Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(19):4106-4116. doi:10.7150/jca.21024 This issue Cite
Research Paper
Long Non-coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429
1. Department of Vascular Surgery, the First Hospital of Jilin University, Changchun, 130021, P.R. China;
2. Department of Clinical Medicine, Norman Bethune Health Science Center of Jilin University, Changchun, 130021,P.R. China;
3. Department of Pediatric Emergency, the First Hospital of Jilin University, Changchun, 130021, P.R. China;
4. Department of Otolaryngology Head and Neck surgery, the First Hospital of Jilin University, Changchun, 130021, P.R. China;
5. Department of Cell Biology, College of Life Science, Zhejiang Chinese Medical University, Hangzhou, 310053, China;
6. Department of Regenerative Medicine, School of Pharmaceutical Science, Jilin University, Changchun, 130021, P.R. China.
Received 2017-5-15; Accepted 2017-10-19; Published 2017-11-6
Abstract
Glioma is a worldwide malignancy, which displays significantly active metastasis and angiogenesis. Interaction between long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) has been shown to play crucial role in regulating tumor properties. However, the potential of lncRNA X-inactive specific transcript (XIST) to function as a miRNA regulator and its relevance in glioma tumorigenicity and angiogenesis have so far remained unclear. Expression analysis of lncRNA XIST in glioma cells revealed its significant up-regulation. Interestingly, silencing of XIST repressed both metastatic and pro-angiogenic ability in vitro as well as in vivo. Subsequent studies revealed that lncRNA XIST expression inversely correlated with miR-429 expression in glioma cells; miR-429 modulated XIST expression by directly targeting the XIST gene sequence. In addition, miR-429 inhibitor restored metastatic and pro-angiogenic ability of gliomas abolished by silencing XIST. Our data provide insight into the key roles of the lncRNA-miRNA functional network in gliomas, which can aid in developing new therapeutic strategies for gliomas through clinical trials.
Keywords: lncRNA XIST, miR-429, glioma, HBMVECs, angiogenesis
Introduction
Noncoding RNAs (ncRNAs) are a functionally versatile class of transcripts, which are not translated into a protein but play crucial biological roles at the RNA level [1]. Following the completion of human genome sequencing, more and more ncRNAs with different targets and functions have been identified [2, 3]. ncRNAs are divided into short and long ones depending on their length; among these, microRNAs (~22nts in length, miRNAs), which regulate post-transcriptional gene silencing by controlling translation and RNA stability [4, 5], and long non-coding RNAs (more than 200nts in length, lncRNAs), which exert their biological function through X chromosome silencing, genomic imprinting, chromatin modification, transcriptional regulation, and post-transcriptional regulation [6-9], have recently gained significant attention. Both of these ncRNAs have been demonstrated to be pivotal players in the regulation of a wide range of cancers [10-13]; thus, ncRNAs exhibit the potential to be developed into biomarkers for diagnosis and prognosis.
Glioma is the most common and aggressive primary brain tumor [14]. Its invasive growth makes complete tumor resection very difficult, leading to high rate of lethality [2]. The mean life expectancy of patients with gliomas undergoing conventional treatment is less than 15 months [15]. However, the molecular mechanisms underlying glioma formation still remain elusive. Identification of critical molecules involved in glioma tumorigenicity and angiogenesis may provide breakthroughs for glioma treatment. Previous studies have found that aberrant expression of lncRNA and microRNA is a characteristic feature of gliomas [16-18], and lncRNA-miRNA interaction plays a significant role in glioma angiogenesis [19].
LncRNA X-inactive specific transcript (XIST), a product of the XIST gene, is essential for transcriptional silencing of one X-chromosome during mammalian female development [20]. It has emerged as a critical regulator of human cell differentiation, proliferation, and genome maintenance [21]. More recently, it has been recognized that abnormal expression of lncRNA XIST may have a pathological role in cancer, including gliomas [22]. However, the specific molecular mechanisms involved are still under investigation. Using an online software program starbase v2.0, we noticed that lncRNA XIST might be an lncRNA target of miR-429. In this study, we aimed to elucidate the function of lncRNA XIST in promoting glioma tumorigenicity and angiogenesis, and to identify the relationship between lncRNA XIST and miR-429 in the regulation of tumorigenicity and angiogenesis of glioma.
Materials and Methods
Cell lines and animals
The Human astrocytes HA1800, human A172 and U251 glioblastoma cell lines, and human brain microvascular endothelial cells (HBMVECs) were purchased from the Cell Resource Center of Shanghai Institute of Life Sciences. The cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco BRL Co. Ltd., USA), supplemented with 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 mg/mL) at 37°C in 5% CO2. BALB/c nude mice (male, 5-week-old, 16-20 g) were purchased from Shanghai Slack Laboratory Animal Co. Ltd. (animal license number: SCXK (Shanghai) 2012-0002). All the experimental procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals of Zhejiang Chinese Medical University, Hangzhou, China, and they complied with the National Institutes of Health Guide for Care and Use of Laboratory Animals (NIH Pub. No. 85-23, revised 1996).
Human tissue samples
Glioma gene expression datasets are deposited at NCBI's Gene Expression Omnibus (GEO) public database (http://www.ncbi.nlm.nih.gov/geo/, GEO accession number, GSE4290 and GSE2223). GSE4290 contains 180 samples including 23 non-tumor, 26 astrocytoma (7, Grade II; 19, Grade III), 50 oligodendroglioma (38, Grade II; 12, Grade III), and 81 glioblastoma samples. GSE2223 contains 4 normal brain and 50 glial brain tumor samples.
Cell transfection
miR-429 mimic, miR-429 inhibitor, and its negative control were purchased from Biomics Biotechnologies Co., Ltd., Nantong, China. For the transfection, cells were grown until their number reached 105 6‑well plates and were then transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
Lentivirus production and transduction
Lentiviral small hairpin RNA targeting XIST (sh-lncRNA-XIST) and sh-NC (negative control) were designed and cloned into the pLVshRNA-Puro vector according to manufacturer's instructions (Inovogen Tech. Co., Beijing, China). The viruses were packaged in HEK293T cells following standard protocols, and the virus particles were harvested 72 h later. Glioma cells were infected with virus particles and selected with puromycin. Glioma cells stably expressing sh-lncRNA-XIST are denoted as sh-lncRNA-XIST cells and those expressing sh-NC as sh-NC cells.
RNA isolation and quantitative real-time PCR analysis (qPCR)
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For qPCR, 1 μg of total RNA was reverse transcribed to complementary DNA (cDNA) using HiFi‑MMLV cDNA kit (Beijing ComWin Biotech Co., Ltd., Beijing, China) and qPCR was performed using the UltraSYBR Mixture (Beijing ComWin Biotech Co., Ltd.). To amplify miR‑429 cDNA, specific RT primers were used based on its sequence and the U6 RT primer used was the same as the U6 reverse PCR primer. The primers for XIST were 5'‑CCATTGAAGATACCACGCTGC‑3' (forward) and 5'‑GGTTGTTGCCCAGGGTAGTG‑3' (reverse) and for β-actin were 5'‑GGCACCACACCTTCTACAAT‑3' (forward) and 5'‑GTGGTGGTGAAGCTGTAGCC‑3' (reverse). The RT primer for miR-429 was 5'‑GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACGGTTGA‑3', and the PCR primers were 5'‑CGGTAATACTGTCTGGTAA‑3' (forward) and 5'‑GTGCAGGGTCCGAGGT‑3' (reverse). U6 primers were 5'‑CTCGCTTCGGCAGCACA‑3' (forward) and 5'‑AACGCTTCACGAATTTGCGT‑3' (reverse). The relative expression of each gene was calculated and normalized using the 2-ΔΔCt method relative to β-actin or U6.
MTT assay
To determine cell proliferation, sh-lncRNA-XIST cells and sh-NC cells were seeded at a density of 5×103 cells/well in a 96-well plate. After 24, 48, and 72h of incubation, MTT (5 mg/mL) was added to each well, followed by an incubation of 4 h. After the supernatants were removed, dimethyl sulfoxide (DMSO) was added to each well and the absorbance value was measured at 490 nm using a SpectraMaxM3 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Colony formation assay
For colony formation assay, sh-lncRNA-XIST cells and sh-NC cells were seeded into 6-well plates at a density of 5×102 cells/well and cultured for another fortnight. Then, cells were washed twice with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 30 minutes, and stained with Giemsa for 15 minutes. Colonies were counted under a microscope (Nikon Eclipse Ti-S, Japan).
Flow cytometry analysis (FCM)
sh-lncRNA-XIST cells and sh-NC cells were seeded in 35 mm-petri plates at a density of 1x105cells/mL. After 24 h of incubation, cells were harvested and fixed in 70% ethanol, then stained with propidium iodide (Nanjing KeyGen Biotech Co. Ltd., Nanjing, China) containing RNase A (1 mg/ml; Takara Biotechnology Co., Ltd.) for 30 min at 37˚C. Subsequently, 500 μL of cells was filtered through 200‑μm mesh sieves and the cell cycle profiles were assayed using a Guava easyCyte 8 Flow Cytometer (EMD Millipore, Billerica, MA, USA).
HBMVEC recruitment assay
A 24-well plate with 8-μm pore size polycarbonate membrane (Corning, USA) was used for the endothelial recruitment assay. sh-lncRNA-XIST cells and sh-NC cells were placed in the lower compartments. Then, HBMVECs were resuspended in 100 μL serum-free medium and seeded in the upper compartments of the chambers. The cells remaining on the upper surface of the membrane were removed after incubation at 37°C for 12 h. The cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 30 minutes and stained with Giemsa for 15 minutes.
Collection of conditional medium (CM)
The supernatant of the cells was collected and centrifuged at 2,000×g for 20 minutes at 4°C to remove cell debris 48h after the transfection. Then, aliquots of the CM were stored at -80°C until further use.
HBMVEC capillary tube formation assay
HBMVECs (2.5×104 cells) were grown in the absence or presence of 100% CM for 6 h at 37°C in a 96-well plate coated with Matrigel (BD Biosciences, USA). The formation of capillary-like structures was observed under a light microscope. The branch points of the formed tubes, which represented the degree of angiogenesis in vitro, were scanned and quantitated under a light microscope (Nikon Eclipse Ti-S, Japan).
Luciferase reporter assay
For the luciferase reporter assay, A172/U251 cells were co‑transfected with miRNA (miR‑429 mimic or miR-429 mimic NC) and reporter vectors (pmirGLO‑WT or pmirGLO‑MUT) using Lipofectamine 2000. Luciferase activity was assayed 48 h after transfection using a Dual‑Luciferase Reporter Assay system (Beyotime Institute of Biotechnology, Haimen, China). The values were normalized to those obtained for miRNA negative control transfection. All transfection experiments were performed in triplicates.
Nude mouse xenograft studies
A172-shNC or A172-sh-XIST cells (1 × 107) were injected into the dorsal flanks of 5-week-old male BALB/c nude mice. The length and width of tumor xenografts were measured every 3 days starting from the 10th day after inoculation. Tumor volume was calculated using the formula V = W2 × L × 0.5.When the tumors grew to a size of 500 mm3, mice were sacrificed and the xenografts were dissected and weighed.
Immunohistochemistry (IHC)
The dissected tumors were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and then sectioned at a width of 5 μm. The sections mounted on the glass slides were deparaffinized and rehydrated. For immunofluorescence assay, the sections were incubated with antibodies for proliferation marker Ki-67 (Calbiochem, Cambridge, MA) and endothelial cell marker CD34 (Santa Cruz, CA), followed by incubation with secondary antibodies. After the cell nuclei were labeled with hematoxylin, images were captured using a microscope (XSP-C204, Chongqing Optical Instrument Co., Ltd.).
Results
XIST is up-regulated in glioma tissues and cells
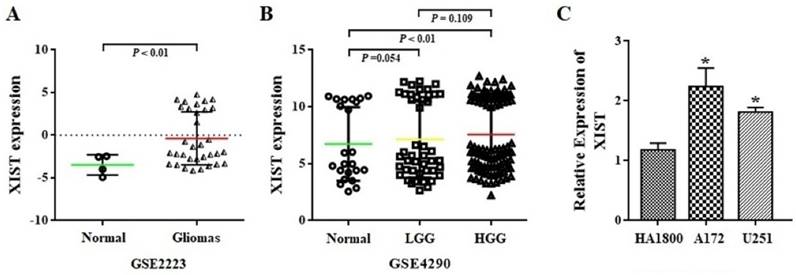
The expression level of XIST was first analyzed in whole-gene profiles of human glioma tissues in GSE2223 acquired from the GEO datasets. As shown in Figure 1A, glioma tissues demonstrated a significant increase in XIST transcript levels, compared with that observed in normal tissues (P<0.01). Next, we employed another GEO dataset GSE4290 to examine the association between XIST expression levels and glioma grade. We found that XIST expression was significant higher in glioma high-grade glioma (HGG) tissues than in normal tissues (P<0.01), but no significant difference in XIST expression levels was observed between HGG and LGG (Figure 1B). Furthermore, we compared the expression level of XIST between human glioma and normal glial cells using qPCR analysis. It was found that XIST expression showed a significant increase in the glioma A172 and U251 cells in comparison with human normal glial HA1800 (Figure 1C). These findings suggest that XIST is up-regulated in glioma cells and tissues but may not fully correlate with glioma grade.
Silencing XIST inhibits tumorigenicity and angiogenesis of glioma cells
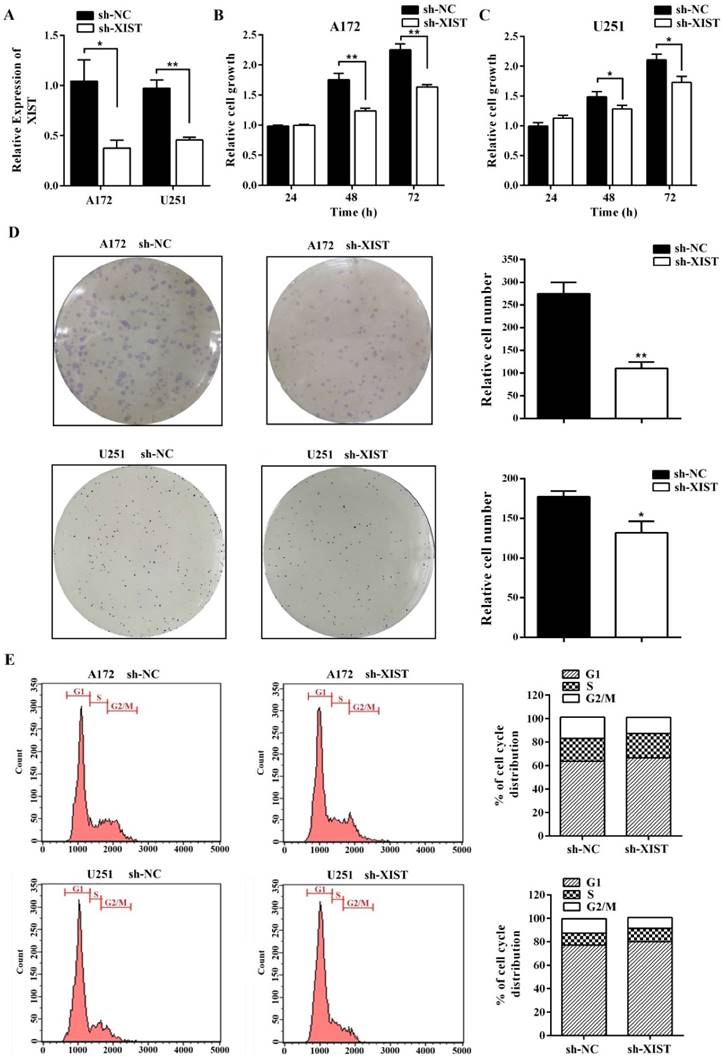
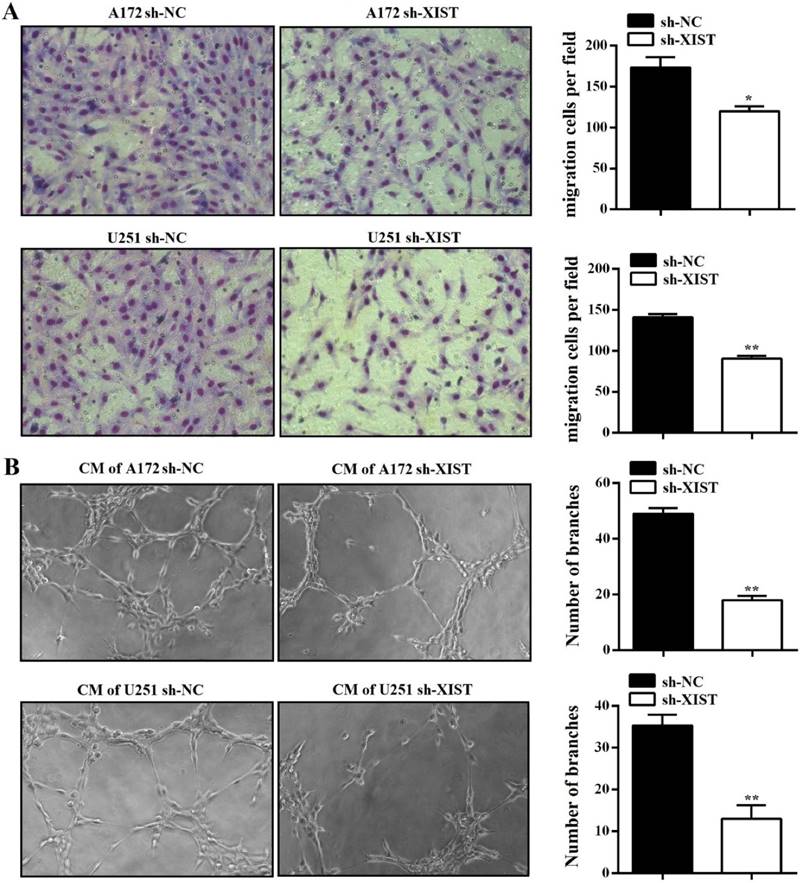
To further explore the function of XIST in glioma, we established XIST stable knockdown in A172 and U251 cells using lentiviral constructs sh-lncRNA-XIST. The knockdown of XIST expression was verified by qPCR in sh-XIST cells (Figure 2A). Next, we performed a series of experiments to explore the effect of XIST silencing on glioma tumorigenicity and angiogenesis. MTT assay revealed that sh-XIST cells had a significantly lower cell proliferation rate after 48 and 72h of incubation compared with sh-NC cells (Figure 2B&C). In parallel, Both A172 and U251 cells formed fewer colonies after silencing XIST, shown by colony formation assay (Figure 2D). We also performed FCM to examine whether XIST promoted glioma cell proliferation by altering cell cycle progression. On analysis, we observed that silencing XIST significantly increased the frequency of G1 and S phase cells and decreased the frequency of G2/M phase cells (Figure 2E). Moreover, compared to sh-NC cells, the ability of sh-XIST cells to promote the migration of HBMVECs was significantly suppressed both in A172 and U251 (Figure 3A). On the other hand, the conditional medium of A172/U251 sh-XIST cells and A172/U251 sh-NC cells collected to incubate with HBMVEC cells separately showed a dramatically suppressive effect of tube formation in HBMVECs treated with the conditional medium of sh-XIST cells (Figure 3B). Taken together, these data suggested that XIST promoted glioma tumorigenicity and angiogenesis.
lncRNA XIST knockdown reduces tumorigenicity and angiogenesis in vivo
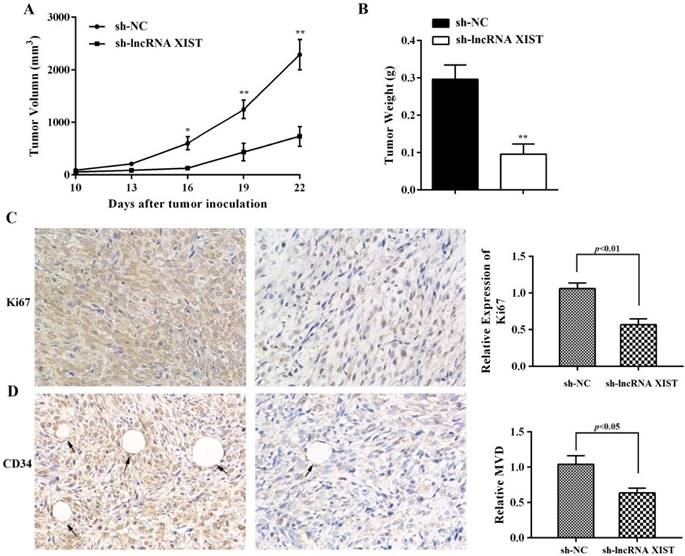
The ability of XIST to promote glioma tumorigenicity and angiogenesis was examined using a nude mouse xenograft tumor model. We observed that, in comparison with sh-NC cell-derived xenograft tumors, sh-XIST cell-derived xenograft tumors revealed a significant reduction in tumor volumes (Figure 4A). The mean weight of sh-XIST cell-derived xenograft tumors was significantly less compared with that of the sh-NC cell-derived xenograft tumors (Figure 4B). Further, the tumor sections were stained for Ki-67 expression to quantitatively assess the proliferation index in xenograft tumors. Here, we observed that the sh-XIST cells displayed a lower Ki-67 proliferation index compared with tumors derived from sh-NC cells (Figure 4C). Meanwhile, the level of microvascular decompression (MVD) was quantified by immunohistochemical staining using endothelial cell marker CD34. As shown in Figure 4D, significantly higher level of CD34 was detected in sh-NC cell-derived xenograft tumors than sh-XIST cell-derived xenograft tumors.
XIST expression in glioma tissues and cells. (A) XIST levels were analyzed in glioma tissues of GSE2223 datasets. (B) XIST levels were analyzed in glioma tissues of GSE4290 datasets. (C) Expression level of XIST in HA1800, A172 and U251 cell lines. Abbreviations: LGG, low-grade glioma; HGG, high-grade glioma. *P < 0.05.
XIST inhibition decreased glioma cell tumorigenicity. (A) XIST expression in A172 and U251 cells transduced with control shRNA vector (sh-NC) or XIST shRNA vector (sh-XIST). (B, C) Silencing XIST significantly decreased cell proliferation in sh-XIST cells compared with sh-NC cells. (D) Colony formation assay demonstrated that sh-XIST cells formed fewer colonies than sh-NC cells (both in A172 and U251). (E) sh-XIST cells displayed a significantly higher frequency of cells at the G1 and S phase and a lower frequency of cells at G2/M phase (both in A172 and U251). The results were reproduced in three independent experiments, and representative images are shown. CM: conditional medium; **P < 0.01, *P < 0.05.
XIST inhibition decreased glioma angiogenesis. (A) Silencing XIST in A172 and U251 significantly decreased the migration capability of HBMVECs. (B) Silencing XIST significantly decreased the tube formation capability of HBMVECs. The results were reproduced in three independent experiments, and representative images are shown. CM: conditional medium; **P < 0.01, *P < 0.05.
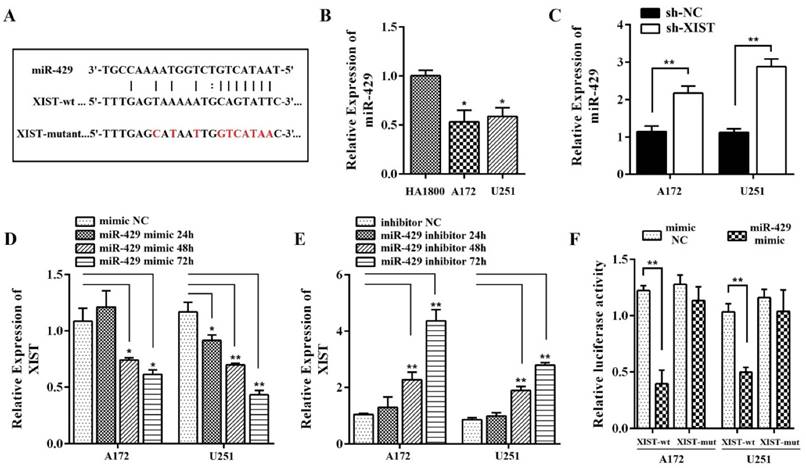
Reciprocal repression exists between XIST and miR-429 in glioma cells
Using the online software program starBase v2.0, we found that miR-429 formed complementary base pairing with XIST (Figure 5A), implying that XIST might be an endogenous sponge of miR-429, exerting its editing function by regulating miR-429 expression. Therefore, we focused on the expression of miR-429 in human glioma and normal glial cells. The qPCR results showed that the level of miR-429 was significantly lower in A172 and U251 cells compared with normal glial cells, HA1800 (Figure 5B). We also found that the level of miR-429 in sh-XIST cells was up-regulated compared with that of sh-NC cells both in A172 and U251, indicating silencing XIST promotes miR-429 expression (Figure 5C). Next, we analyzed whether miR-429 has any negative regulation on XIST expression using miR-429 mimic and inhibitor. As shown in Figure 5D, we observed that miR-429 mimic reduced the XIST expression by approximately 68% after 48h of transfection and approximately 56% after 72h of transfection in A172 cells, by approximately 60% after 48h of transfection and approximately 37% after 72h of transfection in U251 cells. Furthermore, miR-429 inhibitor increased the expression of XIST (Figure 5E).
To further verify whether XIST is a functional target of miR-429, we cloned the predicted miR-429 binding sequences of XIST (XIST-wt) or a mutant miR‑429 target binding sequences (XIST-mutant) into the pmirGLO luciferase vector. Following co‑transfection with the miR‑429 mimic in A172/U251 cells, a Dual‑Luciferase assay was performed to determine the luciferase activity. The results showed that cells co-transfected with pmirGLO XIST-wt and miR-429 mimic had significantly lower luciferase activity compared with that co-transfected with pmirGLO XIST-mutant and miR-429 mimic (Figure 5F). Co-transfection of miR-429 mimic NC with pmirGLO XIST-wt or pmirGLO XIST-mutant did not change the luciferase activity. All these data together demonstrated that miR-429 can directly bind to XIST and negatively regulate XIST expression.
miR-429 mediates the effect of XIST on tumorigenicity and angiogenesis in gliomas
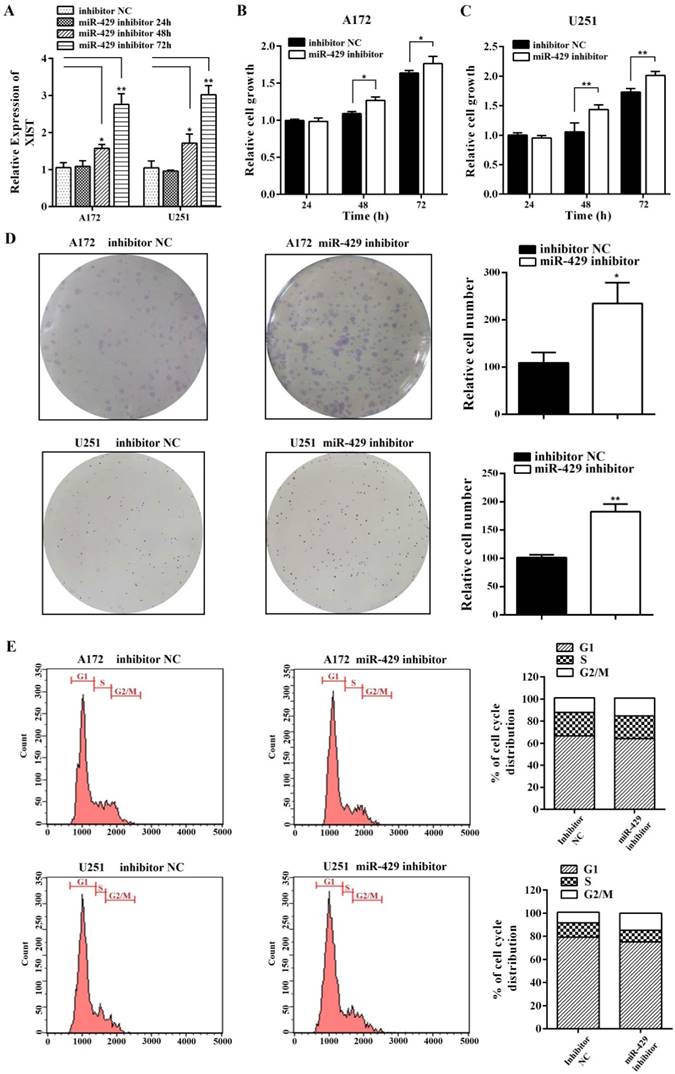
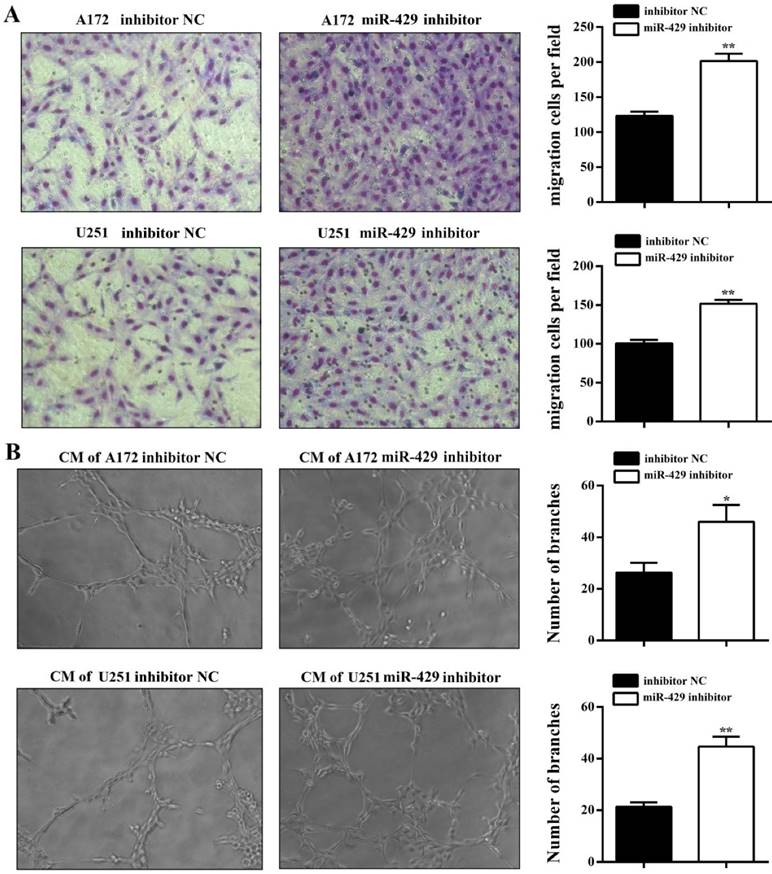
In order to analyze the effect of XIST on glioma tumorigenicity and angiogenesis mediated by miR-429, miR-429 inhibitor and inhibitor NC were transfected into A172/U251 sh-XIST cells separately. As shown in Figure 6A, miR-429 inhibitor strongly restored the expression level of XIST in sh-XIST cells, especially after 72h transfection. The MTT and colony formation assays showed that miR-429 inhibitor significantly increases the growth of sh-XIST cells (Figures 6B, C & D). Meanwhile, the alterations in cell cycle distribution caused by silencing XIST, was also rescued by miR-429 inhibitor (Figure 6E). Moreover, miR-429 inhibitor enhanced the ability of sh-XIST cells to induce HBMVECs migration and tube formation (Figure 7A & B).
IHC analysis of Ki67 expression and tumor vascularization in a glioma xenograft model. (A-B) The mean volume and weight of the xenograft tumors in sh-lncRNA XIST (XIST knockdown) group was smaller than those of sh-NC (negative control) group. (C) The cell proliferation of xenograft tumors was analyzed by Ki67 IHC. The intensity of Ki67-positive cells was calculated with Image J software (×400). (D) Analysis of MVD based on CD34 staining of tumor tissue by Image J software showed that vessel numbers significantly reduced in the sh-lncRNA XIST group (×400) and arrows indicate blood vessels. *P < 0.05, **P < 0.01.
Reciprocal repression of XIST and miR-429 in glioma cells. (A) miR‑429 binding sequence in XIST. The red letters in the XIST sequence show mutations. (B) The expression of miR-429 increased in A172/U251 cells compared with HA1800. (C) miR-429 expression showed increase in sh-XIST cells compared with sh-NC cells. (D-E) Effect of miR-429 on XIST expression. A172/U251 cells were transfected with miR-429 mimic, miR-429 inhibitor, or NC. (F) Luciferase reporter assays in A172/U251 cells. Co-transfection of miR-429 mimic and XIST-wt decreased the luciferase activity, while co-transfection of miR-429 mimic and XIST-mutant did not change the luciferase activity. Co-transfection of mimic NC with XIST-wt or XIST-mutant did not change the luciferase activity. mut: mutant;*P < 0.05, **P < 0.01.
Discussion
XIST is a ~17kb lncRNA, which has a prominent role in regulating X-chromosome inactivation during early embryogenesis [20]. In recent years, dysregulation in the expression of lncRNA XIST was observed in numerous human cancers such as breast cancer, hepatocellular carcinoma, gastric cancer, and non-small cell lung carcinoma [23-26]. Yao et al. [27] demonstrated that knockdown of XIST inhibits cell proliferation, migration, and invasion in human glioblastoma stem cells. In this study, XIST was found to be markedly up-regulated in human glioma cells and tissues. Silencing XIST expression reduced glioma growth both in vitro and in vivo. Because human gliomas are blood vessel-rich tumors, their growth and proliferation depends considerably on angiogenesis [28, 29]. Hence, the identification of critical molecules involved in angiogenesis is also important in glioma treatment. In this regard, here, we present evidence showing that silencing XIST strongly inhibits glioma cells induced HBMVEC migration and tube formation. Furthermore, the level of MVD was reduced in XIST-silenced glioma cells in a nude mice model.
A number of studies suggest that lncRNA is involved in human glioma progression through competitive inhibition of miRNAs by functioning as a molecular sponge. For example, CCAT1 has been shown to act as a molecular sponge for microRNA-410, promoting glioma cell proliferation via inhibition of microRNA-410 [30]. HOTAIR act as an endogenous sponge for miR-148b-3p; miR-148b-3p inhibits malignant biological behaviors of glioma cells induced by high HOTAIR expression [31]. In the present study, we predicted a putative binding site of miR-429 in lncRNA XIST. Further, we focused on the expression of miR-429 and found a negative correlation between lncRNA XIST and miR-429 in glioma A172 cells. Knockdown of XIST significantly induced the expression of miR-429. On the other hand, miR-429 mimic reduced XIST expression and miR-429 inhibitor increased XIST expression, providing supporting evidence that XIST and miR-429 may form a reciprocal repression feedback loop. Furthermore, the luciferase assays indicated that the miR-429 binding site on XIST is vital for its interaction with XIST.
miR-429 inhibitor restores XIST silencing effects of reduced glioma cell tumorigenicity. (A) miR-429 inhibitor transfection led to the restoration of XIST in sh-XIST cells. (B, C) The MTT assay showed that miR-429 inhibitor increased cell proliferation both at 48 and 72h. (D) Colony formation assay showed that colony forming ability of sh-XIST cells was active after treatment of miR-429 inhibitor. (E) FCM assay showed that miR-429 inhibitor transfection increased the frequency of sh-XIST cells at G2/M. **P < 0.01, *P < 0.05
miR-429 inhibitor restores XIST silencing effects of reduced glioma angiogenesis. (A) HBMVEC recruitment assay showed that the ability of sh-XIST cells to promote HBMVECs migration was rescued by miR-429 inhibitor. (B) miR-429 inhibitor enhances sh-XIST induced tube formation in HBMVECs. The results were reproduced in three independent experiments and representative images are shown. **P < 0.01, *P < 0.05.
miR-429 functions either as an oncogene or a tumor suppressor depending on the tumor type; it has been reported to participate in tumor cell proliferation, differentiation, invasiveness, apoptosis, and drug resistance [32-36]. A previous study has demonstrated that miR-429 inhibits glioma invasion through BMK1 suppression [37]. Similarly, we found that miR-429 was down-regulated in glioma cells. To the best of our knowledge, we demonstrated the relationship between miR-429 and lncRNA for the first time in the present study. We further figured out whether miR-429 mediated XIST-induced glioma tumorigenicity and angiogenesis. Our data indicated that while silencing XIST decreased glioma cell proliferation, the inhibition of miR-429 could rescue the effects exerted by XIST silencing. Moreover, further study implied that miR-429 largely abrogated HBMVECs migration and tube formation capabilities abolished by silencing XIST.
In conclusion, our studied showed that lncRNA XIST directly regulates miR-429 expression by acting as a molecular sponge. lncRNA XIST plays a “trigger” role in the tumorigenicity and angiogenesis of gliomas by down-regulating “onco-miRNA” miR-429. Exploring the key roles of other lncRNA-miRNA interactions will provide new therapeutic strategies for gliomas in clinic trail.
Abbreviations
CM: Collection of conditional medium;
DMEM: Dulbecco's modified Eagle's medium;
DMSO: dimethyl sulfoxide;
FCM: Flow cytometry analysis;
HBMVECs: human brain microvascular endothelial cells;
IHC: Immunohistochemistry;
lncRNAs: long non-coding RNAs;
miRNAs: microRNAs;
MVD: microvascular decompression;
ncRNAs: Noncoding RNAs;
PBS: phosphate buffered saline;
qPCR: RNA isolation and quantitative real-time PCR analysis;
XIST: X-inactive specific transcript;
Acknowledgements
This study was supported in part by research fund for National Natural Science Foundation of China (no. 81602624), Project of Science and Technology Department of Zhejiang Province (no. 2015C37012), The Science and Technology Creative Activity Plan for University Students in Zhejiang Province (no. 2016R410037).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hüttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289-97
2. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17-9
3. Santosh B, Varshney A, Yadava PK. Non-coding RNAs: biological functions and applications. Cell Biochem Funct. 2015;33:14-22
4. Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582-600
5. Jing Q, Huang S, Guth S. et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623-34
6. Zhu L, Xu PC. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochemical and Biophysical Res Commun. 2013;432:612-7
7. Hrdlickova B, de Almeida RC, Borek Z. et al. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842:1910-22
8. GeislerS CollerJ. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699-712
9. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313-6
10. Naidu S, Garofalo M. MicroRNAs: An Emerging Paradigm in Lung Cancer Chemoresistance. Front Med (Lausanne). 2015;2:77
11. Patel JS, Hu M, Sinha G. et al. Non-coding RNA as mediators in microenvironment-breast cancer cell communication. Cancer Lett. 2016;380:289-95
12. Ragusa M, Barbagallo C, Statello L. et al. Non-coding landscapes of colorectal cancer. World J Gastroenterol. 2015;21:11709-39
13. Guo LL, Song CH, Wang P. et al. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680-7
14. Wen PY, Kesari S. Malignant gliomas in adults. N Engl JMed. 2008;359:492-507
15. Tanaka S, Louis DN, Curry WT. et al. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat Rev Clin Oncol. 2013;10:14-26
16. Kiang KM, Zhang XQ, Leung GK. Long Non-Coding RNAs: The key Players in Glioma Pathogenesis. Cancers (Basel). 2015;7:1406-24
17. Shi J, Dong B, Cao J. et al. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017;8:27582-92
18. Barciszewska AM. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol. 2016;54:369-74
19. Jia P, Cai H, Liu X. et al. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016;381:359-69
20. Brown CJ, Ballabio A, Rupert JL. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38-44
21. Weakley SM, Wang H, Yao Q. et al. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751-6
22. Wang Z, Yuan J, Li L. et al. Long non-coding RNA XIST exerts oncogenic functions in human glioma by targeting miR-137. Am J Transl Res. 2017;9:1845-55
23. Ma L, Zhou Y, Luo X. et al. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget. 2017;8:4125-35
24. Zhang YL, Li XB, Hou YX. et al. The lncRNA XIST exhibits oncogenic properties via regulation of miR-449a and Bcl-2 in human non-small cell lung cancer. Acta Pharmacol Sin. 2017;38:371-81
25. Huang YS, Chang CC, Lee SS. et al. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediate drepression of PHLPP1 expression. Oncotarget. 2016;7:43256-66
26. Mo Y, Lu Y, Wang P. et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999
27. Yao Y, Ma J, Xue Y. et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating mir-152. CancerLett. 2015;359:75-86
28. Curry RC, Dahiya S, Alva Venur V. et al. Bevacizumab in high-grade gliomas: past, present, and future. Expert Rev Anticancer Ther. 2015;15:387-97
29. Ding H, Shen J, Yang Y. et al. Saw Palmetto Extract Inhibits Metastasis and Antiangiogenesis through STAT3 Signal Pathway in Glioma Cell. Evid Based Complement Alternat Med. 2015;2015:926946
30. Wang ZH, Guo XQ, Zhang QS. et al. Long non-coding RNA CCAT1 promotes glioma cell proliferation via inhibiting microRNA-410. Biochem Biophys Res Commun. 2016;480:715-20
31. Wang G, Li Z, Tian N. et al. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol Lett. 2016;12:879-86
32. Wang P, Cao J, Liu S. et al. Upregulated microRNA-429 inhibits the migration of HCC cells by targeting TRAF6 through the NF-κB pathway. Oncol Rep. 2017;37:2883-90
33. Han Y, Zhao Q, Zhou J. et al. miR-429 mediates tumor growth and metastasis in colorectal cancer. Am J Cancer Res. 2017;7:218-33
34. Zou J, Liu L, Wang Q. et al. Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am J Transl Res. 2017;9:1357-68
35. Peng Y, Chen FF, Ge J. et al. miR-429 Inhibits Differentiation and Promotes Proliferation in Porcine Preadipocytes. Int J Mol Sci. 2016;17:E2047
36. Wu CL, Ho JY, Chou SC. et al. MiR-429 reverses epithelial-mesenchymal transition by restoring E-cadherin expression in bladder cancer. Oncotarget. 2016;7:26593-603
37. Chen W, Zhang B, Guo W. et al. miR-429 inhibits glioma invasion through BMK1 suppression. J Neurooncol. 2015;125:43-54
Author contact
![]() Corresponding author: Dr Yi Wang, Department of Regenerative Medicine, School of Pharmaceutical Science, Jilin University, No.1266, Fujin Road, Changchun, P.R. China. E-mail address: wangyiedu.cn Tel.: +86-431-8561-9715 Fax: +86-431-8561-9715
Corresponding author: Dr Yi Wang, Department of Regenerative Medicine, School of Pharmaceutical Science, Jilin University, No.1266, Fujin Road, Changchun, P.R. China. E-mail address: wangyiedu.cn Tel.: +86-431-8561-9715 Fax: +86-431-8561-9715

 Global reach, higher impact
Global reach, higher impact