Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(16):3309-3317. doi:10.7150/jca.20159 This issue Cite
Research Paper
Heparanase Contributes To Trans-Endothelial Migration of Hepatocellular Carcinoma Cells
1. Department of Hepatobiliary Surgery, Yijishan Hospital of Wannan Medical College, Wuhu 241001, China;
2. Department of General Surgery, Maanshan People's Hospital, Maanshan 243000, China;
3. Department of General Surgery, Affiliated Chaohu Hospital, Anhui Medical University, Hefei 238000, China;
4. Department of Pathology, Yijishan Hospital of Wannan Medical College, Wuhu 241001, China.
Received 2017-3-20; Accepted 2017-8-30; Published 2017-9-16
Abstract
The overall outcome of patients with hepatocellular carcinoma (HCC) is still very poor due to its high metastasis and recurrence rate. During metastasis, trans-endothelial migration (TEM) of HCC cells is a key step. Heparanase (HPSE) is an endo-beta-glucuronidase and exerts prometastatic properties for normal and tumor-derived cells. However, it is remains unclear that HPSE contributes to TEM of HCC cells. In this study, human umbilical vein endothelial cells-C (HUVEC-C) was used to simulate vascular endothelial cells (VECs), and the HCCLM3 cells with high HPSE expression were chosen and used for in vitro TEM assay and in vivo experiment. As results, we found that HCCLM3 cells showed higher TEM rate compared with other HCC cells. Downregulation or inhibition of HPSE activity resulted in suppression of TEM of HCC cells both in vitro and in vivo. Our findings suggest that HPSE contributes to TEM of HCC cells, which may be a new biological function of HPSE.
Keywords: heparanase, hepatocellular carcinoma, vascular endothelial cell, trans-endothelial migration.
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related mortality worldwide [1]. The high mortality of HCC is mainly due to the occurrence of intrahepatic metastasis [2-4]. During metastasis, it is necessary that HCC cells move from the portal vein (PV) to new liver parenchyma by traversing the endothelial cell layer of the blood vessel. Therefore, trans-endothelial migration (TEM) is a key step in the metastatic dissemination of HCC. However, it is unclear what molecules contribute to TEM, and the precise pathogenesis remains to be determined.
Heparanase (HPSE) is an endo-beta-glucuronidase. By cleaving heparan sulfate (HS) side chains of heparan sulfate proteoglycans (HSPGs) on cell surfaces, extracellular matrices (ECM) and basement membrane (BM), HPSE is capable of loosening the ECM and BM barrier and delivering some cytokines such as basic fibroblast growth factor (bFGF) and vascular endothelial cell growth factor (VEGF) to promote metastasis and angiogenesis [5-9]. Our previous studies also demonstrated that HPSE expression was associated with tissue differentiation, HCC stages, angiogenesis, metastatic recurrence and prognosis [10, 11]. Further research showed that HPSE could exert pro-migrative properties for normal and tumor-derived cells by clustering of membrane HSPGs (i.e., syndecans) and activation of signaling molecules such as Akt, Src, and Rac in a HS-dependent and -independent manner [12, 13]. Our recent study confirmed that HPSE could mediate the migration of HCC cells, which could be inhibited by RNAi of HPSE [14]. However, it is unclear whether HPSE contributes to the TEM of cancer cells. TEM is conceptually different from the general migration of cancer cells in ECM and BM. Increased HPSE expression has been found in dicaryocytes, and contributes to passing through the blood vessels in acute immunological rejection of xenotransplantation [15]. The similar phenomenon has also been found in lymphocytes [16]. Cancer cells in the blood stream cross the endothelial cell layer of the blood vessel to enter new parenchyma of the target organ in a manner similar to the extravasation of leukocytes [17]. On the basis of premise, we hypothesized that HPSE could contribute to TEM of HCC cells.
In the present study, human umbilical vein endothelial cells-C (HUVEC-C) and HCC cells with high levels of HPSE were used for in vitro TEM assay and in vivo test. We demonstrate that downregulation of HPSE activity results in suppression of TEM and liver metastasis of HCC cells. Our study suggests HPSE contributes to TEM and promotes intrahepatic dissemination of HCC cells.
Materials and methods
Materials
HUVEC-C, normal liver cell line LO-2 and HCC cell lines (HepG2 and BEL-7402) were from Cell Bank of National Academy of Science of China (Shanghai, China). Human highly metastatic liver cancer cell line HCCLM3 was from Liver Cancer Institute of Zhongshan Hospital, Fudan University (Shanghai, China). Recombinant escherichia coli containing RNAi sequence of HPSE (siHPSE-504, siHPSE-683, siHPSE-852 and siHPSE-3158) and negative control plasmid were from Jikai Genechem Co., Ltd. (Shanghai, China) (table 1). Trizol solution, reverse transcription kits were from MBI Fermentas Corporation (Pittsburgh. Pennsylvania, USA). Markers, dNTP, primers were from Sangon Biotech Co., Ltd. (Shanghai, China). EvaGreen qPCR Master Mix were from Bio-Rad Laboratories (Hercules, California, USA). Lipofectamine 2000, DMEM medium, 10% fetal bovine serum (FBS) and RPMI-1640 medium were from Gibco Company (Invitrogen, Carlsbad, California, USA). PCR purification kits and plasmid extraction kit were from Axygen Scientific Inc. (Carlsbad, California, USA). Polyvinylidene fluoride (PVDF) membrane was from Millipore Corporation (Bedford, Massachusetts, USA). Bicinchoninic acid (BCA) protein assay kit and enhanced chemiluminescence (ECL) Western blotting kit were from Beyotime Biotech Co., Ltd. (Shanghai, China). Rabbit-anti-HPSE polyclonal antibody was from Abcam Inc. (Cambridge British). Rabbit-anti-phosphoglyceraldehyde dehydrogenase (GAPDH) polyclonal antiserum and secondary antibody were from Biogot Biotechnology Co., Ltd. (Louis Park, Minnesota, USA). Rose Bengal was from Sigma Chemical Company (San Francisco, California, USA).
Forty male nude mice (BALB/C-nu/nu) of 4‑week‑old, weighing 16‑20 g were obtained from the Comparative Medicine Center of Yangzhou University (Yangzhou, Jiangsu, China). Heparin sodium injection was from Qianhong Biochemical Pharmaceutical Co., Ltd (Changzhou, Jiangsu, China).
Sequences of RNA interference.
| Name | Direction | Sequence coding |
|---|---|---|
| siHPSE-504 | Forward Reverse | 5' -CCAGGAUAUUUGCAAAUAUTT -3' 5' -AUAUUUGCAAAUAUCCUGGTT- 3' |
| siHPSE-683 | Forward Reverse | 5'- GCUCUGUAGAUGUGCUAUATT- 3' 5'- UAUAGCACAUCUACAGAGCTT- 3' |
| siHPSE-852 | Forward Reverse | 5' -GUCGCAGUUAGGAGAAGAUTT- 3' 5' -AUCUUCUCCUAACUGCGACTT- 3' |
| siHPSE-3158 | Forward Reverse | 5' -GGCGAGGAGAUUCUGUAAATT- 3' 5'- UUUACAGAAUCUCCUCGCCTT -3' |
| Negative control | Insertion sequence | TTCTCCGAACGTGTCACGT |
Cell culture
Human HUVEC-C cells, HCC cells and normal liver cell line LO-2 were cultured in DMEM medium supplemented with 10% FBS. Cells were maintained at 37°C in a humidified incubator with 5% CO2. When cells reached 80-90% confluence (usually in 2 or 3 days), they were harvested using trypsin (0.25%) with 0.01% ethylenediamine tetraacetic acid (EDTA) and seeded (1:2) into new culture flasks with complete DMEM. The media were replaced every day.
Measurement of HPSE expression in HCC cells
HPSE expression at mRNA levels in all cells were determined by real-time qRT-PCR. The primers of HPSE: sense: 5′-GCACAAACACTGACAATCCAAG-3′; antisense: 5′-AAAAGGATAGGGTAACCGCAA-3′. The primers of GAPDH: sense: 5′-GTGGTCTCCTCTGACTTCAACA-3′; antisense: 5′-CCACCACCCTGTTGCTGTAG-3′. The total RNA from HCC cells was isolated and transcribed to cDNA according to the manufacturer's protocol. The quantitative real-time PCR reactions were performed in a total volume of 25 µl containing 1ug cDNA, 5 μM of each primer, and 12.5 µl 2 × PCR Master Mix. The cycling parameters for PCR were as follows: denaturation 95°C 4 min, followed by 40 cycles of denaturation at 94°C 15 sec, annealing at 55°C 30 sec, and extension at 72°C 30 sec. GAPDH was used as an endogenous control. Data were analyzed with the standard 2-∆∆Ct method and values are expressed as the average of triplicates.
HPSE expression at protein levels was measured by Western blot. After harvesting the total protein from cultured cells, the concentration of protein was detected by BCA protein assay kit. An equivalent protein in each sample was separated on the 10% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) electro-phoresis, and electrotransferred to PVDF membranes. After blocking with 5% (w/v) milk in TBS-0.05% Tween, the membranes were incubated with primary antibody against HPSE (1:500) or GAPDH (1:2,000) overnight at 4˚C. Followed by incubation with horseradish peroxidase conjugated secondary antibodies(1:1,000) for 1 h, the immunoblots were visualized using ECL kit according to the recommended procedure. The protein bands were quantified using NIH ImageJ software. The experiments were performed for three times. According to the results, the HCCLM3 cell with highest expression of HPSE was selected for following experiments.
Transient transfection
The HCCLM3 cells were transiently transfected with above 4 recombinant plasmids, respectively, using lipofectamine 2000 according to the manufacturer's protocol. Control groups were transiently transfected with negative control plasmid (NC) or transfection reagent (blank control, BC). The cells were observed to count the efficiency by using fluorescence microscope 48 h after transfection.
Determination of HPSE expression in transfected HCC cells
The transfected HCCLM3 cells were harvested at 48 h post-transfection. The harvested cells were washed and lysed with lysis buffer. The HPSE expression was determined with real-time qRT-PCR and western blot using the same method as mentioned in the previous section. According to the interference efficiency, the optimal RNAi plasmid siHPSE-3158 was chosen for TEM assay.
In vitro TEM assay
TEM assays were performed in transwell plates of 6.5 mm diameter with 8-μm pore filters. Above HUVEC-C cells were seeded into the matrigel-coated transwell filters of apical chambers at 200 µl per well. When compact monolayer is formed, transfected HCCLM3 cells (72 hours) were adjusted to a concentration of 6 × 105/ ml, and then added to endothelial cell monolayer at a density of 1.2×105 cells per well in 200 ml medium. 100 µl DMEM medium containing 20% FBS was added to the basal chambers. Blank control group, negative control group, untransfected HCCLM3 cell group and pure HUVEC-C group were used for control. There were 3 wells in every group. The transwell plate was incubated at 37 °C in 5% CO2 for 24 h, and then the medium in each well was abandoned. The cells in apical chambers were wiped with sterile cotton buds. Rose Bengal solution was added to the basal chamber at 400 µl per well. The staining solution was drawn off and washed thrice 5 min later. In the end, destaining solution (95% alcohol: PBS, 1:1) was added at 400 µl per well and kept at room temperature for 30 min. 200 μl staining solution in each well was in order transferred into another 96-well plate, and the OD value at 570 nm (OD 570nm) was measured by a microplate reader. TEM rate of HCCLM3 cells = OD 570nm of experimental group - OD 570nm of pure HUVEC-C group. The polycarbonate films were observed using an inverted microscope. Each assay was performed in triplicate.
In vivo TEM experiments
All animal experimental procedures in this study were approved by the institutional ethical committee of Yijishan Hospital of Wannan Medical College. The feeding conditions were as follow: temperature 25±2°C, humidity (60±10)%, and illumination time 12 h. Besides male nude mice, the HCCLM3, HepG2 cells and HPSE inhibitor heparin sodium injection were used for in vivo experiments. First, pre-experiments were performed. The nude mice were intraperitoneally injected with three kinds of different concentration of cultured HCC cells (1×106/300 µl, 3×106/300 µl and 5×106/300 µl ) or subcutaneously injected with three kinds of doses of heparin sodium injection (125, 250 and 500 IU), respectively. The general condition, bleeding symptom of animal and tumor formation rates of live were observed. Based on the results of pre-experiments, the HCC cell concentration of 3×106/300 µl and the heparin dosage of 250 IU were chosen for formal test.
Thirty nude mice were randomly divided into 3 groups (control, HCCLM3 cell and HepG2 cell group) with each group 10 mice. After the lower abdomens were sterilized with 75% (v/v) ethanol, 300µl normal sodium (NS) was injected into the lower peritoneal cavity of mice in control group, and equal amounts of HCC cells (3×106) were injected in other two groups, respectively.
All of the above processes were performed under sterile environment. Five mice in each group were sacrificed by the end of the third and fifth week, respectively; omentum and liver tumors were excised, fixed in 10% (v/v) formalin and embedded in paraffin. Tumor sections (4μm thick) were then stained with haematoxylene and eosin (H&E) and examined under an optical microscope. According to the results, HCC cells (HCCLM3 cells) were chosen for establishing a liver metastasis model for human HCC.
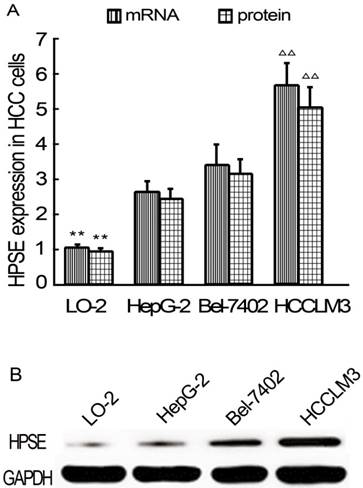
Relative expression levels of HPSE in LO-2 and HCC cells (Real-time qRT-PCR and western blot). A, HPSE mRNA and pritein expression levels. B Western blot. ** P < 0.01 vs HepG2, Bel-7402 and HCCLM3 groups; ∆∆ P < 0.01 vs HepG2 and Bel-7402 groups.
Another 20 nude mice were randomly divided into 2 groups (control and heparin group) with each group 10 mice. The mice in both groups were intraperitoneally injected with 300 µl of HCCLM3 cells (3×106). Since the beginning of second week of cell inoculation, the mice in heparin group were subcutaneously injected with a dose of heparin (250 IU) every 3 day for 2 weeks, and the mice in control group were injected with equal amounts of NS. All of the above processes were also performed under sterile environment. Same observations were performed by the end of the third and fifth week of cell inoculation.
Statistical analysis
The results were expressed as the mean±standard deviation. Comparisons of continuous variables were performed with the one-way ANOVA test and q test. Comparisons of categorical variables were performed with chi-square test or Fisher's exact test. All analyses were performed using SigmaStat 3.5 and P < 0.05 in a two-side test was considered statistically significant.
Results
HPSE expression in HCC cells
HPSE expressions in HCC cells were detected using real-time qRT-PCR and western blot analysis at the beginning of the study. As results, HPSE expression at mRNA levels in HepG2, BEL-7402 and HCCLM3 cells were significantly higher than that in LO-2 cells (P<0.01) (Figure 1A). Of all 3 kinds of HCC cells, HCCLM3 cells showed highest HPSE expressions (P<0.01). HPSE protein expression is consistent with mRNA (Figure 1A-B). Based on these findings, HCCLM3 cell line was selected for following transfection experiments.
Photofluorograms of 4 kinds of RNAi transfected HCCLM3 cells (×100).
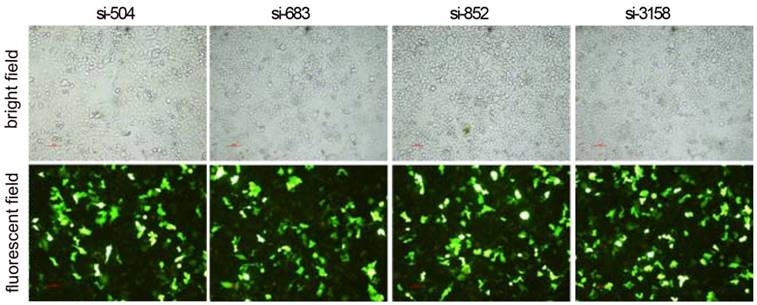
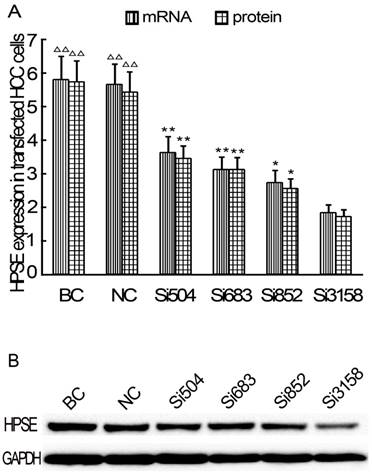
RNAi decreases HPSE expression in HCCLM3 cells
After RNAi plasmids were transiently transfected into HCCLM3 cell line, bright green fluorescence could be observed in 4 kinds of RNAi plasmids transfected HCCLM3 cells using fluorescence analysis 48 h later. The cell outlines were clear (Figure 2). Approximately 25%-35% transfection efficiency was obtained without significant difference among them (P>0.05). HPSE mRNA expressions in RNAi transfected HCCLM3 cells significantly decreased compared with control groups (P<0.01), and lowest expression was found in siHPSE-3158 transfected HCCLM3 cells (P<0.05) (Figure 3A). HPSE protein expressions showed similar feature (P<0.01), and lowest protein expression was also found in siHPSE-3158 transfected cells (P<0.05) (Figure 3A-B). Therefore, siHPSE-3158 plasmid was chosen for subsequent in vitro TEM assay.
Relative expression levels of HPSE in RNAi plasmids transfected HCCLM3 cells (Real-time qRT-PCR and western blot). A, HPSE mRNA and pritein expression levels. B Western blot. BC: Blank control group. NC: Negative control group. * P < 0.05, ** P < 0.01 vs si-3158 group. ∆∆ P < 0.01 vs siHPSE-504, siHPSE-683, siHPSE-852 and siHPSE-3158 groups.
RNAi decreases TEM ability of HCCLM3 cells
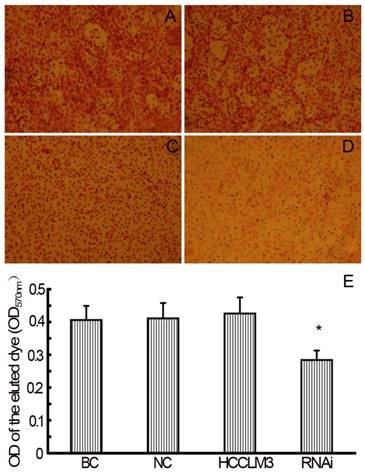
In the TEM assay, staining solution of RNAi (siHPSE-3158) transfected HCCLM3 cells showed lightest staining, suggesting less HCC cells trans-endothelially migrated to the basal chamber. Staining solution of untransfected HCCLM3 cells showed deepest staining, indicating most HCCLM3 cells migrated to the basal chamber (Figure 4A-D). The OD values of basal chamber in RNAi group was significantly lower than those in other 3 groups [(0.284±0.029) vs (0.406±0.043), (0.411±0.047) and (0.426±0.049), all P<0.05)] (Figure 4E).
RNAi decreases theTEM ability of HCCLM3 cells. A, BC group (×200). B, NC group (×200). C, HCCLM3 cell group (×200). D, RNAi group (×200). E, Optical density of all groups. * P < 0.05 vs BC, NC and HCCLM3 cell groups.
2.5 Heparin reduces liver metastasis of HCCLM3 cells
In the in vivo experiments, we evaluated the difference of HepG2 and HCCLM3 cells mediated hepatic metastasis, and established a TEM or/and liver metastasis model. All nude mice survived after intraperitoneal inoculation of HCC cells. The general condition, diet and activity of mice continued without obvious change. The body weights of all 3 groups by the end of the third week had a significant increase compared with those at the beginning of test (all P<0.05), but they had no obvious difference at at the same time (P > 0.05).
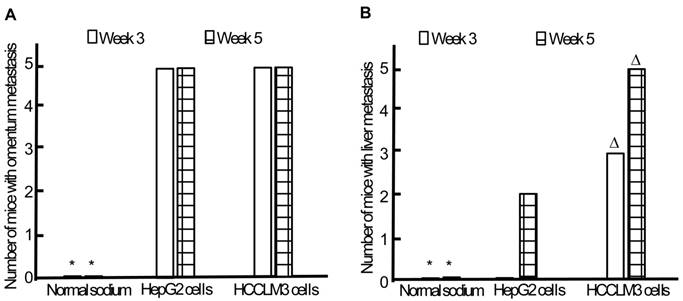
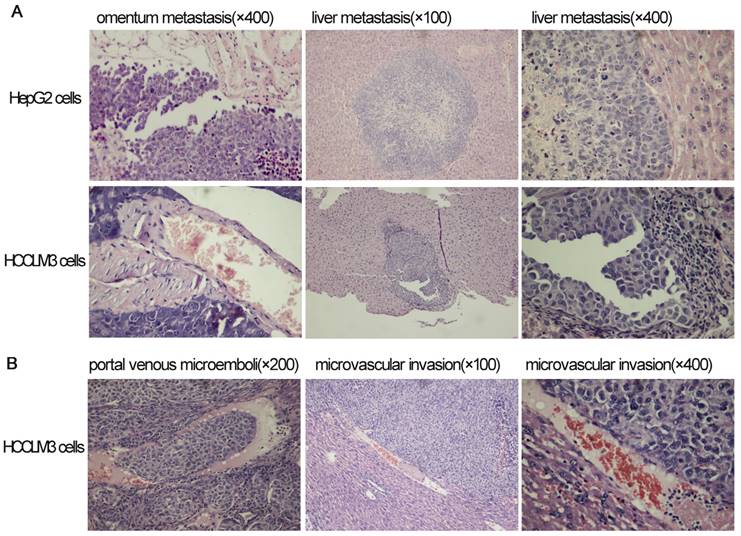
Omentum metastasis could be found in all mice of both HepG2 and HCCLM3 cell groups (5/5,5/5) by the end of the third and fifth week after intraperitoneal inoculation (Figure 5A, Figure 6A), and no cancerous node was found in the omentum of mice in NS group (0/5,0/5). Liver metastasis rates of HCCLM3 cell group were obviously higher than those of HepG2 cell group by the end of the third and fifth week (2/5 and 5/5 vs 0/5 and 3/5, both P<0.05) (Figure 5B, Figure 6A). In addition, portal venous microemboli (PVME) and microvascular invasion were common in HCCLM3 cell group (Figure 6B). No node was observed in the liver of mice in NS group. These findings suggested intraperitoneally inoculated HepG2 and HCCLM3 cells could grow well in the omentum of mice, but only HCCLM3 cells could be better absorbed into the bloodstream of PV, and trans-endothelially migrate into new liver parenchyma resulting in more liver metastasis. Therefore, HCCLM3 cells were chosen for establishing animal model of TEM or/and hepatic metastasis.
Omentum and liver metastasis rates after intraperitoneal inoculation of HCC cells. A, Omentum metastasis. B, Liver metastasis. * P < 0.05 vs HepG2 and HCCLM3 cell groups, ∆ P < 0.05 vs HepG2 cell group.
The results of hematoxylin and eosin staining for tumors in omentum and liver after intraperitoneal inoculation of HCC cells.
Effect of heparin on omentum and liver metastasis. A, Omentum metastasis. B, Liver metastasis. * P < 0.05 vs control group.
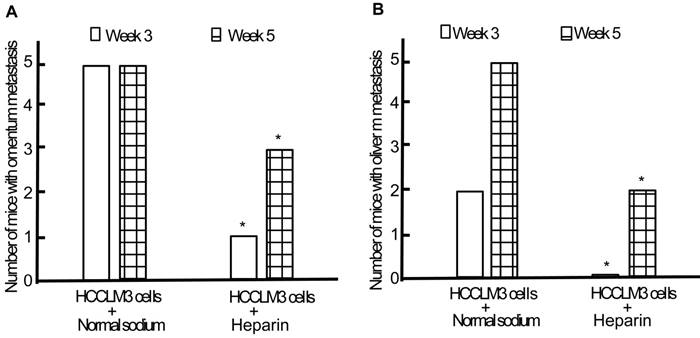
Following establishment and replication of animal model, we observed the effect of heparin on the TEM or/and liver metastasis. Minor subcutaneous hemorrhage could be found in the injection site of heparin in tumor-bearing nude mice without hemorrhoea and death. By the end of the third week of intraperitoneal inoculation, the omentum and liver metastasis rates of heparin group were significantly lower than those of control group (1/5 and 0/5 vs 5/5 and 2/5, both P < 0.05) (Figure 7). By the end of the fifth week, cancerous nodes in omentum and liver developed again with withdrawal of heparin, but the metastasis rates were still lower than that of control group (3/5 and 2/5 vs 5/5 and 5/5, both P < 0.05) (Figure 7). PVME and microvascular invasion in heparin group was less than those in control group.
Discussion
HCC cells that enter portal circulation must trans-endothelially migrate into new liver parenchyma before forming substantive metastasis. Therefore, TEM of HCC cells is one of key steps of liver metastasis. Previous studies showed that HPSE expression is increased in most cancers, and exerts pro-migrative properties for cancer cells [12-14]. However, no further researches prove whether HPSE participates TEM of cancer cells by now.
In order to observe the effect of HPSE on TEM, HUVEC-C cells were seeded into the apical chamber of transwell to simulate endothelial cell in the in vitro TEM assay. When the compact monolayer is formed, siHPSE-3158 transfected and untransfected HCCLM3 cells were added. The TEM ability was detected by observing the staining degree and detecting the OD value of HCCLM3 cells moving to the basal chamber. As results, we found that the TEM rate of HCCLM3 cells in RNAi group significantly decreased compared with other groups. The findings prove that HPSE can induce the TEM of HCC cells and cancer cells crossing endothelial cell monolayer of the blood vessel is similar with the extravasation of leukocytes [17]. To our knowledge, this is the first report that HPSE plays a promoting role in TEM of HCC cells. HPSE can degrade HSPGs on the surface of VEC, BM and ECM. Integrity of blood vessel endothelium barrier is destroyed, which provides a “breakthrough” for HCC cells migrating into target tissue through VEC [5, 12, 19, 20]. Released HS on the surface of VEC can augment the migration ability of cancer cells and angiogenesis [12, 20]. In addition, HPSE participates in the regulation of some signal ways, and augments the migration ability of cancer cells by activating downstream signal molecules such as p38MAPK, Akt, Src and VEGF [21, 22].
To verify the effect of HPSE on TEM, it is necessary to establish an animal model of liver metastasis of HCC. The model should mimic well the process of TEM. At present, there are numerous experimental models of HCC developed by using drug induction, subcutaneous or orthotopic transplantation and genetic engineering [23-27]. However, these models can not simulate TEM of HCC cells. Some reserches reported cancer cells could be injected into spleen or PV, and directly absorded via PV and transported to the liver resulting experimental liver metastasis [28-30]. However, we can not find any liver tumor when HCC cells were injected into spleen or PV in our previous study. After repeated experiment and comparison, we finally established a liver metastatic model using intraperitoneal injection of HCC cells. When the HCC cells are injected into the abdominal cavity of nude mice, they will proliferate in omentum and be absorded via PV and transported to the liver resulting liver metastasis. Therefore, we consider the model could mimic the process of TEM. Compared with HepG2 cells, HCCLM3 cells-induced model can better reflect the characteristics of TEM because it produced more metastatic lesions in the liver of mice besides PVME and microvascular invasion in the liver tumor site. Because HPSE expression level in HCCLM3 cells was also higher than that in HepG2 cells, we think HPSE could promotes the TEM of HCC cells.
In order to verify the effect of HPSE further, we used heparin to inhibit the activity of HPSE in vivo test and found that heparin significantly reduced HCCLM3 cells-induced liver metastasis, PVME and microvascular invasion. These findings further suggest HPSE promotes the TEM of HCC cells and following liver metastasis in nude mice.
In conclusion, HPSE could effectively promote the TEM of HCC cells in vitro and in vivo. Our research discovers a new biological function of HPSE, and provides a cytological and histological evidence for following study of the mechanism.
Abbreviations
HCC: hepatocellular carcinoma; VEC: vascular endothelial cells; PV: portal vein; TEM: trans-endothelial migration; HPSE: heparanase; HSPGs: heparan sulfate proteoglycans; ECM: extracellular matrices; BM: basement membrane; bFGF: basic fibroblast growth factor; VEGF: vascular endothelial cell growth factor; HUVEC: human umbilical vein endothelial cells; RT-PCR: reverse transcriptase-polymerase chain reaction; GAPDH: phosphoglyceraldehyde dehydrogenase; PVME: portal venous microemboli.
Acknowledgements
We thank Dr. Kong Lv, Dr. Mengying Zhang, Dr. Min Zhong, Dr. Qifeng Suo and Dr. Xueqin Li from Central Laboratory, Affiliated Yijishan Hospital of Wannan Medical College for technical assistance. We thank Dr. Gang Feng from Clinical Laboratory, Affiliated Yijishan Hospital of Wannan Medical College for his help in language translation. This study was supported by the National Natural Science Foundation of China (Grant No. 81272412). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30
2. Shirabe K, Wakiyama S, Gion T. et al. Clinicopathological risk factors linked to recurrence pattern after curative hepatic resection for hepatocellular carcinoma-results of 152 resected cases. Hepatogastroenterology. 2007;54:2084-7
3. Nakashima O, Kojiro M. Recurrence of hepatocellular carcinoma: multicentric occurrence or intrahepatic metastasis? A view point in terms of pathology. J Hepatobiliary Pancreat Surg. 2001;8:404-9
4. Chan AC, Chan SC, Chok KS. et al. Treatment Strategy for Recurrent Hepatocellular Carcinoma: salvage transplantation, repeated resection, or radiofrequency ablation? Liver Transpl. 2013;19:411-9
5. Vlodavsky I, Friedmann Y, Elkin M. et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5:793-802
6. Hulett MD, Freeman C, Hamdorf BJ. et al. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:803-9
7. Kussie PH, Hulmes JD, Ludwig DL. et al. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261:183-7
8. Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274:24153-60
9. McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1-14
10. Chen XP, Liu YB, Rui J. et al. Heparanase mRNA expression and point mutation in hepatocellular carcinoma. World J Gastroenterol. 2004;10:2795-9
11. Chen B, Chen XP, Wu MS, Cui W, Zhong M. Expressions of heparanase and upstream stimulatory factor in hepatocellular carcinoma. Eur J Med Res. 2014;19:45-52
12. Roy M, Marchetti D. Cell surface heparan sulfate released by heparanase promotes melanoma cell migration and angiogenesis. J Cell Biochem. 2009;106:200-9
13. Gingis-Velitski S, Zetser A, Flugelman MY. et al. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536-41
14. Chen XP, Luo JS, Tian Y. et al. Downregulation of heparanase expression results in suppression of invasion, migration and adhesion abilities of hepatocellular carcinoma cells. Biomed Res Int. 2015;2015:241983
15. Dempsey LA, Plummer TB, Coombes S. et al. Platelet heparanase in vascular responses to xenotransplantation. Transplant Proc. 2000;32:972
16. Bartlett MR, Underwood PA, Parish CR. Comparative analysis of the ability of leucocytes, endothelial cells and platelets to degrade the subendothelial basement membrane: evidence for cytokine dependence and detection of a novel sulfatase. Immunol Cell Biol. 1995;73:113-24
17. Zabel BA, Lewén S, Berahovich RD. et al. The novel chemokine receptor CXCR7 regulates trans-endothelial migration of cancer cells. Mol Cancer. 2011;10:73
18. Dong W, Zhao H, Zhang C. et al. Gene silencing of heparanase results in suppression of invasion and migration of hepatoma cells. World J Surg Oncol. 2014;12:85
19. Pisano C, Vlodavsky I, Ilan N. et al. The potential of heparanase as a therapeutic target in cancer. Biochem Pharmacol. 2014;89:12-9
20. Barash U, Cohen-Kaplan V, Dowek I. et al. Proteoglycans in health and disease: new concepts for heparanase function in tumor progression and metastasis. FEBS J. 2010;277:3890-3
21. Riaz A, Ilan N, Vlodavsky I. et al. Characterization of heparanase-induced phosphatidylinositol 3-kinase-AKT activation and its integrin dependence. J Biol Chem. 2013;288:12366-75
22. Secchi MF, Masola V, Zaza G. et al. Recent data concerning heparanase: focus on fibrosis, inflammation and cancer. Biomol Concepts. 2015;6:415-21
23. Futakuchi M, Hirose M, Ogiso T. et al. Establishment of an in vivo highly metastatic rat hepatocellular carcinoma model. Jpn J Cancer Res. 1999;90:1196-202
24. Sawada S, Murakami K, Yamaura T. et al. Therapeutic and analysis model of intrahepatic metastasis reflects clinical behavior of hepatocellular carcinoma. Jpn J Cancer Res. 2002;93:190-7
25. Sun FX, Tang ZY, Lui KD. et al. Establishment of a metastatic model of human hepatocellular carcinoma in nude mice via orthotopic implantation of histologically intact tissues. Int J Cancer. 1996;66:239-43
26. Chen Q, Wang X, Wu H. et al. Establishment of a dual-color fluorescence tracing orthotopic transplantation model of hepatocellular carcinoma. Mol Med Rep. 2016;13:762-8
27. Runge A, Hu J, Wieland M. et al. An inducible hepatocellular carcinoma model for preclinical evaluation of antiangiogenic therapy in adult mice. Cancer Res. 2014;74:4157-69
28. Yamada N, Chung YS, Arimoto Y. et al. Establishment of a new human extrahepatic bile duct carcinoma cell line (OCUCh-LM1) and experimental liver metastatic model. Br J Cancer. 1995;71:543-8
29. Ishizu K, Sunose N, Yamazaki K. et al. Development and characterization of a model of liver metastasis using human colon cancer HCT-116 cells. Biol Pharm Bull. 2007;30:1779-83
30. Giavazzi R, Jessup JM, Campbell DE. et al. Experimental nude mouse model of human colorectal cancer liver metastases. J Natl Cancer Inst. 1986;77:1303-8
Author contact
![]() Corresponding authors: Dr. Xiaopeng Chen, Department of Hepatobiliary Surgery, Affiliated Yijishan Hospital of Wannan Medical College, No. 2 Zheshan West Road, Wuhu 241001, China; E-Mail: drchenxpedu.cn; Tel: (86) 553-5739625; Fax: (86) 553-5739038. Or Dr. Linming Lu, Department of Pathology, Yijishan Hospital of Wannan Medical College, Wuhu 241001, China; E-Mail: llm7172com; Tel: (86) 553-5739788; Fax: (86) 553-5739038
Corresponding authors: Dr. Xiaopeng Chen, Department of Hepatobiliary Surgery, Affiliated Yijishan Hospital of Wannan Medical College, No. 2 Zheshan West Road, Wuhu 241001, China; E-Mail: drchenxpedu.cn; Tel: (86) 553-5739625; Fax: (86) 553-5739038. Or Dr. Linming Lu, Department of Pathology, Yijishan Hospital of Wannan Medical College, Wuhu 241001, China; E-Mail: llm7172com; Tel: (86) 553-5739788; Fax: (86) 553-5739038

 Global reach, higher impact
Global reach, higher impact