3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(18):3887-3896. doi:10.7150/jca.19112 This issue Cite
Research Paper
PFKFB3 promotes proliferation, migration and angiogenesis in nasopharyngeal carcinoma
1. Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Nantong University, Nantong
2. Department of Otorhinolaryngology Head and Neck Surgery, Nantong Tumor Hospital, Nantong
*Both authors contributed equally to this work.
Received 2017-1-8; Accepted 2017-7-19; Published 2017-10-23
Abstract
Nasopharyngeal carcinoma (NPC) is a squamous epithelial cancer, arising from the nasopharynx epithelium. It has high morbidity and mortality. PFKFB3 as a glycolytic activator has been implicated in the progression of multiple types of tumor. PFKFB3 can be contributed to the progression and metastasis of cancer. However, whether PFKFB3 is associated with the progression of NPC remains unknown. We postulated that PFKFB3 promotes proliferation, migration and angiogenesis in nasopharyngeal carcinoma. In this study, we found that PFKFB3 was significantly up-regulated in NPC tissues and cell lines compared with normal control. Our study proved that PFKFB3 can regulate the proliferation, metastasis and apoptosis of NPC. By the way, the NPC-derived exosomes come from and CNE2-derived exosomes are enriched in PFKFB3. The enrichment of PFKFB3 played a crucial functional role in promotes HUVECs proliferation, migration and angiogenesis. And tumor angiogenesis is closely related to the proliferation and metastasis of tumor. In conclusion, our findings demonstrate that PFKFB3 could act not only as a clinical biomarker for angiogenesis but also as a therapeutic target to overcome angiogenesis, enhancing the clinical benefits of angiogenesis therapy in NPC patients.
Keywords: PFKFB3, proliferation, metastasis, angiogenesis, nasopharyngeal carcinoma, exosome
Introduction
Nasopharyngeal carcinoma (NPC) is the most common malignancies of the head and neck[1, 2]. NPC, a squamous epithelial cancer, is arising from the nasopharynx epithelium[3, 4]. Within the boundaries of the nasopharynx, the tumour epicentre is frequently seen at the fossa of Rosenmüller, and the tumour invades adjacent anatomical spaces or organs from this[1]. Distinct differences exist between NPC and other epithelial tumours in the head and neck region. Greater than 95% of NPCs are undifferentiated carcinomas[5]. NPC has the highest preponderance for neck lymph node metastasis and distant metastases[6]. Local recurrence and distant metastasis (especially neck lymph node) are the leading causes of poor prognosis among NPC patients[7, 8]. However, the molecular mechanisms of metastasis in NPC are not very clear. Therefore, it is of great clinical value to further reveal the underlying molecular mechanisms of metastasis in NPC carcinogenesis.
Tumor angiogenesis is closely related to the proliferation and metastasis of tumor[9-11]. This could lead to a fast regrowth of the tumor with potentially enhanced metastasis as a likely reason of vessel leakiness and tumor angiogenesis[12]. But, how does tumor affect vascular endothelial cells (HUVECs)? We found that exosomes may play an important role in this processes.
Among the multiple factors underlying metastasis, the adaptation of primary tumor microenvironment by cancer to facilitate cancer cell dissemination and distant engraftment plays an important pro-metastatic role[13]. Exosome is closely associated with the metastasis of tumor micro-environment[14]. Tumor cells actively produce, release, and utilize exosomes to promote the proliferation, migration and angiogenesis of tumor[15]. These exosomes worked as information carriers, transporting genetic and molecular messages from tumor cells to other cells residing at close or distant sites[16]. Target cells absorb these exosome and alter functional attributes and phenotypic. In this article, we found that after taking this exosomes, vascular endothelial cells was reprogrammed into contributors to angiogenesis. However, it is still unclear that which molecular or genetic factors in exosomes play a role in angiogenesis. Therefore, it is of great clinical value to further reveal the molecular mechanisms of NPC carcinogenesis and develop effective therapeutic strategies.
PFKFB-3 (Enzymes 6-phosphofructo-2-kinase/ fructose- 2,6- bisphosphatase-3), a member of PFKFB [17], can encode for inducible 6-phosphofructo-2-kinase. It is a glycolysis-regulatory enzyme. Some studies have shown that PFKFB-3 play a significant role in the regulation of glycolysis in cancer cells as well as its proliferation and survival[18]. PFKFB3 increase the production of Fru-2,6- P2 and lactate, and this glycolytic enhancement results in cell cycle active, apoptosis decreased and addition of colony formation[19], promoted vessel sprouting[20, 21], and the growth of tumor[22, 23]. PFKFB3 has been reported to be up-regulated and exhibit tumor-promotive roles in pancreatic cancer gastric, breast, lung, and colon malignant tumors[21, 24, 25]. PFKFB3 can change in their metabolism that contribute to the proliferation and survival of cancer cells[18]. Despite the implications of PFKFB3 have been studied in many tumors, the role of PFKFB3 in NPC proliferation, metastasis and angiogenesis remains unexplored. Can PFKFB3 transferred by exosomes from NPC to vascular endothelial cells?
In this study, we recently identified a key role for the glycolytic activator PFKFB3 in NPC by regulating proliferation, migration and angiogenesis. Our research shows that overexpression of PFKFB-3 is observed in NPC. PFKFB3 suppression results in cell cycle arrest, apoptosis and migration. The exosomes from CNE2 cells can be transferred to vascular endothelial cells and increase angiogenesis. This process may be caused by PFKFB3 in exosomes. These results suggested that PFKFB3 can promotes proliferation, migration and angiogenesis in nasopharyngeal carcinoma.
Materials and methods
Specimens and cell lines
After diagnosed via histopathological evidence, Nasopharyngeal carcinoma tissue specimens were obtained from the Affiliated hospital of Nantong university and Nantong Tumor Hospital. The patients didn't receive antitumor therapy prior to biopsy. Samples were frozen in liquid nitrogen immediately after surgical removal. The control tissues were came from nasal operation (without nasopharyngeal carcinoma). All human tissues were collected using the protocols approved by the Ethics Committee of the Affiliated Hospital of Nantong university.
4 kinds of human NPC cell lines (CNE2, CNE1, 5-8F, 6-18B) from our laboratory were cultured in RPMI medium 1640 (GibCo BRL, Grand Islan, NY) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Islan, NY). CNE-2 (low differentiation), CNE-1 (high differentiation), 5-8F (high tumorigenesis and metastasis) and 6-10B (low tumorigenesis and metastasis). 5-8F and 6-10B cells were cultured in completed medium with penicillin and streptomycin (Shanghai Genebase Gen-Tech Co., Ltd., Shanghai, China). The immortalized normal nasopharyngeal epithelial cell line NP69 was cultured in Keratinocyte-SFM medium supplemented with epidermal growth facter (EGF) (Invitrogen, Carlsbad, USA). Human umbilical vein endothelial cells (HUVEC cells) were maintained in DMEM with low glucose (DMEMLG, Gibco, Grand Islan, NY) supplemented with 10% fetal bovine serum (FBS, GibCo, Grand Islan, NY). All cells were cultured at 37˚C under 5% CO2.
Quantitative real-time PCR (qRT-PCR)
PFKFB3 expression in cells and tissues was measured with qRT-PCR. Total RNA was isolated from specimens or cell lines using TRIzol reagent (Invitrogen, Shanghai, China). qRT-PCR analysis was performed using SYBR Green. The primers were designed and synthesized by Guangzhou RiboBio Co, Ltd. (Guangzhou, China). The expression of each gene was normalized by expression of GAPDH. Each assay was performed in triplicate, and the average was calculated.
Western blot analysis
Lysates were prepared from frozen samples and cultured cells using the RIPA lysis buffer (Beyotime Institute of Biotechnology, Nantong, China). Membranes were incubated with the primary antibodies. For western blot analysis, we used the following antibodies: anti-PFKFB3 (Abcam; 1:3,000 dilution), anti-AKT (Santa Cruz Biotechnology; 1:2000 dilution), anti-ERK (Santa Cruz Biotechnology; 1:500 dilution), anti-E-cadherin (Santa Cruz Biotechnology; 1:500 dilution), anti-N-cadherin (Santa Cruz Biotechnology; 1:500 dilution),anti-Flotillin-1 (1:1000, Santa Cruz Biotechnology), anti-CD63 (1:1000, Sangon Biotech, Shanghai, China), and anti-CD9 (1:1000, Sangon Biotech, Shanghai, China), and mouse anti-β-actin (Santa Cruz Biotechnology; 1:2500 dilution) as a loading control. The expression levels of each protein were normalized by β-actin. The maximum intensity of each band was quantified using Image J software. The values are representative of at least three independent experiments.
Small interfering RNAs (siRNAs) and transfection
Cells were transfected with specific siRNAs for PFKFB3 or negative controls siRNA (RiboBio, Guangzhou, Shanghai, China) by Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Following culture with 48h, those siRNA changed the phenotype of CNE2. Cells were collected for various experiments.
Immunohistochemical staining
The slides dried at 60˚C for 2h. Tissue sections were deparaffinized by xylene, and re-hydrated with sequential washes of 100%, 75%, and 50% ethanol, and PBS. Firstly, slides epitope retrieval carried out using citrate buffer (Vector Lab, CA) in 100˚C, boiled for 30 min. Endogenous peroxidase activity was inhibited with 3% hydrogen peroxidase in PBS. The sections were blocked with 10% goat serum for 1 h, then incubated with primary antibody (anti- PFKFB3, 1 : 50, Abcam) overnight at 4˚C. Secondly, incubated with goat anti-rabbit secondary antibody (1 : 300, Shenggong, Shanghai, China). Then the sections were developed with 3,3'-diaminobenzidine tetrahydrochloride (Shenggong, Shanghai, China) for 2 min and nuclei counterstained with hematoxylin (Shenggong, Shanghai, China) for 2 min. PBS washes were performed between all steps. After dehydration with washes of 95% and 100% ethanol and xylene, tissue sections with permanent mounting medium were covered with glass coverslips, and viewed by light microscope.
Immunofluorescence microscopy
Cells were cultured with 4% paraformaldehyde and then blocked with 1% normal donkey serum. After that, it incubated with primary antibodies overnight at 4˚C. Staining was visualized using anti-mouse IgG Alexa Flour 488 (Molecular Probes, USA), counterstained with Hoechst and observed with a florescence microscope.
Cell viability assay (Cell Counting kit-8)
Cells were seeded into 96-well plates for 12h. Then transfected with different siRNA and were cultured for different time. 10μl per well of CCK-8 Kit reagents were added and incubated for 2h incubation at 37°C. The absorbance of each well was read at 450nm on a microplate reader. All of the experiments were independently repeated at least three times.
Flow cytometric analysis of proliferation and apoptosis
Cells, grown in plates, were transfected with siRNA. After 48h, we collected those cells by trypsinization. After cells double staining with an Annexin V-FITC apoptosis detection kit (BD Biosciences, Oxford, UK), cells were analyzed by flow cytometry (FCM) analysis using BD CELL Quest software (BD Biosciences, USA).
Cells were obtained by trypsinization and pooled after 48h transfection. Then cells centrifuged at 2000 rpm for 5min. After that the cells were treated by cell cycle detection kit (Key Gene). The samples were analyzed by the FACS calibur flow cytometer (BD Bioscience) and BD Cell Quest software.
Wound healing assay
Cells were seeded in 6-well plates and incubated to confluence. A symmetrical wound was created on the monolayer by a sterilized 200µl micropipette tip when the cells reached 90% density. Then changed the medium to 1640 containing 2% FBS. Duplicate wells for each condition were examined, and each experiment was repeated in triplicate.
Matrigel invasion and migration assays
After different treatment, cells were plated in a serum-free medium in the top chamber. The membrane without a coated (24-well insert; 8 µm pore size; Millipore, USA) and a medium supplemented with 10% serum with or without exosomes was in the lower chamber. After 12 to 48 h, the bottom of the chamber insert was stained with methanol and 0.1% crystal violet, imaged. The Matrigel invasion assay was done with the diluted BD biocoat Matrigel Invasion Chamber (pore size, 8 mm, 24-well; BD Biosciences, CA, USA). Each Matrigel invasion assay was conducted in at least 3 replicates.
Exosome isolation and purification
Exosomes were obtained after 300×g for 5mins, 3,000×g for 20 mins, 6,000×g for 40 mins and 10,000×g for 60 mins. Cell-free supernatants were ultracentrifuged at 100,000×g for 1h at 4°C (Beckman 90 Ti rotor). After washed by PBS, exosomes submitted to a second ultracentrifugation in the same conditions. Then quantify the protein content of exosomes by BCA protein assay kit (PIERCE, Rockford, IL, USA). Eosomes were stored at -80°C.
Statistical analysis
Data are presented as mean ± SE, the Student's t-test was used for assessing the difference between individual groups and P ≤ 0.05 was considered significant.
Results
PFKFB3 overexpression in human NPC tissues and cell lines
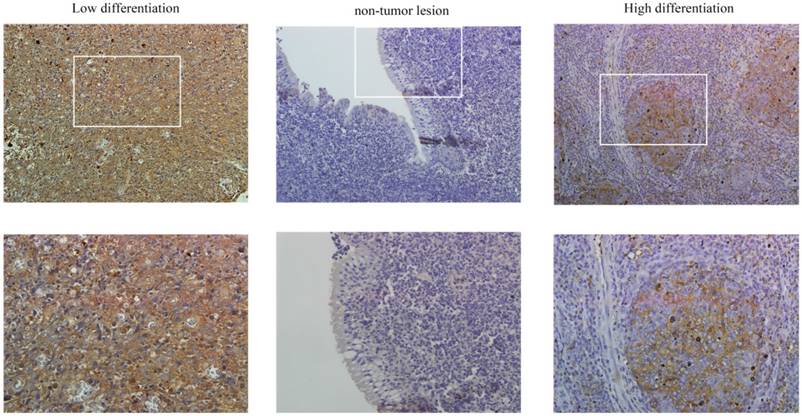
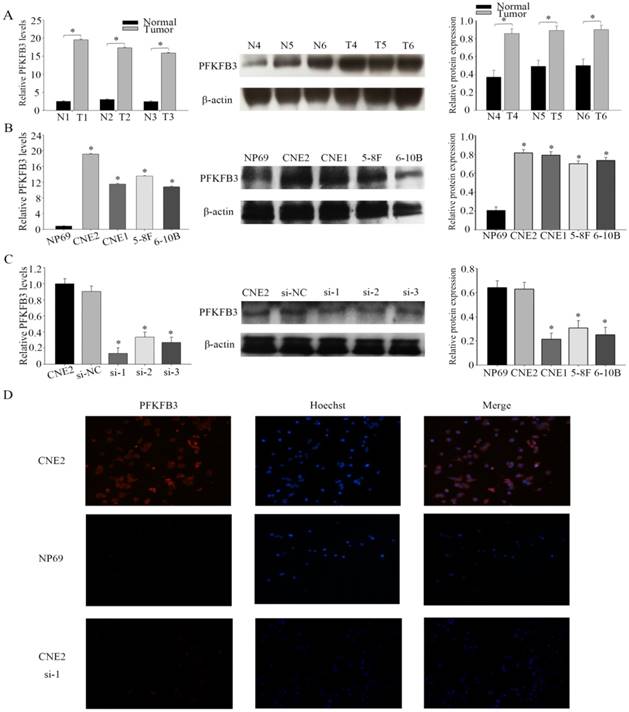
PFKFB3 is overexpression in several cancers have been implicated by some researches. The immunohistochemical analysis was used to detect the expression of PFKFB3 in NPC and non-cancerous nasopharyngeal tissues. The positive expression of PFKFB3 in NPC was higher than that in the non-tumor tissue (Figure 1). Both low differentiation and high differentiation NPC tissues are all overexpression (Figure 1). To confirm these results, we examined the expression of PFKFB3 in NPC tissues and non-tumor nasopharyngeal tissues by qRT-PCR and western blot. In 6 pairs of freshly obtained samples, we found that the expression of PFKFB3 was higher in cancerous tissues compared with the normal tissues (Figure 2a). To confirm the findings in cells, we examined the expression of PFKFB3 in NPC cells (CNE2, CNE1, 5-8F, 6-10B) and normal nasopharyngeal epithelial cell line (NP69). The qRT-PCR and western blot results show that PFKFB3 level was markedly increased in NPC cells (Figure 2b). Taken together, these findings suggested that PFKFB3 was overexpression in NPC.
In four different NPC cell lines, the highest expression level of PFKFB3 was appeared in CNE2. Thus, CNE2 was selected for subsequent experiments. To investigate the role of PFKFB3 in NPC, NPC cells were transfected with siRNA (target to PFKFB3). After that the effect of siRNA was confirmed by qRT-PCR and western blot (Fig. 2c). As we expected, the level of PFKFB3 was markedly reduced after transcribed with siRNA PFKFB3 compared with the scrambled negative controls. Between 4 kinds of siRNA, siRNA-1 was chosen as the most effective siRNA because of the result of fig 2c. Immunofluorescence microscopy was also used to detect the expression of PFKFB3 in CNE2 and NP69. Researches showed that PFKFB3 level was higher in CNE2 than NP69 and CNE2 transfected with siRNA-1 (Fig. 2d).
PFKFB3 is highly expressed in NPC. The immunohistochemical analysis was used to detect the expression of PFKFB3 in NPC and non-cancerous nasopharyngeal tissues. The positive expression of PFKFB3 in NPC was higher than that in the non-tumor tissue. Both low differentiation and high differentiation NPC tissues are all overexpression.
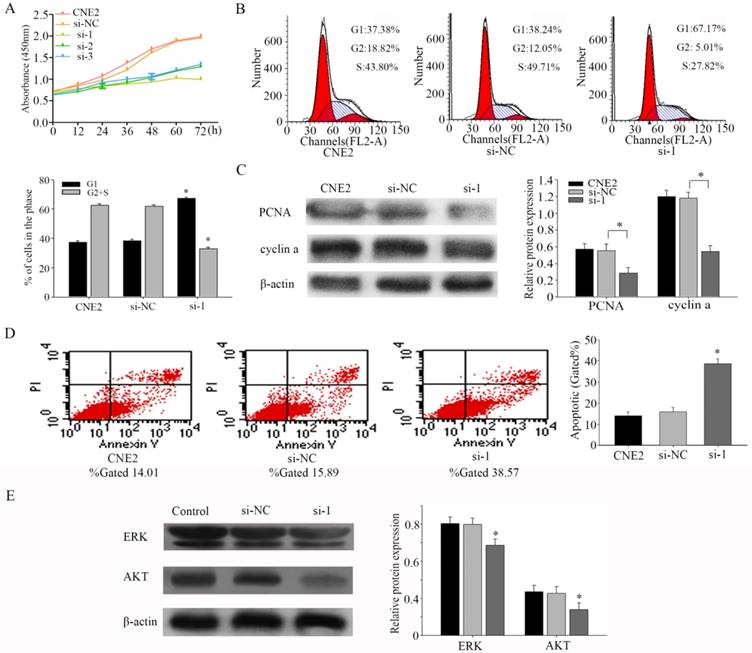
PFKFB3 promotes proliferation and reduces apoptosis of NPC
To assess the effects of PFKFB3 on proliferation, we performed a CCK8 experiment firstly. Our results indicated that silencing PFKFB3 expression decreased cell proliferation in CNE2 (Fig. 3a). Flow cytometer was carried out to further confirm the effect of PFKFB3 on cell proliferation. As shown in figure 3b, Inhibiting the expression of PFKFB3 in CNE2 increased the proportion of G1 phase (from 38.24% to 67.17%), and decreased the proportion of S phase and G2 phase population (from 61.76% to 32.83%). These results may indicate that PFKFB3 had significant effects on cell cycle. To research the mechanism of this process, we detected the expression of PCNA and cyclin a by western blot. Knocking down PFKFB3 clearly reduced the expression of PCNA and cyclin a compared to control (Fig. 3c). These results suggested that PFKFB3 can promote cell proliferation through regulating cell cycle pathway in NPC.
To study the effects of PFKFB3 on apoptosis in CNE2, flow cytometry was implemented. The results showed that decreasing PFKFB3 increased the total apoptosis rate of CNE2 cells from 15.89% to 38.57% (Fig. 3d). These results suggested that PFKFB3 depress the apoptosis of CNE2. Our evidences indicated that ERK and p-AKT were down-regulated after transfected with siRNA-1 (Fig. 3e). This result suggested that PFKFB3 may regulate the process of apoptosis in NPC by activating ERK and p-AKT.
PFKFB3 overexpression in human NPC tissues and cell lines (A) The expression levels of PFKFB3 in NPC and non-cancer tissue were measured by qRT-PCR and western blot. T (human NPC tissues), N (non-cancer tissue). (B) qRT-PCR assays and western blot for the expression of PFKFB3 in NPC cell lines (CNE2, CNE1, 5-8F and 6-10B) compared with the nasopharyngeal epithelia cell NP69 were detected. (C) CNE2 cells were transfected with none-siRNA (CNE2), si-NC (scrambled siRNA) and siRNA PFKFB3. Then qRT-PCR and western blot for PFKFB3 was performed. (D) Immunofluorescence microscopy was used to detect the expression of PFKFB3 in CNE2 , CNE2 transfected with siRNA-1 and NP69. Data are mean ± SE. The same experiments were performed in triplicate. *P < 0.05.
PFKFB3 promotes proliferation and reduces apoptosis of NPC (A) CCK8 assays were used to determine the viability of CNE2 transfected with different siRNA (both of them are targeted to PFKFB3). (B) Flow cytometry assays were performed to analysize the cell cycle progression after 24h when CNE2 cells transfected with siRNA. (C) The expression of PCNA and cyclin a were detected by Western blot analysis. (D) Flow cytometry assays were performed to analysis the cell apoptosis after 48h when CNE2 cells transfected with siRNA. Data are mean ± SE. The same experiments were performed in triplicate. *P < 0.05.
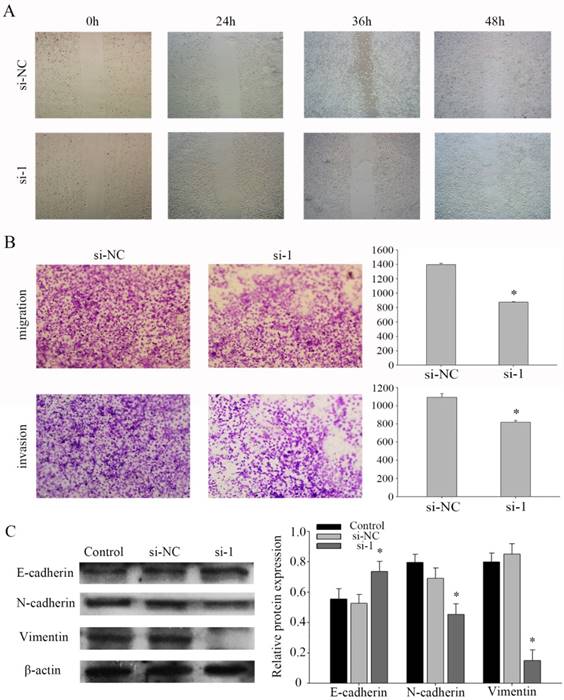
PFKFB3 promotes NPC migration and invasion by influence the EMT program
Based on a series of experiments, we found that PFKFB3 takes part in regulating the migration and invasion in NPC cells. To evaluate the pathway of regulation that PFKFB3 is was critical for the migration and invasion of NPC, cell migration and invasion assays were performed in CNE2 cells transfected with different siRNAs. Further, the wound healing assay showed that knocking down PFKFB3 inhibited the migration of CNE2 cells (Fig. 4a). At the same time, transwell assay indicated that transfected with siRNA-1 significantly reduced the migration and invasion ability in CNE2 compare to the negative control (Fig. 4b). To verify the mechanism of invasion, the epithelial and mesenchymal markers were detected by western blot. As a result, the epithelial markers E-cadherin were up-regulated after CNE2 transfected with siRNA-1. And the mesenchymal markers vimentin and N-cadherin were significantly down-regulated (Figures 4c). Taken together, these results suggest that PFKFB3 promotes NPC migration and invasion by influence the EMT program.
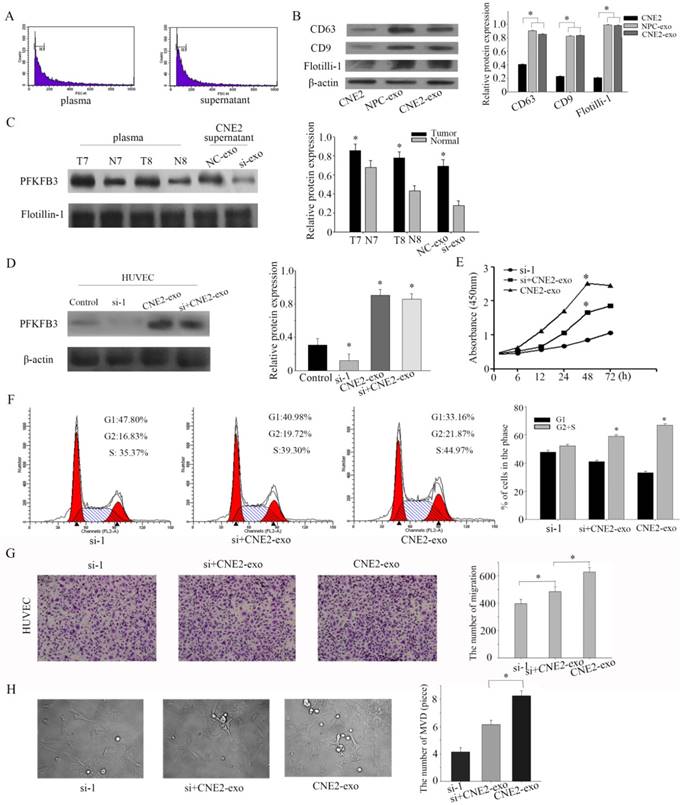
NPC-derived exosomes and CNE2-derived exosomes are enriched in PFKFB3 and promoted HUVECs proliferation, migration and angiogenesis
Exosomes, the microvesicles originate from multivesicular bodies (MVBs), are released into the extracellular environments and play an important role in adjacent cells[26]. Particular populations of proteins are selectively packaged in exosomes and transferred to tissue fluid or blood[27]. First of all, exosomes were isolated by ultracentrifugation, and our task group had successful ensure the isolation by TEM (transmission electron microscope) in several pages [28, 29]. Then, the collected exosomes were examined by flow cytometry sorting (diameter concentrated in the 50-120cm) (Figure 5a). CD63 and CD9 were highly enriched in exosomes according to the references. To ensure successful isolation, the exosomal markers CD63 and CD9 were detected by western blot. The exosomal markers (CD63 and CD9) and flotillin-1 were highly experienced in isolated exosomes than CNE-2 cells (Figure 5b). Based on pilot researches, we found that PFKFB3 is enriched in exosomes from NPC patients compared to healthy volunteers. PFKFB3 is also highly enriched in exosomes from CNE2 cells transfected with control siRNA than transfected with siRNA-1 (Figure 5c). The expression of PFKFB3 in HUVEC was highly improved after cultured with CNE2-exosome (Figure 5d). We knockdown the PFKFB3 in HUVEC, then cultured with exosomes to prove that HUVEC can absorb and take advantage of the PFKFB3 form exosomes. CCK8 evaluate that the exosome from CNE2 cells was critical for modulating the proliferation of HUVECs (Figure 5e). The proliferation of HUVECs was obviously promoted after stimulated by exosomes from CNE2 cells. Flow cytometer was also carried out to evaluate the effect (Figure 5f). By the way, the migration of HUVECs was increased after stimulated by exosomes from CNE2 cells (Figure 5g). The angiogenesis of HUVECs was enhanced after stimulated by exosomes from CNE2 cells (Figure 5h). As a result, CNE2-derived exosomes are enriched in PFKFB3 and promotes HUVECs proliferation, migration and angiogenesis.
PFKFB3 promotes NPC migration and invasion. (A) Wound-healing assay was detected to assess the cell mobility of PFKFB3 in CNE2 cells. (B) The effect of PFKFB3 on migration and invasion ability was measured by transwell assays. (C) The protein expression of E-cadherin, N-cadherin, Vimentin in CNE2 cells transfected with siRNA was analyzed by western blot. All the experiments were performed in triplicate. Data are mean ± SE. *P < 0.01.
NPC-derived exosomes and CNE2- derived exosomes are enriched in PFKFB3 and promotes HUVECs proliferation, migration and angiogenesis. (A) The collected exosomes were examined by flow cytometry sorting (diameter concentrated in the 50-120cm). (B) The exosomal markers CD63 and CD9 were detected by western blot in CNE2 cells, exosome from NPC patients' serum and exosome from CNE2 cells. (C) The protein expression of PFKFB3 in exosome from NPC patients' serum, healthy control and CNE2 cells' supernatant (with or without knockdown PFKFB3) were analyzed by western blot. (D) The protein expression of PFKFB3 was analyzed by western blot in HUVECs. Control: without treated. Si-1: transfected with siRNA-1. CNE2-exo: cultured with exosome from CNE2 cells. Si+CNE2-exo: after transfected with siRNA-1, cultured with exosome from CNE2. (E) CCK8 assays were used to determine the viability of HUVECs after stimulated by exosomes. (F) Flow cytometry assays were performed to analysize the cell cycle progression when HUVECs stimulated by exosomes. (G) The effect of PFKFB3 on HUVECs migration ability was measured by transwell assays. (H) The angiogenesis of HUVECs was measured by tubule formation. The same experiments were performed in triplicate. Data are mean ± SE. *P < 0.01.
Discussion
Mounting evidence indicates that the biological processes of NPC is complicated and characterized by uncontrolled proliferation and migration[1]. It is crucial to identify the mechanisms of NPC's biological processes and to develop novel therapeutic strategies. Neoplastic cells metabolize more glucose relative to normal cells in order to satisfy the need of increased energetic and anabolic[30, 31]. PFKFB3 has been found as a glycolytic activator and highly expressed in many tumors[32]. In this article, we found that PFKFB3 is highly expressed in human NPC tissues and cell lines. The immunohistochemical analysis confirmed that PFKFB3 is significantly over-expressing in different types of NPC. This finding let us to surmise that PFKFB3 may play an important role in the biological processes of NPC. Both CCK8 and flow cytometer found that inhibition of PFKFB3 diminished CNE2 cells proliferation. This result allowed us to further consider PFKFB3's specific mechanisms of proliferation in NPC. The cell cycle marker of PCNA and cyclin a was detected to try to explain the mechanisms that PFKFB3 had significant effects on NPC proliferation by regulating cell cycle. In addition, apoptosis also plays an important role in these processes. Inhibition of PFKFB3 diminished the migration and invasion of CNE2 cells. The change of epithelial and mesenchymal markers means EMT pathway take part in the migration and invasion in CNE2 by PFKFB3. Taking together, these results suggest that PFKFB3 may act as a tumor promoter through promoting cell migration and invasion. Thus molecular parameters suggested that PFKFB3 may be able to drive the CNE2 cells from an epithelial to mesenchymal status. Together, these results suggest that PFKFB3 may be a novel the epithelial-mesenchymal transition (EMT) inducer, and it has a role in NPC progression. Some researches recently identified that the glycolytic activator PFKFB3 play a key role in vessel sprouting by regulating cytoskeleton remodeling, migration and tip cell competitiveness[33]. PFKFB3 has been reported to regulated the proliferation, migration and vessel sprouting of HUVECs[34]. Pathological angiogenesis is a characteristic marker of cancer[35]. Pathological angiogenesis can carry tumor cells to distant metastases by special pathway[36]. Exosomes can be secreted from multiple types of cells and participate in intercellular communication by transmitting intracellular cargoes, such as proteins and nucleic acids[37, 38]. In this report, we found that NPC-derived exosomes and CNE2-derived exosomes are enriched in PFKFB3. This means PFKFB3 in exosomes can become a characteristic signs to NPC. Recently, some studies have suggested that exosomes could potentially affect therapeutic response though the transfer of proteins [39, 40]. Knockdown the expression of PFKFB3 in CNE2 can significantly influence the expression of PFKFB3 in CNE2-derived exosomes. By the way, even knockdown the expression of PFKFB3 in HUVECs, they still can absorb PFKFB3 by foreign exosomes. CNE2-derived exosomes release PFKFB3 to HUVECs and promotes HUVECs proliferation, migration and angiogenesis. Flow cytometry sorting and exosomal markers were detected to ensure successful isolation of exosomes.
In conclusion, our research demonstrated that PFKFB3 has functions in regulating the proliferation, migration and angiogenesis of NPC. It is more important that PFKFB3 can be a crucial angiogenesis site, and can reduce the migration of NPC. It may be critical for the biological activity and to develop novel therapeutic strategies. Therefore, this study extends our knowledge on the oncogenesis of NPC.
Acknowledgements
This study was supported by grants from the Chinese National Natural Science Foundation (nos. 81672682, 81602385), the Natural Science Foundation of Jiangsu Province (SBK2015022581); Social development fund of Nantong Municipal Science and Technology Bureau (MS22015108).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet. 2015
2. Lee NY, Zhang Q, Pfister DG, Kim J, Garden AS, Mechalakos J, Hu K, Le QT, Colevas AD, Glisson BS. et al. Addition of bevacizumab to standard chemoradiation for locoregionally advanced nasopharyngeal carcinoma (RTOG 0615): a phase 2 multi-institutional trial. The Lancet Oncology. 2012;13(2):172-180
3. Li L, Gu M, You B, Shi S, Shan Y, Bao L, You Y. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer science. 2016;107(9):1215-1222
4. Vokes EE, Liebowitz DN, Weichselbaum RR. Nasopharyngeal carcinoma. Lancet. 1997;350(9084):1087-1091
5. Yan M, Zhang Y, He B, Xiang J, Wang ZF, Zheng FM, Xu J, Chen MY, Zhu YL, Wen HJ. IKKα restoration via EZH2 suppression induces nasopharyngeal carcinoma differentiation. Nature communications. 2014;5(33):6227-6230
6. Ho FC, Tham IW, Earnest A, Lee KM, Lu JJ. Patterns of regional lymph node metastasis of nasopharyngeal carcinoma: a meta-analysis of clinical evidence. BMC cancer. 2012;12:98
7. Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012-1024
8. Ropponen KM, Kellokoski JK, Pirinen RT, Moisio KI, Eskelinen MJ, Alhava EM, Kosma VM. Expression of transcription factor AP-2 in colorectal adenomas and adenocarcinomas; comparison of immunohistochemistry and in situ hybridisation. Journal of clinical pathology. 2001;54(7):533-538
9. Jung KH, Zheng HM, Jeong Y, Choi MJ, Lee H, Hong SW, Lee HS, Son MK, Lee S, Hong S. et al. Suppression of tumor proliferation and angiogenesis of hepatocellular carcinoma by HS-104, a novel phosphoinositide 3-kinase inhibitor. Cancer letters. 2013;328(1):176-187
10. Kinkade R, Dasgupta P, Carie A, Pernazza D, Carless M, Pillai S, Lawrence N, Sebti SM, Chellappan S. A small molecule disruptor of Rb/Raf-1 interaction inhibits cell proliferation, angiogenesis, and growth of human tumor xenografts in nude mice. Cancer research. 2008;68(10):3810-3818
11. Mukai H, Muramatsu A, Mashud R, Kubouchi K, Tsujimoto S, Hongu T, Kanaho Y, Tsubaki M, Nishida S, Shioi G. et al. PKN3 is the major regulator of angiogenesis and tumor metastasis in mice. Scientific reports. 2016;6:18979
12. Rivera LB, Bergers G. CANCER. Tumor angiogenesis, from foe to friend. Science. 2015;349(6249):694-695
13. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer cell. 2014;25(4):501-515
14. Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119(6):1159-1167
15. Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Scientific reports. 2016;6:20254
16. Suchorska WM, Lach MS. The role of exosomes in tumor progression and metastasis (Review). Oncology reports. 2016;35(3):1237-1244
17. Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer research. 2002;62(20):5881-5887
18. Minchenko OH, Tsuchihara K, Minchenko DO, Bikfalvi A, Esumi H. Mechanisms of regulation of PFKFB expression in pancreatic and gastric cancer cells. World journal of gastroenterology: WJG. 2014;20(38):13705-13717
19. Calvo MN, Bartrons R, Castano E, Perales JC, Navarro-Sabate A, Manzano A. PFKFB3 gene silencing decreases glycolysis, induces cell-cycle delay and inhibits anchorage-independent growth in HeLa cells. FEBS letters. 2006;580(13):3308-3314
20. De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G. et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651-663
21. Cruys B, Wong BW, Kuchnio A, Verdegem D, Cantelmo AR, Conradi LC, Vandekeere S, Bouche A, Cornelissen I, Vinckier S. et al. Glycolytic regulation of cell rearrangement in angiogenesis. Nature communications. 2016;7:12240
22. Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, Hue L. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. The Biochemical journal. 2004;381(Pt 3):561-579
23. Sakata J, Abe Y, Uyeda K. Molecular cloning of the DNA and expression and characterization of rat testes fructose-6-phosphate,2-kinase:fructose-2,6-bisphosphatase. The Journal of biological chemistry. 1991;266(24):15764-15770
24. Simon-Molas H, Calvo-Vidal MN, Castano E, Rodriguez-Garcia A, Navarro-Sabate A, Bartrons R, Manzano A. Akt mediates TIGAR induction in HeLa cells following PFKFB3 inhibition. FEBS letters. 2016;590(17):2915-2926
25. Domenech E, Maestre C, Esteban-Martinez L, Partida D, Pascual R, Fernandez-Miranda G, Seco E, Campos-Olivas R, Perez M, Megias D. et al. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nature cell biology. 2015;17(10):1304-1316
26. Th Xe C, ry. Exosomes: secreted vesicles and intercellular communications. F1000prime Rep. 2011;3:15
27. Soldevilla B, Rodríguez M, San Millán C, García V, Fernández-Periañez R, Gil-Calderón B, Martín P, García-Grande A, Silva J, Bonilla F. Tumor-derived exosomes are enriched in ΔNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Human molecular genetics. 2014;23(2):467-478
28. You B, Cao X, Shao X, Ni H, Shi S, Shan Y, Gu Z, You Y. Clinical and biological significance of HAX-1 overexpression in nasopharyngeal carcinoma. Oncotarget. 2016;7(11):12505-12524
29. Shi S, Zhang Q, Xia Y, You B, Shan Y, Bao L, Li L, You Y, Gu Z. Mesenchymal stem cell-derived exosomes facilitate nasopharyngeal carcinoma progression. American journal of cancer research. 2016;6(2):459-472
30. Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High expression of inducible 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer research. 2002;62(20):5881-5887
31. Yalcin A, Telang S, Clem B, Chesney J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatases in cancer. Experimental and molecular pathology. 2009;86(3):174-179
32. Riera Ls, Manzano A, Navarro-Sabaté A, Perales JC, Bartrons R. Insulin induces PFKFB3 gene expression in HT29 human colon adenocarcinoma cells. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research. 2002;1589(2):89-92
33. Bock KD, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquière B, Cauwenberghs S, Eelen G. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651-663
34. Schoors S, De BK, Cantelmo AR, Georgiadou M, Ghesquière B, Cauwenberghs S, Kuchnio A, Wong BW, Quaegebeur A, Goveia J. Partial and transient reduction of glycolysis by PFKFB3 blockade reduces pathological angiogenesis. Cell Metabolism. 2014;19(1):37-48
35. Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249-257
36. Organizer DJC, Goode JA. Chemokines: Angiogenesis and Metastases in Lung Cancer. John Wiley & Sons, Ltd. 2008
37. Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer cell. 2014;26(5):707-721
38. Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar CM. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature medicine. 2012;18(6):883-891
39. van de Velde CJ, Boelens PG, Borras JM, Coebergh J-W, Cervantes A, Blomqvist L, Beets-Tan RG, van den Broek CB, Brown G, Van Cutsem E. EURECCA colorectal: multidisciplinary management: European consensus conference colon & rectum. European journal of cancer. 2014;50(1):1. e1-1. e34
40. Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. Journal of the National Cancer Institute. 2015;107(7):djv135
Author contact
![]() Corresponding authors: Yiwen You, Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province 226000, China. Tel ⁄Fax: +86-0513-8116-1025; E-mail: youyiwen_nantongcom and Qicheng Zhang, Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province 226000, China. Tel ⁄Fax: +86-0513-8116-1025; E-mail: qczhang_entcom
Corresponding authors: Yiwen You, Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province 226000, China. Tel ⁄Fax: +86-0513-8116-1025; E-mail: youyiwen_nantongcom and Qicheng Zhang, Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province 226000, China. Tel ⁄Fax: +86-0513-8116-1025; E-mail: qczhang_entcom

 Global reach, higher impact
Global reach, higher impact