Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(18):3785-3794. doi:10.7150/jca.21151 This issue Cite
Research Paper
Polymorphisms of CCNB1 Associated With the Clinical Outcomes of Platinum-Based Chemotherapy in Chinese NSCLC Patients
1. Department of Thoracic Surgery, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, P.R. China;
2. Department of Respirology and Critical Care Medicines, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, P.R. China;
3. Central Laboratory, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, P.R. China;
4. Department of Thoracic Surgery, Shanghai Chest Hospital, Shanghai Jiaotong Universtiy, Shanghai, P.R. China.
Received 2017-5-23; Accepted 2017-8-15; Published 2017-10-17
Abstract
As a crucial cell cycle regulator and G2/M phase promotor, CCNB1 played an essential role in progression of chemotherapy related cell death. Platinum-based chemotherapy is still the first-line chemotherapy regimen for most advanced NSCLC patients. We aim to investigate the correlation of CCNB1 polymorphisms to the efficiency of platinum-based chemotherapy in Chinese advanced NSCLC patients. We enrolled 972 patients with advanced NSCLC, and extracted DNA from their peripheral blood for genotyping CCNB1 four tagSNPs which selected from the Hapmap database. We analyzed the association of CCNB1 four tagSNPs with efficiency of platinum-based chemotherapy. We found that rs2069429 and rs2069433 of CCNB1 were associated with the OS of advanced NSCLC patients. Patients with GG genotype of rs2069429 had longer OS than non-GG patients (HR=0.81, 95%CI=0.68-0.95, p=0.009); and patients with AA genotype of rs2069433 had longer OS than non-AA patients (HR=0.78, 95%CI=0.61-0.98, p=0.036). And the haplotype GAAA of CCNB1 was a putative factor in subgroup patients with clinical stage IV. The association of CCNB1 polymorphisms and toxicities after platinum-based chemotherapy was assessed. Rs2069433 and rs350104 were related with gastrointestinal toxicity of platinum-based chemotherapy. The patients with GG genotype of rs2069433 (p=0.013) and/or non-GG genotype of rs350104 (p=0.042) may have a severe gastrointestinal toxicity after chemotherapy, and then clinician may can reduce the dosage of chemotherapy agents to avoid sever toxicities in these patients. In summary, CCNB1 polymorphisms may contribute to the clinical efficiency of platinum-based chemotherapy in advanced NSCLC patients, and it is helpful for the personalized treatment.
Keywords: Platinum, Chemotherapy, CCNB1, Single nucleotide polymorphism, Lung cancer.
Introduction
With high morbidity and mortality, 48.3/105 and 39.3/105, respectively, lung cancer is a leading cause of cancer related death disease in the worldwide, including China [1]. Lung cancers are divided into two subtypes according to histology, non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is account for 85% in whole lung cancer patients. No matter what the histology of lung cancer is, the patients are diagnosed in advanced clinical stag (stage III or IV) at the first time, with losing chance of surgical resection [2]. And then chemotherapy, targeted therapy, and immune therapy are becoming promising treatment for advanced lung cancer patients. However, the patients harboring positive driver mutation are only accounting for merely 10% and 2%‒7%, respectively (for EGFR driver mutation and ALK rearrangement). Based on 2015 NCCN guidelines for NSCLC, the patients without typical mutations still rely on chemotherapy, and the platinum-based double chemotherapy is the first-line chemotherapy regimen in advanced lung cancer patients [3].
The efficiency of chemotherapy is not identical among patients and this individual differences may can be explained by single nucleotide polymorphisms (SNP), a single base differences among people in human genome which amount to approximately 15 million [4]. To better understanding the links between disease and genome, many databases about SNP were created by sequencing, like the international HapMap project, 1000 Genomes Project. Many research have been reported lung cancer susceptibility was affected by genome variants at different populations [5-8]. To find the dominantly polymorphisms of some gene which can predict the clinical outcomes of cancer are essential for patients with NSCLC.
As a cell cycle regulator, CCNB1 (also called cyclin B1), combined with CDK1 to be a complex, is a crucial factor for cell cycle, controlling the cell cycle G2/M phase [9]. The tyrosine 15 site phosphorylated of CCNB1-CDK1 can make cells entry into mitosis in normal status, leading to cell proliferation. The activation of CCNB1-CDK1 complex contributes to switch-like all or none behavior which decide to devote to mitosis. It is a positive feedback loop make sure that the activated CCNB1-CDK1 complex is perpetual. CCNB1 was reported with overexpressed in many cancers [10-12] (such as lung cancer, breast cancer, gastric and so on), and associated with chemotherapy resistance germ cell tumors [13]. Research has been reported that CCNB1 polymorphisms were critical for disease, not only for cancers. For example, Silvestre-Roig et al. found that the genetic variants of CCNB1 rs350099, rs350104, and rs164390 increased the risk of coronary in-stent restenosis [14]. We can see that CCNB1 was played an important role in diseases' formation. CCNB1 was also treated as a tumor antigen which holds promise as a putative cancer vaccine [15]. A part of functions and features of CCNB1 were listed in Table S1. We aim to investigate the relationship between CCNB1 tagSNPs/haplotypes and efficiency of platinum-based chemotherapy in advanced lung cancer, we enrolled 972 advanced NSCLC patients treated with platinum-based double regimen in our study and sequenced their tagSNPs (rs352626, rs2069429, rs2069433, and rs350104), which choose from the hapmap database based on criteria: 1) minor allele frequency (MAF) cutoff equals 0.05 and 2) correlation coefficient (r2) threshold equals 0.8 (details see Material and Methods), and then analyzed their association using log-rank test, univariate and multivariate cox's regression analysis. In our study, we found that CCNB1 polymorphisms contribute to NSCLC patients' clinical benefits.
Material and Methods
Ethical approval
The procedures involving human beings performed in our study were complying with ethical standards of research committee in Shanghai Pulmonary Hospital, Shanghai Changhai Hospital, Shanghai Chest Hospital, and Shanghai Zhongshan Hospital. It is in accordance with 1964 Helsinki declaration and its later amendments ethical standards. We informed all the advanced NSCLC patients in this study and got their written consent form. No animals were used for research in our study by any of authors. All experiments involving in our study were performed under approved guidelines and regulations.
Study cohorts
Our study recruited 972 patients with advanced (stage III or IV) NSCLC who were treated by first-line platinum-based double chemotherapy regimen from March 2005 to February 2010. All the patients received surgery, radiotherapy were excluded when enrolled. Whole patients' Eastern Cooperative Oncology Group performance status (ECOG PS) were in 0-2 points. The chemotherapy toxicities were assessed with hematologic, cardiovascular, hepatic and gastrointestinal. The association of CCNB1 tagSNPs with clinical outcomes in advanced NSCLC patients treated by platinum-based chemotherapy was evaluated.
Treatment
972 advanced NSCLC patients in our study were received platinum-based chemotherapy regimen, including double-DNA damage agents (cisplatin or carboplatin combined gemcitabine), platinum-tubulin targeting agents (cisplatin or carboplatin combined navebline, paclitaxel or docetaxel), and other combination (cisplatin or carboplatin combined etoposide or bevacizumab). They accepted above regimen for two to six cycles. The dosage for cisplatin and carboplatin was 75 mg/m2 and AUC 5 respectively on day 1 every three weeks. 1250 mg/m2 on days 1 and 8 every three weeks was for gemcitabine. The usages for tubulin-targeting agents, navelbine, paclitaxel and docetaxel were 25 mg/m2 on days 1 and 8 every three weeks, 175 mg/m2 on day 1 every three weeks, 75 mg/m2 on day 1 every three weeks respectively.
Clinical outcomes
Clinical outcomes after platinum-based chemotherapy with advanced NSCLC patients contained overall survival (OS), progression-free survival (PFS), disease control rate (DCR), and objective response rate (ORR) were evaluated by Response Evaluation Criteria in Solid Tumors. OS was predominantly for evaluating the efficacy of chemotherapy, started from the first day of receiving chemotherapy to death or to the final follow-up in our study. PFS reflecting the short-term response of chemotherapy, was defined from the day accepted chemotherapy to disease progression or the day of death (whichever occurred first) or the last follow-up for progression-free. DCR was consist with complete response (CR), partial responses (PR), and stable disease (SD). ORR was comprised of CR and PR.
Toxicity
Chemotherapy toxicities were evaluated twice weekly based on the National Cancer Institute (NCI) Common Terminology Criteria for Adverse Events (CTCAE) version 3.0. We collected the worst toxic records at the beginning two cycles of therapy for analysis. Toxicities in our study dominantly assessed hematologic (neutropenia, thrombocytopenia, and anemia), cardiovascular, hepatic and gastrointestinal (vomiting, nausea). Grades 3 or 4 was defined to severe toxic effects; grade 5 meant death. However, no grade 5 was observed in this study.
CCNB1 tagSNPs and haplotypes
Four tagSNPs of CCNB1, (rs352626, rs2069429, rs2069433 and 350104) were selected from CHB data of the phase 2 HapMap SNP database (http://www.hapmap.org/) for study. Then we extracted the patients' DNA from their peripheral blood using QIAamp DNA Maxi Kit (Qiagen GmbH, Hilden, Germany). All the experiments were according to the manufacturer's guidelines for genotyping purpose. We genotyped the patients' tagSNPs with iSelect HD Bead-Chip (Illumina, San Diego, CA, USA), and the quality-control criteria are following: 1) Minor allele frequency (MAF) > 0.05; 2) Genotyping call rate > 0.95, and 3) GenCall score > 0.2 (Figure S1). The concordance of replicates was over 99.9%. The personal haplotype frequencies of CCNB1 four tagSNPs were calculated by PHASE 2.0 Program (version 2.0.2) based on the Bayesian algorithm (Table S2).
Statistical analysis
We did statistical analysis with the SPSS version 20.0 software (SPSS INc. Chicago, IL, USA). The Kaplan-Meier curves and univariate Cox regression analysis were used for illustrating relationship between patients' OS/PFS and four tagSNPs of CCNB1. The independent significant factors were adjusted by multivariate Cox proportional hazard model, including clinical factors and CCNB1 tagSNPs related to OS/PFS of advanced NSCLC patients. For studying the relationship between the significant tagSNPs of CCNB1 and DCR or CCR, we used X2 test. Correlation between CCNB1 tagSNPs/haplotypes and severe toxicities was assessed by univariate logistic analysis. All statistical analysis considered the differences of p < 0.05 as statistically significant and defined two sides.
Results
Study Patients and Patients' clinical characteristics
The study patients were recruited from Shanghai Changhai Hospital, Shanghai Zhongshan Hospital, Shanghai Chest Hospital and Shanghai pulmonary Hospital. Our study included 972 patients, which consisted with 690 male patients and 282 female patients. The patients' clinical characteristics were involving age, sex, smoking status, clinical stage, histology of tumor, ECOG PS, and chemotherapy regimen. The detail of the patients' clinical characteristics and OS/PFS were listed in the Table 1. All patients were diagnosed with advanced NSCLC, which clinical stage was III or IV. The patients' median age was 57.8 years old. The youngest patient was 26 years old in this study, and patients with 82 years were the oldest. For tumor histology, we classified it with three types, adenocarcinoma, squamous carcinoma, and others (included adenosquamous carcinoma, mixed-cell, neuroendocrine, or undifferentiated carcinoma). The patients with adenocarcinoma type were most than the other types, account for 63.4 percent in 972 patients. Every patient in our study received platinum-based chemotherapy regimen. About 70.9 percent of the total patients, accepted the platinum-tubulin-targeting regimen chemotherapy. Patients who received the double DNA damage agent chemotherapy accounted for 24.3 percent. The rest patients used other combination regimen. Then we analyzed the association between clinical characteristics and patients' OS/PFS using log-rank test (Table 1).
We found that age, sex, smoking status, and tumor histology of the clinical characteristics were associated with patients' OS. For patients' age, the younger patients (<60 years) had longer OS than older one (>=60 years) (X2=8.95, p=0.003). Female patients may had a beneficial survival time when compared with the male patients (X2=8.87, p=0.003). Patients who smoke a lot had shorter OS than non-smoking patients (X2=6.30, p=0.012). Patients with lung adenocarcinoma can be survive longer than other histology's patients (X2=6.40, p=0.04). The other clinical characteristics, such as clinical stage, ECOG PS, and chemotherapy regimen, were not significantly related to the patients' OS. Only one clinical characteristic, ECOG PS, was significantly associated with PFS (p=0.001). No differences were observed when analyzed relationship between other clinical factors and PFS (Table 1).
CCNB1 rs2069429 and rs2069433 were associated with OS
We have chosen four tagSNPs (rs352626, rs2069429, rs2069433, and rs350104) of CCNB1 from the HapMap database, which followed some criteria (see Material and Methods). The genotype frequencies and MAF (minor allele frequency) of CCNB1 four tagSNPs were calculated among the current data or Han Chinese in Beijing, China (CHB), Utah residents with Northern and Western European ancestry from the CEPH collection (CEU), and Yoruban in Ibadan, Nigeria (YRI). The data of CHB, CEU, and YRI population were collected from the NCBI website (https://www.ncbi.nlm.nih.gov/). The results were shown in table 2. The MAF of CCNB1 four tagSNPs in our study were close to the MAF of CHB, CEU population from HapMap data (data not show). Our MAF of CCNB1 can be represented for the CHB and CEU population in some extent, not for the YRI population.
Then the association between CCNB1 tagSNPs and OS/PFS was analyzed by log-rank test, univariate and multivariate COX' regression analysis. The four tagSNPs of CCNB1 were classified to three types, genotype, and dominant/recessive model. We find that rs2069429 genotype and rs2069433 dominant (G) model were significantly associated with OS through univariate analysis. The patients with GG genotype (mOS=19.5, 95%CI=17.9-21.2) of rs2069429 had longer OS than patients with AG genotype (mOS=17.9, 95%CI=15.5-20.3) of rs2069429 (AG vs. GG, HR=1.2, 95%CI=1.02-1.41, p=0.025) (Figure 1). Another tagSNP, rs2069433, was associated with patients' OS too. Patients with AA genotype of rs2069433 had longer months (3.2 month) than non-AA genotype's patients (HR=0.75, 95%CI=0.60-0.94) (Figure 1, Table 3). However, the other tagSNPs, rs352626, rs350104 were not correlated with OS of NSCLC patients through univariate analysis.
The clinical characteristics of 972 advanced NSCLC patients treated with platinum-based chemotherapy.
| Variables | N =972 | OS | PFS | |||||
|---|---|---|---|---|---|---|---|---|
| mOS (95%CI) | X2 | Pl-g | mPFS(95%CI) | X2 | Pl-g | |||
| Median age, y (range) | 57.8(26-82) | |||||||
| <60 | 502(51.8%) | 21.3(19.3-23.4) | 8.95 | 3E-3* | 7.7(6.2-9.2) | 0.249 | 0.618 | |
| ≥60 | 467(48.2%) | 17.2(15.3-19.0) | 9.5(8.0-11.0) | |||||
| Censored | 3 | |||||||
| Sex | ||||||||
| Male | 690(71.0%) | 18.1(16.5-19.6) | 8.87 | 3E-3* | 9.4(8.0-10.8) | 1.322 | 0.250 | |
| Female | 282(29.0%) | 22.5(19.0-26.0) | 7.7(5.8-9.5) | |||||
| Smoking status | ||||||||
| Nonsmoker | 405(41.8%) | 21.2(19.0-23.4) | 6.30 | 0.012* | 7.8(6.3-9.3) | 0.614 | 0.433 | |
| Ever Smoker | 563 (58.2%) | 17.9(16.1-19.7) | 9.5(7.9-11.1) | |||||
| Censored | 4 | |||||||
| Clinical stage | ||||||||
| IIIa | 76 (7.9%) | 23.0(14.6-31.) | 4.40 | 0.111 | 12.6(6.9-18.3) | 4.371 | 0.112 | |
| IIIb | 283(29.3%) | 19.1(17.1-21.1) | 9.8(7.2-12.4) | |||||
| IV | 608(62.8%) | 19.1(16.9-21.2) | 8.0(6.8-9.2) | |||||
| Censored | 1 | |||||||
| ECOG PS | ||||||||
| 0-1 | 879(91.7%) | 19.4(18.0-20.8) | 3.078 | 0.079 | 9.3(8.1-10.6) | 10.775 | 0.001* | |
| 2 | 80(8.3%) | 17.9(9.6-26.2) | 5.4(3.4-7.5) | |||||
| Censored | 3 | |||||||
| Tumor histology | 0.230 | 0.891 | ||||||
| adenocarcinoma | 612(63.4%) | 20.2(18.4-22.1) | 6.40 | 0.040* | 9.1(7.7-10.6) | |||
| Squamous carcinoma | 213(22.0%) | 16.6(13.1-20.2) | 9.5(6.8-12.1) | |||||
| othersa | 141(14.6%) | 18.3(14.8-21.8) | 8.0(6.2-9.8) | |||||
| Censored | 6 | |||||||
| Doublet regimen | 5.119 | 0.077 | ||||||
| DNA damaging agents | 236(24.3%) | 19.8(17.2-22.3) | 1.95 | 0.378 | 9.3(7.6-11.0) | |||
| Platinum-tubulin-targeting drugsb | 689(70.9%) | 19.1(17.3-20.9) | 9.1(7.6-10.7) | |||||
| Other combinationc | 47(4.8%) | 20.2(15.6-24.8) | 5.1(2.7-7.6) | |||||
a Others NSCLC included adenosquamous carcinoma, mixed-cell, neuroendocrine carcinoma, or undifferentiated carcinoma.
b Tubulin-targeting drugs includes paclitaxel, docetaxel or navelbine.
cOther combination included etoposide or bevacizumab.
*p<0.05. Abbreviations: mOS: median overall survival, CI: confidence interval
Genotype frequencies and MAF of CCNB1 tag SNPs in current data, or in CHB, CEU and YRI from HapMap SNP database.
| Tag SNPs | Location | Allele frequencies | ||||
|---|---|---|---|---|---|---|
| Current Data | CHB | CEU | YRI | p | ||
| Rs352626 | 5'-near gene | |||||
| MAF | 0.470(A) | 0.463(A) | 0.447 (A) | 0.456(G) | 0.016* | |
| A/A | 207(21.3%) | 16(19.5%) | 44(19.5%) | 68(30.1%) | ||
| A/G | 499(51.3%) | 44(53.7%) | 114(50.4%) | 110(48.7%) | ||
| G/G | 266(27.4%) | 22(26.8%) | 68(30.1%) | 48(21.2%) | ||
| Rs350104 | intron | |||||
| MAF | 0.313(G) | 0.367(G) | 0.392(G) | 0.158(G) | <0.0001 | |
| A/A | 462(47.5%) | 32(35.6%) | 42(35.0%) | 82(68.3%) | ||
| A/G | 412(42.4%) | 50(55.6%) | 62(51.7%) | 38(31.7%) | ||
| G/G | 98(10.1%) | 8(8.89%) | 16(13.3%) | 0 | ||
| Rs2069429 | 5'-near gene | |||||
| MAF | 0.180 (A) | 0. 233(A) | NA | NA | <0.0001 | |
| A/A | 31(3.2%) | 0(0%) | 0 | 0 | ||
| A/G | 287(29.5%) | 42(46.7%) | 0 | 0 | ||
| G/G | 654(67.3%) | 48(53.3%) | 120(100%) | 120(100%) | ||
| Rs2069433 | intron | |||||
| MAF | 0.055(G) | 0.056(G) | 0.083(G) | 0.192(G) | <0.0001 | |
| A/A | 869(89.4%) | 80(88.9%) | 100(83.3%) | 76(63.3%) | ||
| A/G | 100(10.3%) | 10(11.1%) | 20(16.7%) | 42(35.0%) | ||
| G/G | 3(0.3%) | 0 | 0 | 2(1.7%) | ||
*p<0.05. Abbreviations: UTR, untranslated region; MAF, Minor allele frequency; CHB, Han Chinese in Beijing, China; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; YRI, Yoruban in Ibadan, Nigeria
Kaplan-Meier curve of association between rs2069429/rs2069433 of CCNB1and OS in advanced NSCLC patients. A: Kaplan-Meier curve of rs2069429 of CCNB1to advanced NSCLC patients' OS B: Kaplan-Meier curve of rs2069433 of CCNB1to advanced NSCLC patients' OS
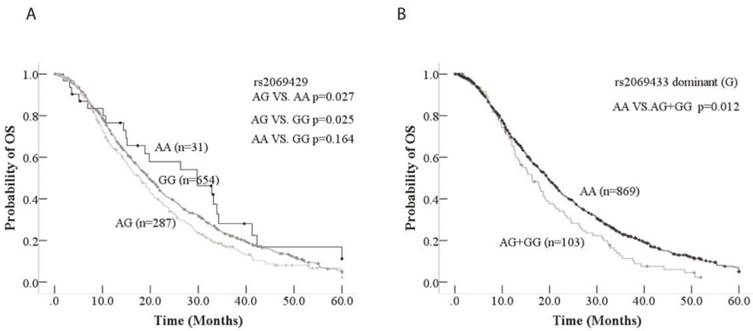
The clinical factors, such as age, sex, TNM, ECOG PS, smoking, histology, treatment, and four tagSNPs were included into multivariate cox's regression analysis. For clinical characteristics, age and sex were significantly associated with advanced NSCLC patients' OS according to multivariate cox's regression analysis (Table 4). Younger patients (<60 years old) obviously had longer OS than older (>=60) (HR=1.23, 95%CI=1.06-1.43, p=0.006); Female had longer OS than male (HR=0.81, 95%CI=0.69-0.96, p=0.015). Among four tagSNPs, rs2069429 dominant (A) and rs2069433 dominant (G) were significantly observed association with OS based on multivariate cox's regression analysis. Patients with GG genotype of rs2069429 had longer OS than non-GG patients (HR=0.81, 95%CI=0.68-0.95, p=0.009); and patients with AA genotype of rs2069433 had longer OS than non-AA patients (HR=0.78, 95%CI=0.61-0.98, p=0.036) (Table 4). Above all, in term of CCNB1 tagSNPs, rs2069429 dominant (A) and rs2069433 dominant (G) were independent predictive factors of advanced NSCLC patients' OS in our study.
When PFS was used for endpoint, only ECOG PS was significantly associated with patients' PFS according to multivariate cox's regression analysis (p=0.003) (Table S4). However, no significantly tagSNPs were observed through univariate and multivariate analysis (Table S3, Table S4).
Univariate analysis of four tagSNPs of CCNB1 with OS in 972 advanced NSCLC patients.
| SNP ID | genotype | N(%) | mOS(95%CI) | X2 | PL-G | HR(95%CI) | PCOX |
|---|---|---|---|---|---|---|---|
| Rs352626 | 0.952 | 0.621 | 0.622 | ||||
| Genotype | AG | 499(51.3%) | 19.1(17.1-21.0) | R | |||
| GG | 266(27.4%) | 19.8(17.1-22.6) | 0.92(0.77-1.09) | 0.338 | |||
| AA | 207(21.3%) | 19.3(16.0-22.2) | 0.99(0.82-1.19) | 0.892 | |||
| Dominant(A) | AA +AG | 706(72.6%) | 19.1(17.3-20.8) | ||||
| GG | 266(27.4%) | 19.8(17.1-22.6) | 0.933 | 0.334 | 0.92(0.78-1.09) | 0.335 | |
| Recessive(A) | AA | 207(21.3%) | 19.1(15.9-22.2) | ||||
| GG+AG | 765(78.7%) | 19.3(17.7-20.8) | 0.036 | 0.850 | 0.98(0.83-1.17) | 0.850 | |
| Rs2069429 | 7.92 | 0.019* | 0.020* | ||||
| Genotype | GG | 654(67.3%) | 19.5(17.9-21.2) | R | |||
| AG | 287(29.5%) | 17.9(15.5-20.3) | 1.20(1.02-1.41) | 0.025* | |||
| AA | 31(3.2%) | 29.8(14.3-45.3) | 0.73(0.48-1.13) | 0.157 | |||
| Dominant(A) | AA+AG | 318(32.7%) | 18.2(16.0-20.5) | ||||
| GG | 654(67.3%) | 19.5(1.9-21.2) | 2.58 | 0.108 | 0.88(0.76-1.03) | 0.109 | |
| Recessive(A) | AA | 31(3.2%) | 29.8(14.3-45.3) | ||||
| GG+AG | 914(96.8%) | 19.1(17.7-20.5) | 2.81 | 0.094 | 1.44(0.94-2.20) | 0.096 | |
| Rs2069433 | 7.52 | 0.023* | 0.025* | ||||
| Genotype | A/A | 869(89.4%) | 19.6(18.2-21.1) | R | - | ||
| A/G | 100(10.3%) | 16.4(13.0-19.8) | 1.32(1.05-1.66) | 0.019 | |||
| G/G | 3(0.3%) | 12.9(10.4-15.4) | 2.3(0.74-7.17) | 0.151 | |||
| Recessive(G) | AA+AG | 969(99.7%) | 19.3(17.9-20.6) | 2.02 | 0.155 | 0.45(0.14-1.40) | 0.166 |
| GG | 3(0.3%) | 12.9(10.4-15.4) | |||||
| Dominant(G) | AA | 869(89.4%) | 19.6(18.2-21.1) | 6.33 | 0.012 | 0.75(0.60-0.94) | 0.012* |
| GG+AG | 103(11.6%) | 16.4(12.9-20.0) | |||||
| Rs350104 | 1.14 | 0.566 | 0.567 | ||||
| genotype | A/A | 462(47.5%) | 18.8(17.0-20.6) | R | - | ||
| A/G | 412(42.4%) | 19.7(17.6-21.7) | 0.93(0.80-1.08) | 0.343 | |||
| G/G | 98(10.1%) | 20.3(16.2-24.5) | 0.91(0.71-1.17) | 0.461 | |||
| Recessive(G) | AA +AG | 874(90.9%) | 19.1(17.7-20.5) | 0.23 | 0.631 | 1.06(0.84-1.35) | 0.632 |
| GG | 98(10.1%) | 20.3(16.2-24.5) | |||||
| Dominant(G) | AA | 462(47.5%) | 18.8(17.0-20.6) | 1.12 | 0.291 | 1.08(0.94-1.25) | 0.292 |
| GG+AG | 510(53.5%) | 19.8(18.0-21.7) | |||||
*p<0.05. Abbreviations: mOS: median overall survival, HR: hazard ratio, CI: confidence interval, NA: Not available, R: Reference
CCNB1 haplotype GAAA was related with OS in patients with clinical stage IV
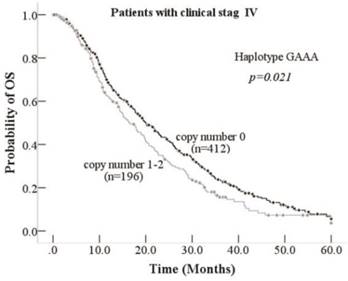
We analyzed the relationship between tagSNPs of CCNB1 and the OS/PFS, and we found rs2069429 dominant (A) and rs2069433 dominant (G) were valuable tagSNPs for advanced NSCLC patients. We explored the haplotype of this four tagSNPs using PHASE 2.0 software and analyzed the association between haplotype of CCNB1 and OS/PFS. Eight haplotypes based on the rs352626, rs350104, rs2069429, rs2069423 were calculated by PHASE 2.0 program. We analyzed three haplotypes which the frequencies were more than 10%. Haplotype GAAA (haplotype 1), haplotype GGGA (haplotype 4), haplotype AAGA (haplotype 7) frequencies were 18%, 31.3%, 41.5% respectively (Table S3). Then we analyzed this three haplotypes and the OS/PFS using univariate and multivariate analysis. No significantly association was observed between three haplotypes and OS/PFS (Table S5). However, stratified analysis according to TNM clinical stag shows that haplotype GAAA was a predictable factor of patients' OS. Patients who was in clinical stage IV with zero copy number GAAA had 3.6 months longer OS than one or two copy number GAAA's patients (p=0.021,Table 5, Figure 2). And stage IV patients with haplotype GGGA one or two copy number was slightly significant than zero copy number's patients (p=0.051, Table 5). This haplotype GAAA may connected with lung cancer metastatic, resulting to the significantly differences in patients with clinical stag IV rather than stage III. However, we did not find any haplotype can be a predictable factor for patients' PFS under different clinical stage (Table S6).
CCNB1 tagSNPs/haplotypes association with toxicities after chemotherapy
Excepting survival time, there were other important factors that may influence patients' treatment efficiency, which was the side effects of treatment. And then we analyzed the links between four tagSNPs, three haplotypes of CCNB1 and toxicities after platinum-based chemotherapy using univariate logistic regression analysis. The results displayed in Table 6, rs2069433 recessive (G) model and rs350104 recessive (G) model were related with the gastrointestinal toxicity after platinum-based chemotherapy. The patients with GG genotype of rs2069433 (p=0.013) and/or non-GG genotype of rs350104 (p=0.042) may have a severe gastrointestinal toxicity after chemotherapy. If the patients were with GG genotype of rs2069433 or non-GG genotype of rs350104, the clinicians can reduce the dosage of platinum and other chemotherapy agents for the patients to avoid sever gastrointestinal toxicities. No haplotypes of CCNB1 four tagSNPs were related with the toxicities in our study by analysis (data not shown).
Discussion
Many studies have been reported that genes' SNP were related with the efficiency of platinum-based chemotherapy in lung cancer, including our previous studies about SNP [16]. Nowadays, many targeted drugs such as EGFR tyrosine kinase inhibitors (TKIs [erlotinib, gefitinib, and afatinib]) is widely used in NSCLC patients, however, the number of patients with EGFR driver mutation is limited, only 17% , and only 7% of lung adenocarcinoma patients having ALK translocation (also driver mutation gene)[17]. For the rest large number of advanced NSCLC patients, chemotherapy remains the first-line treatment, especially platinum-based double regimen. Much research has reported that genes are related to efficacy of chemotherapy drugs, such as WEE1, and ERCC1 for platinum and gemcitabine, respectively, and TUBB3 for paclitaxel [16, 18, 19]. CCNB1 as a critical M-phase promoting factor in cell cycle, may be a next putative gene to predict efficiency of platinum-based chemotherapy for advanced NSCLC patients according to our study. This is the first time that we investigate the association between CCNB1 four tagSNPs/haplotypes (rs352626, rs350104, rs2069429, rs2069433) and OS/PFS in advanced NSCLC patients treated with platinum-based chemotherapy. The results showed that rs2069429 dominant (A) and rs2069433 dominant (G) of CCNB1 were significantly associated with OS of NSCLC patients who treated with platinum-based chemotherapy. The GG genotype of rs2069429 and AA genotype of rs2069433 were positive factors for advanced NSCLC patients, following with longer OS. Our finding is consistent with Li et.al. 's research, they have been reported rs2069429 recessive model of CCNB1 was associated with breast cancer susceptibility, progression and survival, with increasing breast cancer risk [20]. Ma and colleagues studied the relation between 21 cell cycle genes (including CCNB1) and survival time of 586 Chinese NSCLC patients, they also find CCNB1 rs2069429 was associated with the survival time of patients in univariate analysis [21]. We explored the rs2069429 function in online SNP function predictive software (https://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). Rs2069429 is located in the 5'-near gene, it belongs to transcription factor binding site (TFBS), which means an important role in CCNB1 gene expression. Rs2069429 may can control the cell-cycle progression by regulating the expression of CCNB1, and then resulting to affect the efficiency of chemotherapy, or even survival time of NSCLC patients. However, the function of CCNB1 intron rs2069433 is not yet clear. Rs2069433 may influence OS of advanced NSCLC patients through linkage with rs2069429, which can mediate CCNB1 expression indirectly.
Kaplan-Meier curve of association between Haplotype GAAA and OS inpatients with clinical stage IV
Multivariate Cox's regression analysis of clinical factors and CCNB1 tagSNPs for overall survival in the 972 advanced NSCLC patients treated with platinum-based chemotherapy.
| Variables | HR (95% CI) | p |
|---|---|---|
| Age (>=60 vs.<60) | 1.23(1.06-1.43) | 0.006* |
| Sex (male vs. female) | 0.81(0.69-0.96) | 0.015* |
| TNM(IV vs. IIIa vs. IIIb) | 1.11(0.99-1.24) | 0.076 |
| ECOG PS(2 vs.1) | 1.21(0.92-1.57) | 0.171 |
| Smoking (ever vs. never) | 1.04(0.84-1.29) | 0.741 |
| Histology | 0.181 | |
| adenocarcinoma | R | |
| squamous | 1.19(0.97-1.46) | 0.096 |
| adenosquamous | 1.29(0.74-2.24) | 0.378 |
| others | 1.22(0.97-1.54) | 0.087 |
| Treatment | 0.568 | |
| DNA-damaging agents a | R | |
| Platinum-tubulin-targeting drugs b | 0.96(0.80-1.15) | 0.632 |
| Other combination c | 0.83(0.57-1.20) | 0.321 |
| Rs2069429 dominant (A) | 0.81(0.68-0.95) | 0.009* |
| Rs2069429 recessive (A) | 1.54(0.98-2.41) | 0.063 |
| Rs2069433 dominant (G) | 0.78(0.61-0.98) | 0.036* |
| Rs2069433 recessive (G) | 0.69(0.16-2.89) | 0.607 |
| Rs350104 dominant (G) | 0.96(0.79-1.17) | 0.702 |
| Rs350104 recessive (G) | 0.94(0.73-1.21) | 0.641 |
| Rs352626 dominant (A) | 0.92(0.72-1.81) | 0.517 |
| Rs352626 recessive (A) | 0.88(0.61-1.27) | 0.498 |
a Others NSCLC included adenosquamous carcinoma, mixed-cell, neuroendocrine carcinoma, or undifferentiated carcinoma.
b Tubulin-targeting drugs includes paclitaxel, docetaxel or navelbine.
c Other combination included etoposide or bevacizumab.
*p<0.05. Abbreviations: HR: hazard ratio, CI: confidence interval;
Stratified analysis of association between CCNB1 haplotypes with OS in patients with clinical stage III and IV
| Haplotype | Patients with clinical stage III | Patients with clinical stage IV | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N (%) | mOS(95% CI), mo | X2 | P-value | N (%) | mOS(95% CI), mo | X2 | P-value | ||||
| GAAA | |||||||||||
| Copy number 0 | 238(66.3%) | 19.3(17.0-21.5) | - | 0.814 | 412(67.8%) | 20.0(17.4-22.6) | - | 0.021* | |||
| Copy number 1-2 | 121 (33.7%) | 19.8 (16.2-23.4) | 0.055 | 196(32.2%) | 16.4(13.2-19.6) | 5.304 | |||||
| GGGA | |||||||||||
| Copy number 0 | 167 (46.5%) | 19.8(17.2-22.4) | - | 0.424 | 292(48.0%) | 18.0(15.8-20.1) | - | 0.051 | |||
| Copy number 1-2 | 192(53.5%) | 19.3(17.5-21.1) | 0.640 | 316(52.0%) | 20.2(17.3-21.1) | 3.814 | |||||
| AAGA | |||||||||||
| Copy number 0 | 121(33.7%) | 20.4(17.5-23.3) | - | 0.296 | 201(33.1%) | 17.7(15.0-20.4) | - | 0.770 | |||
| Copy number 1-2 | 238(66.3%) | 19.1(16.1-22.0) | 1.093 | 407(66.9%) | 19.5(17.4-21.6) | 0.085 | |||||
*p<0.05. Abbreviations: mOS: median overall survival; mo: months, HR: hazard ratio, CI: confidence interval
Association between CCNB 1 tagSNPs/haplotypes and toxicities in the 972 advanced NSCLC
| Toxicities | SNP/haplotype | grade3/4(%) | OR(95%CI) | p |
|---|---|---|---|---|
| Gastrointestinal | Rs2069433 recessive (G) | |||
| GG | 2/3(66.7%) | - | ||
| AA+AG | 74/933(7.9%) | 0.05(0.00-0.53) | 0.013* | |
| Rs350104 recessive (G) | ||||
| GG | 2/95(2.1%) | - | ||
| AA+AG | 74/841(8.8%) | 4.37(1.06-18.2) | 0.042* |
*p<0.05. Abbreviations: OR: odds ratio, CI: confidence interval
The haplotypes of these four tagSNPs (rs352626, rs350104, rs2069429, rs2069423) were analyzed by PHASE 2.0 software program. Three haplotypes, GAAA, GGGA, and AAGA, which frequencies are over 10%, were chosen for study. No one was significant with OS in whole patients after univariate and multivariate analysis. However, when we stratified with TNM clinical stag, haplotype GAAA was a valuable factor for clinical stag IV patients. For clinical stag IV patients, harboring zero copy number GAAA has 3.6 months longer OS than patients with one or two copy number. This haplotype GAAA may related with NSCLC's metastatic, leading to the significantly differences in patients with clinical stag IV rather than stage III. We can find scientists have been reported that CCNB1 expression is associated with metastasis and tumor differentiation degree of hypopharyngeal squamous cell carcinoma [22]. We can infer that the haplotype of GAAA may be related with CCNB1 expression, because of rs2069429's TFBS function. No significantly association between CCNB1 tagSNPs/haplotypes and patients' PFS. We can see the similar results in treatment studies when drugs can affect immune function. A possible reason why CCNB1 polymorphisms influence OS but not PFS may be that the treatment drugs' role of pharmacogenetics can altered the metabolism, even the immune function [23, 24]. CCNB1 is transiently expressed in the cell cycle, however, CCNB1 was identified to be a new tumor antigen in year of 1997 and then found CCNB1 was overexpressed in many cancers, such as lung, breast, and colorectal cancer and involved in the CD8+ T cell immune progression [25, 26, 27, 28]. The correlation between CCNB1 and the immune system may be the reason why polymorphisms of CCNB1 is associated with OS, but not PFS in our study.
Due to resistance and toxicities of drugs, the 5-year overall survival time of lung cancer patients is 16.1% in China over last 40 years [1]. Toxicities of chemotherapy drugs effect patients' quality life, even survival time. Many researches have reported that polymorphisms can be a predictive factor for patients' toxicities. Qian reported that polymorphisms of Caspase 8 and Caspase 10 was related with advanced NSCLC patients' hematologic toxicity in treatment of platinum-based chemotherapy [29]. The association between CCNB1 tagSNPs/haplotypes and toxicities was assessed in our study. We found that rs2069433 recessive (G) model and rs350104 recessive (G) model were putative factor for gastrointestinal toxicity. We may infer that patients harboring GG genotype of rs2069433 and/or non-GG genotype of rs350104 have a server gastrointestinal toxicity than others. The clinicians can regulate the dosage of chemotherapy drugs in clinical treatment to avoid sever toxicities, and then increase patients' quality of life, even survival time.
In summary, CCNB1 is a crucial cell-cycle regulator and G2/M phase promotor in cell's proliferation and differentiation. Only one article investigated the polymorphisms of CCNB1 in NSCLC before us. They found rs2069429 of CCNB1 was associated with the survival time of patients in univariate analysis, however, no significant CCNB1 SNPs associated with survival time in multivariate analysis, which may can explain with limited patients [21]. We enrolled 972 patients in our study and first come up with that rs2069429 and rs2069433 of CCNB1 were associated with the OS of advanced NSCLC patients. Patients holding GG genotype of rs2069429 had longer OS than non-GG patients (p=0.009); and patients with AA genotype of rs2069433 had longer OS than non-AA patients (p=0.036). We also accessed the haplotype of CCNB1 four tagSNPs and clinical outcomes in 972 advanced NSCLC patients. The haplotype GAAA of CCNB1 was a potential factor in subgroup patients with clinical stage IV in our study. The contribution of CCNB1 SNPs in toxicities after platinum-based chemotherapy were evaluated as well. Rs2069433 and rs350104 were correlated with gastrointestinal toxicity after receiving platinum-based chemotherapy. If validated, rs2069429 and rs2069433 of CCNB1 can be a putative biomarker for NSCLC patients treated with platinum-based chemotherapy. Haplotype GAAA of CCNB1 was predictive for clinical IV stage subgroup patients largely. The polymorphisms of CCNB1 were related with patients' toxicities after treatment. All in all, CCNB1 polymorphisms may contribute to the clinical outcomes of platinum-based chemotherapy in advanced NSCLC. We still need further prospective studies of CCNB1 polymorphisms.
Supplementary Material
Supplementary figures and tables.
Abbreviations
CCNB1: cyclin B1
OS: overall survival
PFS: progression-free survival
NSCLC: non-small-cell lung cancer
ALK: anaplastic lymphoma kinase
EGFR: epidermal growth factor receptor
NCCN: National comprehensive cancer network
SNP: Single nucleotide polymorphisms
CHB: Han Chinese in Beijing
CEU: Utah residents with Northern and Western European ancestry from the CEPH collection
YRI: Yoruban in Ibadan
TKIs: tyrosine kinase inhibitors
ECOG PS: Eastern Cooperative Oncology Group performance status
RECIST: Response Evaluation Criteria in Solid Tumors
DCR: disease control rate
CR: complete response
PR: partial response
SD: stable disease
NCI: National Cancer Institute
CTCAE: Common Terminology Criteria for Adverse Events
Acknowledgements
This work was funded by National Natural Science Foundation of China (No. 81572269, No. 81672279), Science and Technology Commission of Shanghai Municipality (No. 14411966400, No. 134119a3400, No. 15ZR1434500), and Shanghai Health Bureau Foundation (No. 201440397).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Zheng R, Zeng H, Zuo T, Zhang S, Qiao Y, Zhou Q. et al. Lung cancer incidence and mortality in China, 2011. Thoracic cancer. 2016;7:94-9
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
3. Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR. et al. Non-Small Cell Lung Cancer, Version 6.2015. Journal of the National Comprehensive Cancer Network: JNCCN. 2015;13:515-24
4. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA. et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061-73
5. Naykoo NA, Dil A, Rasool R, Shah S, Ahangar AG, Bhat IA. et al. Single nucleotide polymorphisms, haplotype association and tumour expression of the vascular endothelial growth factor (VEGF) gene with lung carcinoma. Gene. 2017;608:95-102
6. Singh A, Singh N, Behera D, Sharma S. Polymorphism in XRCC1 gene modulates survival and clinical outcomes of advanced North Indian lung cancer patients treated with platinum-based doublet chemotherapy. Medical oncology (Northwood, London, England). 2017;34:64
7. Zhou F, Wang Y, Liu H, Ready N, Han Y, Hung RJ. et al. Susceptibility loci of CNOT6 in the general mRNA degradation pathway and lung cancer risk-A re-analysis of eight GWASs. Molecular carcinogenesis. 2017;56:1227-38
8. George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH. et al. Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:1220-6
9. Nakayama Y, Yamaguchi N. Role of cyclin B1 levels in DNA damage and DNA damage-induced senescence. International review of cell and molecular biology. 2013;305:303-37
10. Yuan J, Kramer A, Matthess Y, Yan R, Spankuch B, Gatje R. et al. Stable gene silencing of cyclin B1 in tumor cells increases susceptibility to taxol and leads to growth arrest in vivo. Oncogene. 2006;25:1753-62
11. Kawamoto H, Koizumi H, Uchikoshi T. Expression of the G2-M checkpoint regulators cyclin B1 and cdc2 in nonmalignant and malignant human breast lesions: immunocytochemical and quantitative image analyses. Am J Pathol. 1997;150:15-23
12. Wang A, Yoshimi N, Ino N, Tanaka T, Mori H. Overexpression of cyclin B1 in human colorectal cancers. J Cancer Res Clin Oncol. 1997;123:124-7
13. De Giorgi U, Yuan J, Moroni M, Veronese S, Sartore-Bianchi A, Broggini M. et al. Germ cell tumors overexpress the candidate therapeutic target cyclin B1 independently of p53 function. The International journal of biological markers. 2015;30:e275-81
14. Silvestre-Roig C, Fernandez P, Mansego ML, van Tiel CM, Viana R, Anselmi CV. et al. Genetic variants in CCNB1 associated with differential gene transcription and risk of coronary in-stent restenosis. Circulation Cardiovascular genetics. 2014;7:59-70
15. Egloff AM, Weissfeld J, Land SR, Finn OJ. Evaluation of anticyclin B1 serum antibody as a diagnostic and prognostic biomarker for lung cancer. Annals of the New York Academy of Sciences. 2005;1062:29-40
16. Liu D, Wu C, Jiao Y, Hou L, Lu D, Zheng H. et al. WEE1 kinase polymorphism as a predictive biomarker for efficacy of platinum-gemcitabine doublet chemotherapy in advanced non-small cell lung cancer patients. Scientific reports. 2015;5:11114
17. Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba, II. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. Jama. 2014;311:1998-2006
18. Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M. et al. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8:2286-91
19. Zhang HL, Ruan L, Zheng LM, Whyte D, Tzeng CM, Zhou XW. Association between class III beta-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung cancer (Amsterdam, Netherlands). 2012;77:9-15
20. Li Y, Chen YL, Xie YT, Zheng LY, Han JY, Wang H. et al. Association study of germline variants in CCNB1 and CDK1 with breast cancer susceptibility, progression, and survival among Chinese Han women. PLoS One. 2013;8:e84489
21. Ma H, Chen J, Pan S, Dai J, Jin G, Hu Z. et al. Potentially functional polymorphisms in cell cycle genes and the survival of non-small cell lung cancer in a Chinese population. Lung cancer (Amsterdam, Netherlands). 2011;73:32-7
22. Li W, Dong Q, Li L, Zhang Z, Cai X, Pan X. Prognostic significance of claudin-1 and cyclin B1 protein expression in patients with hypopharyngeal squamous cell carcinoma. Oncology letters. 2016;11:2995-3002
23. Hattinger CM, Serra M. Role of pharmacogenetics of drug-metabolizing enzymes in treating osteosarcoma. Expert opinion on drug metabolism & toxicology. 2015;11:1449-63
24. Pandey JP, Namboodiri AM, Kistner-Griffin E. A genetic variant of FcgammaRIIIa is strongly associatedwith humoral immunity to cyclin B1 in African American patients with prostate cancer. Immunogenetics. 2013;65:91-6
25. Covini G, Chan EK, Nishioka M, Morshed SA, Reed SI, Tan EM. Immune response to cyclin B1 in hepatocellular carcinoma. Hepatology (Baltimore, Md). 1997;25:75-80
26. Kao H, Marto JA, Hoffmann TK, Shabanowitz J, Finkelstein SD, Whiteside TL. et al. Identification of cyclin B1 as a shared human epithelial tumor-associated antigen recognized by T cells. The Journal of experimental medicine. 2001;194:1313-23
27. Vella LA, Yu M, Fuhrmann SR, El-Amine M, Epperson DE, Finn OJ. Healthy individuals have T-cell and antibody responses to the tumor antigen cyclin B1 that when elicited in mice protect from cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14010-5
28. Egloff AM, Vella LA, Finn OJ. Cyclin B1 and other cyclins as tumor antigens in immunosurveillance and immunotherapy of cancer. Cancer Res. 2006;66:6-9
29. Qian J, Qu HQ, Yang L, Yin M, Wang Q, Gu S. et al. Association between CASP8 and CASP10 polymorphisms and toxicity outcomes with platinum-based chemotherapy in Chinese patients with non-small cell lung cancer. The oncologist. 2012;17:1551-61
Author contact
![]() Corresponding authors: Bo Su, Ph.D., Central Laboratory (subogroupcom); Ke Fei, M.D, Department of Thoracic Surgery (ffeikenet); Shanghai Pulmonary Hospital, Tongji University School of Medicine, No. 507, Zheng Min Road, Shanghai, 200433, P.R. China. Phone: +86-21-65115006 Fax: +86-21-65111298 E-mail: subogroupcom; ffeikenet
Corresponding authors: Bo Su, Ph.D., Central Laboratory (subogroupcom); Ke Fei, M.D, Department of Thoracic Surgery (ffeikenet); Shanghai Pulmonary Hospital, Tongji University School of Medicine, No. 507, Zheng Min Road, Shanghai, 200433, P.R. China. Phone: +86-21-65115006 Fax: +86-21-65111298 E-mail: subogroupcom; ffeikenet

 Global reach, higher impact
Global reach, higher impact