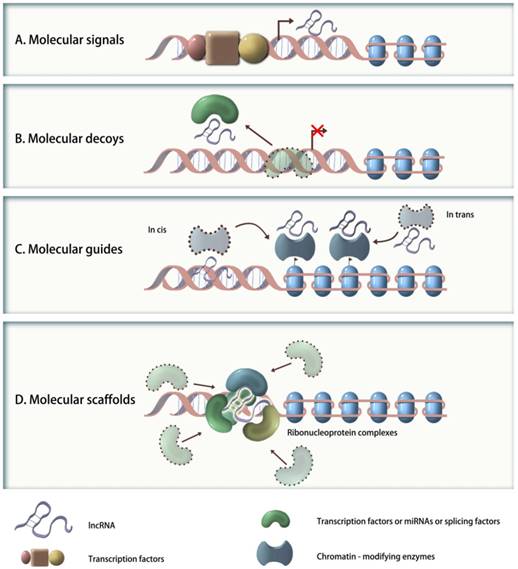
To better understand their functional significance, lncRNAs can be further reclassified based on their molecular mechanisms into following categories (Figure 1): (1) signaling molecules (e.g., Xist, lincRNA-p21, COLDAIR, or HOTAIR), which are transcribed in a time- or space-specific manner in response to diverse stimuli, and reflect the integrated transcriptional activity; (2) molecular decoys (e.g., MALAT1, PTENP1, IPS1 or PANDA), which bind and competitively inhibit transcription factors, miRNAs or other targets from chromatin, but do not exert any additional functions; (3) molecular guides (e.g., Xist, COLDAIR, lincRNA-p21 or HOTAIR), which can recruit chromatin-modifying enzymes to specific target sequences, either in cis or in trans; and (4) molecular scaffolds (e.g., TERC, HOTAIR, ANRIL), which can work as central platforms and assemble proteins together to form architecture of ribonucleoprotein complexes[21]. In fact, several lncRNAs have multiple modes of action that are critical to their biological function[21]. For example, the Hox transcript antisense intergenic RNA (HOTAIR), located at chromosome 12q13.13, is a lincRNA that may function not only as an anatomic signal that guides chromatin-modifying complexes to their targets but also as a molecular scaffold for histone modification complexes[9]. Thus, it is apparent that the molecular mechanisms of genes regulation and cellular functions mediated by lncRNAs may be far more sophisticated than by miRNAs.
Methods of lncRNA detection or quantification
Schematic representation of the molecular mechanisms of four categories of lncRNAs. A. Signaling molecules: such lncRNAs are transcribed in a time- or space-specific manner to respond to diverse stimuli and reflect the integrated actions of transcription factors; B. Molecular decoys: such lncRNAs bind and titrate away transcription factors, miRNAs or other targets from chromatin; C. Molecular guides: such lncRNAs can recruit chromatin-modifying enzymes to specific targets, either in cis or in trans; D. Molecular scaffolds: such lncRNAs can work as central platforms and assemble proteins to form architecture of ribonucleoprotein complexes.
Several techniques are currently available that can be used for lncRNA detection and quantification, including RNA sequencing (RNA-seq), RNA microarray, reverse transcription polymerase chain reaction (RT-PCR), fluorescence in situ hybridization (FISH), and RNA blot analysis[12]. Most lncRNAs operate as RNA-protein complexes, and advanced methods for studying lncRNA-protein interactions include the following: RNA interference (RNAi), RNA Pull-down, RNA-binding protein immunoprecipitation (RIP), chromatin isolation by RNA purification (ChIRP), ChIRP-sequencing (ChIRP-seq), crosslinking-immunopurification (CLIP) and CLIP-sequencing (CLIP-seq)[22].
Meanwhile, the available computational approaches and bioinformatics resources allow researchers to better characterize the functional lncRNAs and to shed additional light on the roles of lncRNAs in CRC[23]. The related database, annotation tools and other bioinformatics resources available are listed in Table 1.
lncRNA expression profiles in CRC
Research focused on the molecular mechanisms of CRC has highlighted the stepwise progression following the adenoma-carcinoma sequence. Accumulated evidence indicates that CRC-associated lncRNAs acts as master gene regulators through different mechanisms. With the aid of tools ranging from global lncRNA expression profiles with RNA-seq or RNA microarray to selected lncRNAs expression examining with qRT-PCR or FISH, several studies have detected a number of consistently and reproducibly altered CRC-associated lncRNAs both in vivo and in vitro[38-42].
The first CRC-related lncRNA was identified in 1996 when Hibi and colleagues[43] reported that the endogenous H19 gene is frequently abundant in CRC specimens, and the overexpression of H19 plays an important role in CRC development. Since that discovery, accumulating reports of dysregulated expression of lncRNAs in CRC have suggested that aberrant lncRNAs are involved in all stages of CRC (Table 2). Recent studies are aimed at determining whether lncRNAs can be used as novel predictors for CRC screening, diagnosis, prognosis and therapeutic outcomes.
The lncRNA bioinformatics resources.
| Bioinformatics Resources | Web Link | Reference |
|---|---|---|
| NONCODE v4 | http://www.noncode.org | [24] |
| H-InvDB rel 8.3 | http://www.h-invitational.jp | [25] |
| LncRNADisease | http://cmbi.bjmu.edu.cn/lncrnadisease | [26] |
| LincSNP | http://bioinfo.hrbmu.edu.cn/LincSNP. | [27] |
| Rfam 11.0 | http://rfam.janelia.org | [28] |
| Human Body Map lincRNAs | http://www.broadinstitute.org/genome_bio/human_lincrnas/ | [29] |
| lncRNAdb | http://www.lncrnadb.org/ | [30] |
| ncFANs | http://www.ebiomed.org/ncFANs/ | [31] |
| Noncoder | http://noncoder.mpi-bn.mpg.de | [32] |
| NRED | http://nred.matticklab.com/cgi-bin/ncrnadb.pl/ | [33] |
| ChIPBase | http://deepbase.sysu.edu.cn/chipbase/ | [34] |
| LNCipedia 2.0 | http://www.lncipedia.org/) | [35] |
| DIANA-LncBase | http://www.microrna.gr/LncBase/ | [36] |
| iSeeRNA | http://www.myogenesisdb.org/iSeeRNA/ | [37] |
LncRNAs associated with CRC.
| LncRNAs and references | Cytoband | Expression level | Types | Mode of action | Potential clinical significance |
|---|---|---|---|---|---|
| CCAL[44] | 3q29 | Elevated | Oncogenic | Activates Wnt signaling by suppressing AP-2α | Prognosis and therapeutics |
| CCAT1[38, 42, 45] | 8q24.21 | Elevated | Oncogenic | N/A | Screening, diagnosis, and prognosis |
| CCAT1-L[41] | 8q24.21 | Elevated | Oncogenic | Regulates chromatin interactions at the MYC locus | N/A |
| CCAT2[46] | 8q24.21 | Elevated | Oncogenic | Up-regulates and Responds to Wnt signaling; underlies metastatic progression and chromosomal instability | Screening, diagnosis, and prognosis |
| CRNDE[47, 48] | 16q12.2 | Elevated | Oncogenic | Responds to EGFR signaling; regulates genes involved in central metabolism | Diagnosis |
| E2F4 antisense[49] | 16q21-22 | Elevated | Oncogenic | Activated by Wnt signaling; contributes to carcinogenesis by reducing the level of the E2F4 cell cycle repressor | NA |
| HOTAIR[39, 50] | 12q13.13 | Elevated | Oncogenic | Correlates with PRC2 function; participates in EMT | Prognosis |
| HULC[51] | 6p24.3 | Elevated | Oncogenic | N/A | Diagnosis |
| KCNQ1OT1/LIT1[52, 53] | 11p15.5 | LOI | Oncogenic | Correlates with epigenetic status at the KvDMR1 | Diagnosis |
| lncRNA-ATB[54] | 14q11.2 | Elevated | Oncogenic | Activated by TGF-β; promotes EMT | Prognosis and therapeutics |
| MALAT1[55] | 11q13.1 | Elevated | Oncogenic | Promotes EMT by regulating ZEB1, ZEB2 and Slug expression; activates Wnt signaling | Diagnosis and prognosis |
| ncNRFR[56] | 1p13.2 | Elevated | Oncogenic | Inhibits the oncosuppressive function of let-7 by binding to target mRNAs | Diagnosis |
| PCAT1[57] | 8q24.21 | Elevated | Oncogenic | N/A | Prognosis |
| PVT1[58] | 8q24 | Elevated | Oncogenic | Generates antiapoptotic activity via TGF-β signaling | Prognosis |
| uc.73A[59] | 2q22.3 | Elevated | Oncogenic | Reduces apoptosis | Diagnosis, prognosis, and therapeutics |
| CUDR[60] | 19p13.12 | Elevated | Oncogenic | N/A | N/A |
| BANCR[61] | 9q21 | Decreased | Oncosuppressive | Suppresses CRC proliferation via targeting p21 | N/A |
| CAHM[62] | 6q26 | Decreased | Oncosuppressive | N/A | Screening and diagnosis |
| lncRNA-LET[63] | 15q24.1 | Decreased | Oncosuppressive | Repressed by histone deacetylase 3; contributes to hypoxia-mediated metastasis | NA |
| lincRNA-p21[64] | 6p21.2 | Decreased | Oncosuppressive | Acts with hnRNP-K as a transcriptional coactivator of p53 | Prognosis |
| Loc285194[65, 66] | 3q13.31 | Decreased | Oncosuppressive | Induced by p53; inhibits tumor cell growth in part through reducing miR-211 level | Prognosis |
| MEG3[67, 68] | 14q32.2 | Decreased | Oncosuppressive | Induces accumulation of TP53 by down-regulating MDM2 | NA |
| ncRAN[69] | 17q25.1 | Decreased | Oncosuppressive | N/A | Prognosis and therapeutics |
| PTENP1[70] | 9p13.3 | Decreased | Oncosuppressive | Regulates cellular levels of PTEN by binding to the PTEN-targeting miRNAs | therapeutics |
| BA318C17.1[71] | 20p12.1 | Decreased | N/A | N/A | N/A |
| H19[40, 72, 73] | 11p15.5 | Elevated or LOI | Oncogenic/ oncosuppressive | Decreases RB expression by acting as the precursor of miR-675; controls the number of polyps in the APC murine model of CRC; promotes EMT via functioning as miR-138 and miR-200a sponges | Prognosis and therapeutics |
| PRNCR1[68] | 8q24 | N/A | N/A | Contains CRC- related SNPs within its locus | Screening |
| XIST[74, 75] | Xq13.2 | Elevated | N/A | N/A | N/A |
Involvement of lncRNAs in CRC pathogenesis
It is widely accepted that CRC tumorigenesis is a multistage process[76]. CRC develops typically from normal mucosal epithelium to an benign adenoma and ultimately progresses to malignant tumor[77]. Understanding the tumorigenic signaling pathways that involve lncRNAs would provide further insights into CRC pathogenesis.
lncRNAs participate in many known signaling pathways involved in CRC pathogenesis, including the Wnt/β-catenin signaling pathway, epidermal growth factor receptor (EGFR)/insulin-like growth factor type I receptor (IGF-IR) signaling pathway (KRAS and phosphatidylinositol-3-kinase (PI3K) pathways), transforming growth factor-beta (TGF-β) signaling pathway, p53 signaling pathway, and the epithelial-mesenchymal transition (EMT) program (Figure 2).
The Wnt/β-catenin pathway
The disruption of the Wnt/β-catenin pathway contributes to CRC initiation[78]. The β-catenin destruction complex, comprising of multiprotein like Axin, Glycogen Synthase Kinase 3β (GSK-3β), APC, casein kinase 1 (CK1), protein phosphatase 2A (PP2A), activator protein 2α (AP-2α), are switch controls for Wnt signaling[79]. Malfunction of this destruction complex leads to stimulation of the Wnt signaling via the translocation of accumulated free β-catenin into the nucleus and subsequent activation of T-cell factor(TCF)/lymphoid enhancing factor (LEF) targets, such as cyclin D1 (CCND1), E2F transcription factor-4 (E2F4), and proto-oncogene c-Myc[80].
Involvement of lncRNAs in the “adenoma-carcinoma sequence”, multi-step model of CRC pathogenesis. LncRNAs participate in many signaling pathways involved in the pathogenesis of CRC, including the activation of Wnt/β-catenin signaling pathway, activation of EGFR/IGF-IR signaling pathway (KRAS and PI3K pathways), activation of TGF-β)/Smad signaling pathway, suppression of the p53 signaling pathway, and activation of EMT program.
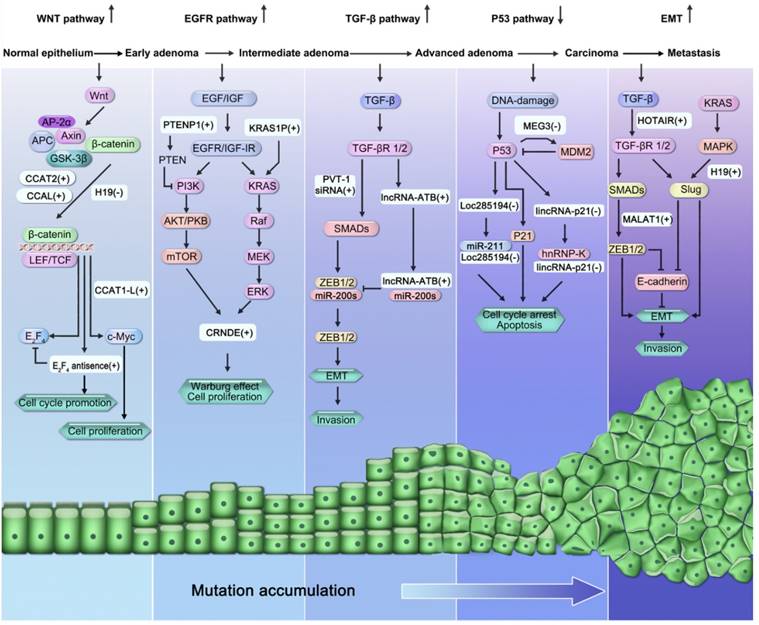
Inactivation of the oncosuppressive gene APC is the main rate-limiting step of Wnt signaling, occurring in more than 80% of sporadic CRCs[78, 81]. The APC gene has been reported to be modulated by a lncRNA called H19[72]. H19 is a paternally imprinted and maternally expressed transcript located at 11p15.5[82]. Oncofetal H19 is elevated during embryogenesis, but is inactivated postnatally and re-expressed during tumorigenesis[83]. Based on their findings that H19 knockout mice never spontaneously develop tumors, Yoshimizu et al. demonstrated that H19 acts as a oncosuppressive gene rather than as an oncogene[72]. Meanwhile, in the absence of H19 expression in the APC mutant CRC murine model, the number of adenomas, especially smaller polyps, were significantly higher than in its presence, suggesting that H19 could control the early event of CRC tumorigenesis[72].
AP-2α belongs to β-catenin destruction complex[84]. In vivo and in vitro studies showed that AP-2α served as a oncosuppressive protein by interacting with APC and β-catenin[84]. One of our recent study has shown that new-found colorectal cancer-associated lncRNA (CCAL) negatively regulated AP-2α and then opened the way to Wnt signaling in CRC tumorigenesis[44].
The E2F family of transcription factors are in charge of the cell cycle (G1/S transition) and the action of oncosuppressive genes[85]. Yochum and colleagues[49] revealed a β-catenin-regulated lncRNA derived from the E2F4 locus termed E2F4 antisense transcript. E2F4 antisense transcript is expressed in HCT116 β-catenin signaling-stimulated CRC cells[49]. Both β-catenin and TCF4 could bind to the 3'-untranslated region (3'-UTR) sequences of E2F4, driving expression of this lncRNA that reduces E2F4 levels and decreases binding of E2F4 to promoters of targets such as proliferating cell nuclear antigen (PCNA) and cyclin A2, ultimately activating the expression of genes repressed by E2F4[49].
The MYC region upstream contains an large tumor-type-specific super enhancer[86]. Xiang et al.[41] showed a super-enhancer located at locus 8q24.21, 515 kb MYC upstream region, that transcribes a human CRC-specific lncRNA CCAT1-L. CCAT1-L is known as an isoform of the recently described lncRNA colon cancer-associated transcript-1 (CCAT1)[87], which is exclusively localized to the nucleus, and its expression is upregulated in CRC cell lines and CRC tissue samples[41]. Furthermore, CCAT1-L participates in a positive control of MYC transcription by interacting with a zinc finger protein, CCCTC-binding factor (CTCF), and facilitating CTCF binding to MYC locus[41]. In addition, Ling et al.[46] concluded that a novel gene transcript named colon cancer-associated transcript-2 (CCAT2), which maps to the 8q24.21 genomic region, is a downstream target of Wnt. CCAT2 is overexpressed in microsatellite-stable (MSS) CRC samples[46]. Higher MYC expression was found in CRC cells overexpressing CCAT2, and transient knockdown of CCAT2 reduced MYC expression[46]. Additionally, there is a complex feedback loop between CCAT2 and Wnt signaling. On the one hand, CCAT2 could broadly enhance Wnt activities by binding to TCF7L2, modulating its transcriptional activity[46]. On the other hand, TCF7L2 knockdown consistently reduced CCAT2 expression, indicating that TCF7L2 is necessary for the maintenance of high CCAT2 expression levels[46].
The observations described above suggest that alteration of lncRNA expression participates in the early event of the etiopathogenesis of CRC.
EGFR/IGF-IR signaling (KRAS and PI3K pathways)
The epidermal growth factor receptor (EGFR) is one of the members of human EGFR family of receptor tyrosine kinases[88]. Functional stimulation of their receptors occurs in a broad spectrum of solid tumors including CRC[88]. Like EGFR, insulin-like growth factor-I receptor (IGF-IR) is also a ubiquitous transmembrane tyrosine kinase that plays crucial roles in colorectal tumorigenesis and progression[89]. Binding of their respective ligands to the EGFR and IGF-IR lead to their autophosphorylation and subsequent activation of the KRAS/Raf/mitogen-activated protein kinase (MAPK) and PI3K/AKT/mammalian target of rapamycin (mToR) signaling cascades, contributing to uncontrolled cellular proliferation, adhesion, angiogenesis, migration, metastasis, and survival[90].
The newly identified IGF driven gene termed colorectal neoplasia differentially expressed (CRNDE), located at locus hCG_1815491 on chromosome 16, is activated early in CRC and potentially interacts with chromatin-modifying complexes to engage in gene expression[47]. Ellis et al.[48] reported that CRNDE is one of the downstream targets of both the PI3K and KRAS signal cascades. Upregulation of CRNDE expression promotes the metabolic shift of neoplastic colon cells from oxidative metabolism to aerobic glycolysis, contributing to the “Warburg effect”, a hallmark of CRC cell metabolism[48].
Another lncRNA that is involved in the pathogenesis of CRC is a pseudogene called phosphatase and tensin homolog pseudogene 1 (PTENP1) located at 9p13.3 [91]. PTENP1 is highly homologous to the crucial tumor-suppressor gene PTEN, an inhibitor of PI3K pathway signaling[70]. Poliseno et al.[70] showed that the bona fide tumor suppressor gene PTENP1, which contains a relatively shorter 3'UTR than that of PTEN, functions as a decoy to bind to the PTEN-targeting miRNAs, allowing for the expression of the PTEN protein. Moreover, deletion of PTENP1 in CRC directly correlates with reduced PTEN expression[70].
The TGF-β signaling pathway
TGF-β is a 25-kDa cytokine that signals via transmembrane serine-threonine kinase receptors(STKR) and intracellular mediators of Smad family[92]. The TGF-β signal pathway plays dichotomous roles in the CRC progression[93]. In the early phase of tumorigenesis, TGF-β signaling acts as a cancer suppressor. In contrast, in the late cancer stages, it accelerates CRC invasiveness. Improved understanding of how the TGF-β signaling cascade mediates interactions with lncRNA will provide important insights into colorectal carcinogenesis, development and progression[94].
Some researchers have demonstrated the involvement of lncRNAs in TGF-β signaling pathways that are associated with various cancer types, including CRC[50]. TGF-β signaling is induced by the aberrant expression of lncRNAs. The human PVT-1 oncogene, which encodes the lncRNA PVT-1, is involved in cancer pathophysiology[95]. Takahashi and colleagues[58] conducted gene expression microarray analyses using PVT-1 knockdown of CRC cell lines RKO and HCT116 and found that the proteins relating to the canonical TGF-β signaling (such as tumor suppressor gene Smad4) and apoptosis were significantly activated by PVT-1 knockdown.
On the other hand, lncRNAs can also be activated by TGF-β. For example, Yuan and colleagues[54] discovered a novel TGF-β-induced lncRNA termed lncRNA-activated by TGF-β (lncRNA-ATB) that promotes the invasion-metastasis cascade in hepatocellular carcinoma through Smad-independent signaling. lncRNA-ATB competitively binds to and sponges miR-200s away from their mRNA targets, zinc-finger E-box binding homeobox family proteins (including ZEB1 and ZEB2), which encode the key EMT-promoting transcription factors, thus promoting EMT[54]. Notably, lncRNA-ATB is also significantly upregulated by TGF-β in the Smad4-deficient CRC cell line SW480, indicating that lncRNA-ATB also promotes CRC progression through the noncanonical TGF-β pathway[50, 54].
Taken together, these findings suggest an existence of a complex feedback loop between the lncRNAs and the TGF-β signaling pathway.
The p53 pathway
The tumor suppressor p53, known as “a cellular gatekeeper”, provides an intrinsically powerful defense against CRC. As a master transcription factor, p53 not only upregulates anti-proliferative and proapoptotic genes such as cyclin-dependent kinase inhibitor 1 (p21) (G1 arrest), cyclin B1 (G2 arrest), p53-upregulated modulator of apoptosis (PUMA; apoptosis) and the plasminogen activator inhibitor 1 (PAI-1; cellular senescence), but it also suppresses the genes involved in control of cell proliferation such as polo-like kinase-1(PLK1) and apoptosis (Bcl-2)[96, 97]. Loss of p53 function, through either mutations in p53 itself or perturbations in pathways signaling to p53, is a frequent alteration in the majority of human cancers, including CRC[61].
Strong evidences have unraveled the link between p53 and the lncRNA network[98]. Several lncRNAs, such as MEG3, MALAT1, p53-eRNAs, and Wrap53, serve as p53 regulators, while other lncRNAs, such as Loc285184, lncRNA-p21, H19,and PANDA, serve as p53 effectors[99]. For example, the human maternally expressed gene-3 (MEG3), with a length of 1595 nucleotides, is an imprinted ncRNA located at human 14q32.3[100]. As a p53 regulator, MEG3 downregulates expression of the well-known negative p53 regulator, mouse double minute 2 homolog (MDM2), selectively regulates p53 targets expression, ultimately inhibiting CRC pathogenesis and progression[67]. Furthermore, the expression of MEG3 is under epigenetic control, and because of hypermethylation in the MEG3 promoter, MEG3 loss is common in many human tumors (including CRC) and tumor cell lines (including colon cell lines HCT116 and H29)[67, 68, 101, 102]. Thus, MEG3 could represent a tumor suppressor gene.
Unlike the p53 regulator MEG3, the lncRNA Loc285194 (also referred to as LSAMP antisense RNA 3) serves as a p53 effector. The Loc285194 gene, located at 3q13.31 and consisting of 4 exons for a total of 2105 nt in length, was originally identified to be downregulated in osteosarcoma samples and subsequently reported to be downregulated in CRC[66]. Through in vitro and in vivo analyses, Liu and colleagues[65] further reported that the Loc285194 upstream region is a putative transcription target of p53. As an endogenous sponge, Loc285194 inhibits CRC cell growth by binding to and repressing oncogenic miR-211[65]. These results imply Loc285194 as a potential CRC suppressor.
In addition to Loc285194 expression, which is triggered by p53, Huarte and colleagues[103] identified a p53-activated lincRNA called lincRNA-p21 that played an important role in triggering apoptosis. lincRNA-p21 is also a direct p53 transcription target near the cyclin-dependent kinase (CDK) inhibitor p21 (CDKN1A) locus, a canonical transcriptional target of p53[104]. Loss of lincRNA-p21 affects many genes that are usually suppressed by p53, whereas expression of this lincRNA functions as a transcriptional repressor by modulating the localization of heterogeneous nuclear ribonucleoprotein K (hnRNP-K), a well-known inhibitor complex in the p53 pathway[103].
In light of these findings, we conclude that lncRNAs also belong to the p53-regulatory network and are involved in CRC tumorigenesis.
The EMT program
Distant metastasis is the primary cause of CRC-related mortality[105]. Recent preclinical or clinical research has implicated the activation of the EMT program and its reverse program in the metastatic process in CRC[106]. The hallmarks of EMT include the downregulation of E-cadherin (CDH1) and upregulation of non-epithelial cadherins, like N-cadherin (CDH2)[1]. Several reasons contribute to the disturbance of controlled epithelial balance, including the altered expression levels of lncRNAs[107].
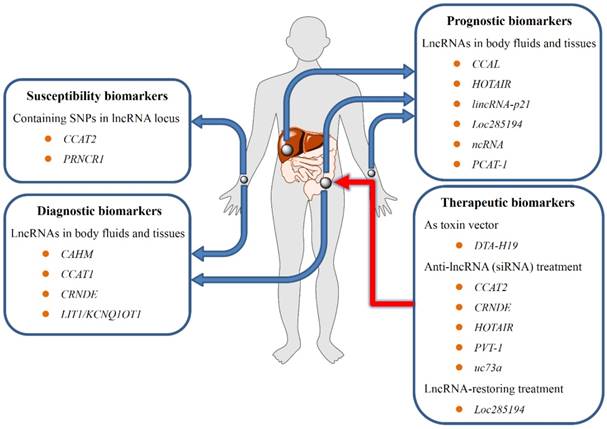
Potential use of lncRNAs in CRC screening, diagnosis, prognosis, and treatment. First, Polymorphisms within lncRNA-encompassing regions may contribute to susceptibility to CRC; second, lncRNAs isolated from blood samples may be substantial biomarkers for early diagnosis and prognosis of CRC; third, the aberrant lncRNA expression in CRC correlates with patient survival or treatment outcome, providing the theoretical rationale for using lncRNAs as promising therapeutic targets.
Several researches have highlighted the roles of lncRNAs in regulating the epithelial phenotype by controlling EMT inducers such as ZEB1, ZEB2 and Snail zinc finger family proteins [including Snail1 and Snail2 (also known as Slug)][108]. For example, metastasis-associated lung adenocarcinoma transcript-1 (MALAT1) has been confirmed to be an important mediator of TGF-β-induced EMT[109]. Upregulated MALAT1 promotes EMT by regulating expression levels of ZEB1, ZEB2 and Slug and activating Wnt signal during the metastasis of bladder cancer[110]. Because the overexpression of MALAT1 was previously confirmed to be involved in CRC progression, we hypothesized that MALAT1 may also play critical roles in CRC cell EMT programs; this hypothesis remains to be validated.
A second lncRNA functioning as a positive moderator of EMT progression in CRC is H19. H19 was found to be upregulated in two EMT model, including Methotrexate (MTX) resistant HT-29 CRC cell, TGF-β1-treated HT-29 CRC cell and TGF-β1-treated SW620 CRC cell[73]. H19 modulated ZEB1 and ZEB2 expression levels by functioning as decoys for miR-138 and miR-200a, promoting EMT transition in CRC[73]. Moreover, lncRNA H19 enhanced the invasive potential of cancer cells by inhibiting E-cadherin via induction of the expression of miR-675-dependent Slug[107].
A third lncRNA participating in EMT-controlled CRC metastasis is called HOTAIR. Wu and colleagues[111] revealed that HOTAIR overexpression promotes CRC cell migration and invasiveness, while depletion of HOTAIR suppresses the EMT program, including upexpression of E-cadherin and loss of vimentin and matrix metalloproteinase-9 (MMP9). These examples demonstrate the functional importance of lncRNAs in invasion and metastasis.
Clinical implications of lncRNAs in CRC
Effect of above-mentioned lncRNAs in CRC etiopathogenesis clearly revealed that dysregulated lncRNAs in CRC may be of significant clinical value (Figure 3). Several lncRNAs have emerged as promising biomarkers for susceptibility screening, diagnosis, prognosis, and lncRNA-based therapies for CRC[12]. Specifically, polymorphisms within lncRNAs encompass regions that may contribute to susceptibility to CRC, and circulating lncRNAs may be promising markers for the early detection of CRC. Meanwhile, the fact that abnormal lncRNA expression in CRC correlates with patient survival or treatment outcome provides the theoretical rationale for utilizing lncRNAs to be promising therapeutical targets.
Polymorphisms within lncRNA-encompassing regions as susceptibility screening markers
Genome-wide association studies (GWAS) combined with single nucleotide polymorphism (SNP) arrays and NGS technology have identified powerful associations of multiple variants, such as SNPs and other genetic abnormalities located on the chromosomal region 8q24 with the onset of CRC[112]. Several highly conserved lncRNAs were discovered to be transcribed from the "gene-desert" region of 8q24[46, 68, 113]. Therefore, oncogenic SNPs located within these lncRNAs transcripts may involve in CRC carcinogenesis.
For instance, the lncRNA CCAT2, which maps to the 8q24.21 region, encompasses a cancer-related rs6983267 SNP that is faithfully linked to CRC oncogenesis, progression, and prognosis through regulating MYC transcription[46]. Using pyrosequencing data on heterogeneous cell lines (DLD1 and KM12SM cells) and CRC samples, Ling et al.[46] found that the G allele of rs6983267 produces more CCAT2 transcripts and induces higher MYC expression than the T allele. The variant genotype of rs6983267 affects the expression levels and functions of CCAT2, providing an alternative explanation of the risk of SNP-conferred CRC.
A second lncRNA involved in cancer-related gene polymorphisms is known as prostate cancer non-coding RNA-1 (PRNCR1). Li et al.[68] identified 4 new functional CRC susceptibility variants (rs13252298, rs7007694, rs16901946, and rs1456315) in the lncRNA PRNCR1 at the 8q24 locus. The authors further demonstrated that the rs13252298 and rs1456315 alleles may contribute to a decreased CRC risk, whereas polymorphisms of the others may promote CRC growth and differentiation [68]. This evidence suggests that SNPs in the lncRNA locus likely increase CRC susceptibility.
Considering the results of these two studies, it is anticipated that with further information from GWAS, more polymorphisms in the lncRNA locus will be discovered, allowing clinicians to better assess the genetic susceptibility to CRC screening.
Potential of lncRNA expression in CRC early diagnosis
Due to the screening of fiber-optic colonoscopies, fecal occult blood test (FOBT), and stool-based DNA assays, 5-year CRC-related mortality rates have slightly declined to date. Nonetheless, colonoscopies are invasive and cost intensive, whereas FOBT and stool-based DNA assays have hyposensitivity and low specificity. Until now, only a few functionally characterized serum candidates, such as carcino-embryonic antigen (CEA) and carbohydrate antigen 19-9 (CA-19-9), have been clinically utilized. However, neither CEA nor CA19-9 meets expectations for early detection or prognosis of CRC. Consequently, there is an urgent need for seeking novel biomarkers which are sensitive or specific enough.
Compared with the protein-coding RNAs or their proteins, the use of a lncRNA as a biomarker is of great advantage because its expression is a better indicator of tumor status[114]. The lncRNA expression patterns, either in tissues or in circulation, have the potential to be diagnostic markers for CRC in clinical routine and could lessen the number of unnecessary colonoscopies follow-up procedures.
For example, Tanaka and coworkers[52] found a lncRNA termed long QT intronic transcript-1 (LIT1/KCNQ1OT1) which was frequently (40%) detected to be loss of imprinting (LOI) in the CRC tissues, whereas negative in peritumoral samples. Coincidentally, Nakano et al.[53] supported the fact that LOI of LIT1 was observed only in CRC tissue specimens at a high frequency (53%), but negative(0%) in adjacent normal samples, thereby making LIT1 a potential for the diagnosis of CRC.
Furthermore, Pedersen et al.[62] reported that the lncRNA called colorectal adenocarcinoma hypermethylated (CAHM) was hypermethylated in 81% of adenomas and 71% of CRC but not in normal tissue (8%); meanwhile, methylated CAHM was observed in the plasma of 55% of CRC compared with 4% of adenomas and 7% of subjects without neoplasia, showing the promise of CAHM as a blood candidate for early detection of CRC.
Another potentially diagnostic lncRNA is CRNDE[47]. Graham and colleagues[47] reported that CRNDE upregulation is an early event in CRC. In tissue samples, the expression levels of the CRNDE transcript were found to be elevated in more than 90% of tissues from colorectal adenomas and adenocarcinomas, whereas levels were very low in normal tissues or inflammatory bowel disease (IBD) samples. In plasma samples, CRNDE transcript was positive in 13 of 15 CRC patients but in only 1 of 15 controls, affording a sensitivity and specificity of 87% and 93%[47].
Moreover, CCAT1 is another promising RNA biomarker candidate[42]. The 2628 nucleotide-long lncRNA CCAT1, located in the vicinity of c-Myc on chromosome 8q24.21, was first identified from blood and fecal samples from CRC patients in 2005[115]. Studies from the same group subsequently examined the CCAT1 levels in human biospecimens and found out that CCAT1 was markedly upregulated across the colon adenoma-carcinoma sequence[45]; meanwhile, peripheral blood samples of CCAT1 was shown to be overexpressed (up to 1700 fold) in approximately 40% of CRC, but not in that of healthy volunteers[42]. The researchers first described the use of a CCAT1-specific peptide nucleic acid (PNA)-based molecular beacons (TO-PNA-MB) to be a powerfulpredictor for specific identification of CRC in vitro, ex vivo, and in vivo[38].
Taken together, the ectopic presences of these lncRNAs in tissues, stools, and blood samples raise hope for using combining optimized lncRNAs for detection of CRC.
Potential of lncRNA expression for CRC prognosis and prediction
Currently, the only comprehensive criteria employed in clinical practice to provide an indication of CRC prognosis or to guide adjuvant chemotherapy choices is the American Joint Committee on Cancer (AJCC) tumor node metastasis (TNM) staging system[116]. Regrettably, the TNM system hasn't provided the clinicians with the optimal staging allocation, but has resulted in substantial under- or over-treatment of certain CRC patients over the past decades[117, 118].
Recent studies suggest that both oncosuppressive and oncogenic lncRNAs may have potential to be used as efficient biomarkers of CRC in clinical routine.
For example, Zhai et al.[64] revealed a contradiction regarding the function of tumor suppressor lincRNA-p21 in CRC progression. On the one hand, the expression levels of lincRNA-p21 in CRC tissue samples were significantly lower compared with the tumor-adjacent samples. On the other hand, as the CRC stage progresses, the expression levels of this lincRNA in tumor tissues were significantly elevated[64]. The finding indicates that the disruption of the p53/lincRNA-p21 network may contribute to CRC development and malignant transformation[64]. Another notable candidate is non-coding RNA expressed in aggressive neuroblastoma (ncRAN). This lncRNA located at 17q25.1 is dramatically downregulated in CRC tissues and CRC cell lines, such as DLD1, LoVo and HCT116[69]. Moreover, decreased expression of ncRAN leads to CRC metastasis and is correlated to poorer overall survival (OS) in CRC patients[69]. A third example of a tumor suppressive lncRNA is Loc285194. Qi et al[66] showed that the prevalent loss of Loc285194 in CRC tumor tissues was positively related to a larger tumor size, a poorer TNM stage, an increased number of distant metastases and a poorer disease-free survival (DFS)[66]. Hence, Loc285194 might function as a novel prognostic indicator in CRC.
Apart from tumor suppressive lncRNA, the group of oncogenic lncRNAs whose expression is increased may also serve as potential prognostic factors for CRC.
For example, Kogo et al.[39] found that the upregulation of HOTAIR was frequent in stage IV CRC tissues with liver metastases. Using gene set enrichment analysis, the authors further revealed that HOTAIR expression was significantly correlated to the polycomb-repressive complex-2 (PRC2) gene function, thus contributing globally to cancer progression[39, 119]. Ge et al[57] reported that the lncRNA prostate cancer-associated ncRNA transcript-1 (PCAT1) gene, located on the chromosome 8q24 725 kb upstream of the c-Myc, was more frequently upregulated in CRC tumor tissues than in normal adjacent tissues. Moreover, upregulation of PCAT1 implied a poorer OS time in patients with CRC[57]. Therefore, this lncRNA may be used to complement the TNM staging system to predict prognosis[57]. Our recent study shown that overexpression of lncRNA CCAL in tumor tissues of patients with CRC was connected with a poorer OS and a worse therapeutic outcome, implying CCAL as good indicator for guideness of clinical chemotherapy.
Compared with single utilization mode, the ensemble features of lncRNAs may have an obvious advantage for CRC prognosis. For example, Hu and colleagues[120] performed a lncRNA expression profiling in large CRC cohorts, identifying a set of six lncRNAs (AK024680, AK123657, CR622106, BX649059, BX648207, and AK026784) that correlated with the DFS of CRC. Based on this finding, the authors successfully classified CRC patients into high- or low-risk subgroup for survival prediction[120]. Furthermore, the six-lncRNA signature combined with the AJCC TNM staging system showed a stronger power for DFS prediction in the ROC analysis, implying that the stage-independent lncRNA signatures may have potential to refine the current prognostic model[120].
Although these results above are promising, further studies remain necessary to make sure that the lncRNAs may provide more information than current standard diagnostic procedures.
lncRNAs as a future therapeutic approach in CRC
In spite of the fact that long-term rate of survival of patients with early-phase CRC has increased dramatically over the past decades, successful therapeutic strategies for patients with advanced CRC remain limited, requiring novel aggressive therapy in addition to surgical intervention.
At present, anticancer strategies that focus on cancer-specific expression of oncogenic lncRNAs as target molecules are under development. For instance, the paternally-imprinted, oncofetal lncRNA H19 has been found to be a promising target for CRC therapy[121]. BC-819 (also termed DTA-H19) is a novel double-stranded DNA plasmid carrying the A fragment of diphtheria toxin (DTA) gene under the control of H19 promoter[122-124]. Intra-tumoral or intra-arterial treatment with DTA-H19 leads to overexpression of DTA in the colorectal tumor, causing a significant suppression of subcutaneous or metastatic colorectal tumor growth in vivo[121, 125].
Alternatively, genetic therapies may be designed to treat CRC by manipulating the expression of lncRNAs. For instance, an RNA duplex of siRNAs exhibits high knockdown efficacy of many oncogenic lncRNAs in CRC cells. Silencing of lncRNA expression via administration of specific siRNA oligonucleotides targeting the oncogenic lncRNA CRNDE[48], HOTAIR[39, 111], CCAT2[46], Pvt1 oncogene (PVT1)[58], and uc.73A[59] sequences have been confirmed to exhibit significant anticancer effects, such as decreasing invasion or proliferation either in vivo or in vitro. In addition to knockdown of the oncogenic lncRNAs, restoring endogenous tumor suppressive lncRNAs function in patients with CRC may also be beneficial. One example is in the case of the p53-induced lncRNA Loc285194. Delivery of Loc285194 by plasmid expression vector into CRC cells in xenograft models or in the p53 wild-type CRC cell line HCT-116 significantly suppresses tumor cell growth[65] Although the nucleic acid-based approach for CRC treatment is in the development phase, the lncRNA-based therapeutics for CRC still has several difficulties to overcome. For example, the challenge of achieving a better delivery system with efficacy, safety and specificity remains to be solved.
To implement these experiment-based research advances into medical practice, future robust and rigorous investigations into the therapeutic potential of lncRNAs are required.
Conclusions
To date, we and others have taken important initial steps on the road to figure out the biological functions of lncRNAs in CRC. Given the exponential growth of newly discovered cancer-associated lncRNAs, future studies must specifically address the super-secondary structure (motifs), secondary structure, or tertiary structure of lncRNAs. As we move forward, we will refine our global view of the gene regulatory network involving lncRNAs and ultimately establish more effective measures or strategies for the field of lncRNA as a potential biomarker for clinical diagnosis, prognosis and therapeutics in patients with CRC. However, further study employing a greater number of clinical samples will be required to confirm the sensitivity and specificity for these lncRNA as clinical biomarkers in CRC patients.
Abbreviations
lncRNA, long noncoding RNA; CRC, colorectal cancer; CCAL, colorectal cancer-associated lncRNA; AP-2α, activator protein 2α; CCAT1, colon cancer-associated transcript-1; MYC, v-myc avian myelocytomatosis viral oncogene homolog; CRNDE, colorectal neoplasia differentially expressed; EGFR, epidermal growth factor receptor; E2F4, E2F transcription factor-4; HOTAIR, Hox transcript antisense intergenic RNA; PRC2, polycomb repressive complex-2; EMT, epithelial-mesenchymal transition; HULC, hepatocellular carcinoma up-regulated long non-coding RNA; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; LIT1, long QT intronic transcript-1; LOI, loss of imprinting; KvDMR1, KCNQ1 opposite strand/antisense transcript 1; TGF-β, transforming growth factor-beta; lncRNA-ATB, lncRNA-activated by TGF-β; MALAT1, metastasis associated lung adenocarcinoma transcript-1; ZEB1, zinc finger E-box binding homeobox-1; ncNRFR, non-coding Nras functional RNA; PCAT1, prostate cancer associated transcript-1; PVT1, Pvt1 oncogene; CUDR, urothelial cancer associated-1; BANCR, BRAF activated non-coding RNA; p21, cyclin-dependent kinase inhibitor 1A; CAHM, colorectal adenocarcinoma hypermethylated; lncRNA-LET, lncRNA low expression in tumor; hnRNP-K, heterogeneous nuclear ribonucleoprotein K; Loc285194, also known as TUSC7, tumor suppressor candidate 7; PTENP1, phosphatase and tensin homolog pseudogene 1; PTEN, phosphatase and tensin homolog; miRNA, microRNA; MEG3, maternally expressed gene-3; TP53, tumor protein p53; MDM2, mouse double minute 2 homolog; ncRAN, non-coding RNA expressed in aggressive neuroblastoma; BA318C17.1, also known as MACROD2-AS1, MACROD2 antisense RNA 1; H19, imprinted maternally expressed transcript; RB, retinoblastoma 1; PRNCR1, prostate cancer non-coding RNA-1; SNPs, single nucleotide polymorphisms; XIST, X-inactive Specific transcript; N/A, Not applicable.
Acknowledgements
This work was sponsored by grants from the National Natural Science Foundation of China (No. 81372615), the Fudan Outstanding Young Talent Training Plan (YJYQ201601), and Sponsored by Shanghai Pujiang Program (17PJD007).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA biology. 2012;9:703-19
2. Skipper M. Genomics: users' guide to the human genome. Nature reviews Genetics. 2012;13:678
3. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145-66
4. Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends in genetics: TIG. 2009;25:120-8
5. Ma Y, Zhang P, Wang F, Zhang H, Yang Y, Shi C. et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nature communications. 2012;3:1291
6. Ma Y, Zhang P, Wang F, Zhang H, Yang J, Peng J. et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut. 2012;61:1447-53
7. Cech TR, Steitz JA. The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell. 2014;157:77-94
8. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA biology. 2010;7:582-5
9. Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA biology. 2013;10:925-33
10. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775-89
11. Xie C, Yuan J, Li H, Li M, Zhao G, Bu D. et al. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic acids research. 2014;42:D98-103
12. Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2014
13. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095-128
14. Arends JW. Molecular interactions in the Vogelstein model of colorectal carcinoma. The Journal of pathology. 2000;190:412-6
15. Ma Y, Yang Y, Wang F, Zhang P, Shi C, Zou Y. et al. Obesity and risk of colorectal cancer: a systematic review of prospective studies. PloS one. 2013;8:e53916
16. Ma Y, Zhang P, Wang F, Yang J, Liu Z, Qin H. Association between vitamin D and risk of colorectal cancer: a systematic review of prospective studies. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3775-82
17. Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479-507
18. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
19. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537-50
20. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651-69
21. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904-14
22. Rowley MJ, Bohmdorfer G, Wierzbicki AT. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods. 2013;63:160-9
23. Da Sacco L, Baldassarre A, Masotti A. Bioinformatics Tools and Novel Challenges in Long Non-Coding RNAs (lncRNAs) Functional Analysis. International journal of molecular sciences. 2012;13:97-114
24. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760-74
25. Yamasaki C, Murakami K, Takeda J, Sato Y, Noda A, Sakate R. et al. H-InvDB in 2009: extended database and data mining resources for human genes and transcripts. Nucleic acids research. 2010;38:D626-32
26. Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X. et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic acids research. 2013;41:D983-6
27. Ning S, Zhao Z, Ye J, Wang P, Zhi H, Li R. et al. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC bioinformatics. 2014;15:152
28. Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP. et al. Rfam 11.0: 10 years of RNA families. Nucleic acids research. 2013;41:D226-32
29. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915-27
30. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic acids research. 2011;39:D146-51
31. Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo H. et al. ncFANs: a web server for functional annotation of long non-coding RNAs. Nucleic acids research. 2011;39:W118-24
32. Gellert P, Ponomareva Y, Braun T, Uchida S. Noncoder: a web interface for exon array-based detection of long non-coding RNAs. Nucleic acids research. 2013;41:e20
33. Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic acids research. 2009;37:D122-6
34. Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic acids research. 2013;41:D177-87
35. Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K. et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic acids research. 2013;41:D246-51
36. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM. et al. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic acids research. 2013;41:D239-45
37. Sun K, Chen X, Jiang P, Song X, Wang H, Sun H. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC genomics. 2013;14(Suppl 2):S7
38. Kam Y, Rubinstein A, Naik S, Djavsarov I, Halle D, Ariel I. et al. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer letters. 2013
39. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer research. 2011;71:6320-6
40. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-8
41. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014
42. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M. et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. International journal of cancer Journal international du cancer. 2012;130:1598-606
43. Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S. et al. Loss of H19 imprinting in esophageal cancer. Cancer research. 1996;56:480-2
44. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P. et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. 2015
45. Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V. et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC cancer. 2013;13:196
46. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS. et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome research. 2013;23:1446-61
47. Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT. et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes & cancer. 2011;2:829-40
48. Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochimica et biophysica acta. 2014;1843:372-86
49. Yochum GS, Cleland R, McWeeney S, Goodman RH. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. The Journal of biological chemistry. 2007;282:871-8
50. Li W, Kang Y. A New Lnc in Metastasis: Long Noncoding RNA Mediates the ProMetastatic Functions of TGF-beta. Cancer cell. 2014;25:557-9
51. Matouk IJ, Abbasi I, Hochberg A, Galun E, Dweik H, Akkawi M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. European journal of gastroenterology & hepatology. 2009;21:688-92
52. Tanaka K, Shiota G, Meguro M, Mitsuya K, Oshimura M, Kawasaki H. Loss of imprinting of long QT intronic transcript 1 in colorectal cancer. Oncology. 2001;60:268-73
53. Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T. et al. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer science. 2006;97:1147-54
54. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A Long Noncoding RNA Activated by TGF-beta Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer cell. 2014;25:666-81
55. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. International journal of oncology. 2011;39:169-75
56. Franklin JL, Rankin CR, Levy S, Snoddy JR, Zhang B, Washington MK. et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochemical and biophysical research communications. 2013;440:99-104
57. Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H. et al. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Medical oncology. 2013;30:588
58. Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H. et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. British journal of cancer. 2014;110:164-71
59. Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. 2007;12:215-29
60. Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. Rna. 2007;13:890-8
61. Shi Y, Liu Y, Wang J, Jie D, Yun T, Li W. et al. Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PloS one. 2015;10:e0122679
62. Pedersen SK, Mitchell SM, Graham LD, McEvoy A, Thomas ML, Baker RT. et al. CAHM, a long non-coding RNA gene hypermethylated in colorectal neoplasia. Epigenetics: official journal of the DNA Methylation Society. 2014:9
63. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F. et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular cell. 2013;49:1083-96
64. Zhai H, Fesler A, Schee K, Fodstad O, Flatmark K, Ju J. Clinical significance of long intergenic noncoding RNA-p21 in colorectal cancer. Clinical colorectal cancer. 2013;12:261-6
65. Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z. et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic acids research. 2013;41:4976-87
66. Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C. et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. Journal of translational medicine. 2013;11:122
67. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R. et al. Activation of p53 by MEG3 non-coding RNA. The Journal of biological chemistry. 2007;282:24731-42
68. Menigatti M, Staiano T, Manser CN, Bauerfeind P, Komljenovic A, Robinson M. et al. Epigenetic silencing of monoallelically methylated miRNA loci in precancerous colorectal lesions. Oncogenesis. 2013;2:e56
69. Qi P, Xu MD, Ni SJ, Shen XH, Wei P, Huang D. et al. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Molecular carcinogenesis. 2014
70. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
71. Davison EJ, Tarpey PS, Fiegler H, Tomlinson IP, Carter NP. Deletion at chromosome band 20p12.1 in colorectal cancer revealed by high resolution array comparative genomic hybridization. Genes, chromosomes & cancer. 2005;44:384-91
72. Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A. et al. The H19 locus acts in vivo as a tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12417-22
73. Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX. et al. The LncRNA H19 promotes epithelial to mesenchymal transition by functioning as MiRNA sponges in colorectal cancer. Oncotarget. 2015
74. Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U. et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. Journal of molecular medicine. 2007;85:293-304
75. Brim H, Lee E, Abu-Asab MS, Chaouchi M, Razjouyan H, Namin H. et al. Genomic aberrations in an African American colorectal cancer cohort reveals a MSI-specific profile and chromosome X amplification in male patients. PloS one. 2012;7:e40392
76. Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079-99
77. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-67
78. Wilkins JA, Sansom OJ. C-Myc is a critical mediator of the phenotypes of Apc loss in the intestine. Cancer research. 2008;68:4963-6
79. Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482-91
80. Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP. et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & development. 2004;18:1385-90
81. Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nature reviews Cancer. 2001;1:55-67
82. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The international journal of biochemistry & cell biology. 2013;45:1895-910
83. Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5523-7
84. Li Q, Lohr CV, Dashwood RH. Activator protein 2alpha suppresses intestinal tumorigenesis in the Apc(min) mouse. Cancer letters. 2009;283:36-42
85. Garneau H, Paquin MC, Carrier JC, Rivard N. E2F4 expression is required for cell cycle progression of normal intestinal crypt cells and colorectal cancer cells. Journal of cellular physiology. 2009;221:350-8
86. Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320-34
87. Younger ST, Rinn JL. 'Lnc'-ing enhancers to MYC regulation. Cell research. 2014
88. Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:1626-34
89. Guo ST, Jiang CC, Wang GP, Li YP, Wang CY, Guo XY. et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32:1910-20
90. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature reviews Cancer. 2005;5:341-54
91. Rigoutsos I, Furnari F. Gene-expression forum: Decoy for microRNAs. Nature. 2010;465:1016-7
92. Drabsch Y, ten Dijke P. TGF-beta signalling and its role in cancer progression and metastasis. Cancer metastasis reviews. 2012;31:553-68
93. Meulmeester E, Ten Dijke P. The dynamic roles of TGF-beta in cancer. The Journal of pathology. 2011;223:205-18
94. Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. TGF-beta signalling in colon carcinogenesis. Cancer letters. 2012;314:1-7
95. Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A. et al. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. Journal of cellular physiology. 2007;213:511-8
96. Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413-31
97. Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609-22
98. Barsotti AM, Prives C. Noncoding RNAs: the missing "linc" in p53-mediated repression. Cell. 2010;142:358-60
99. Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. Journal of molecular cell biology. 2014;6:181-91
100. Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y. et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer research. 2010;70:2350-8
101. Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR. et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. The Journal of clinical endocrinology and metabolism. 2003;88:5119-26
102. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. Journal of molecular endocrinology. 2012;48:R45-53
103. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409-19
104. Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature reviews Molecular cell biology. 2008;9:402-12
105. Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J. et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315-26
106. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature reviews Cancer. 2013;13:97-110
107. Matouk IJ, Raveh E, Abu-Lail R, Mezan S, Gilon M, Gershtain E. et al. Oncofetal H19 RNA promotes tumor metastasis. Biochimica et biophysica acta. 2014;1843:1414-26
108. Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS. et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Molecular cancer. 2014;13:68
109. Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F. et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1531-41
110. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Molecular bioSystems. 2012;8:2289-94
111. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S. et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncology reports. 2014
112. Berndt SI, Potter JD, Hazra A, Yeager M, Thomas G, Makar KW. et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Human molecular genetics. 2008;17:2665-72
113. Meyer KB, Maia AT, O'Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P. et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS genetics. 2011;7:e1002165
114. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:155-65
115. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149-54
116. Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS medicine. 2013;10:e1001453
117. Reimers MS, Zeestraten EC, Kuppen PJ, Liefers GJ, van de Velde CJ. Biomarkers in precision therapy in colorectal cancer. Gastroenterology report. 2013;1:166-83
118. Nitsche U, Maak M, Schuster T, Kunzli B, Langer R, Slotta-Huspenina J. et al. Prediction of prognosis is not improved by the seventh and latest edition of the TNM classification for colorectal cancer in a single-center collective. Annals of surgery. 2011;254:793-800 discussion -1
119. Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-9
120. Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian J. et al. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget. 2014;5:2230-42
121. Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Current opinion in molecular therapeutics. 2010;12:607-16
122. Hasenpusch G, Pfeifer C, Aneja MK, Wagner K, Reinhardt D, Gilon M. et al. Aerosolized BC-819 inhibits primary but not secondary lung cancer growth. PloS one. 2011;6:e20760
123. Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Journal of translational medicine. 2010;8:134
124. Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I. et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. Journal of translational medicine. 2009;7:69
125. Sorin V, Ohana P, Mizrahi A, Matouk I, Birman T, Hochberg A. et al. Regional therapy with DTA-H19 vector suppresses growth of colon adenocarcinoma metastases in the rat liver. International journal of oncology. 2011;39:1407-12
Author contact
![]() Corresponding author: Dr. Yanlei Ma, Department of Colorectal Surgery, Fudan University Shanghai Cancer Center; Department of Oncology, Shanghai Medical College, Fudan University, 270 Dong'an Road, Shanghai, 20032, China, Tel.: +86 21 64175590; Fax: +86 21 54175590, E-mail: yanleimaedu.cn
Corresponding author: Dr. Yanlei Ma, Department of Colorectal Surgery, Fudan University Shanghai Cancer Center; Department of Oncology, Shanghai Medical College, Fudan University, 270 Dong'an Road, Shanghai, 20032, China, Tel.: +86 21 64175590; Fax: +86 21 54175590, E-mail: yanleimaedu.cn

 Global reach, higher impact
Global reach, higher impact