3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(19):4141-4154. doi:10.7150/jca.22076 This issue Cite
Review
Post-transcriptional Regulation of Genes Related to Biological Behaviors of Gastric Cancer by Long Noncoding RNAs and MicroRNAs
1. Tumor Etiology and Screening Department of Cancer Institute and General Surgery, the First Hospital of China Medical University, and Key Laboratory of Cancer Etiology and Prevention (China Medical University), Liaoning Provincial Education Department, Shenyang, 110001, Liaoning Province, P R China;
2. Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, NO. 44 Xiaoheyan Road, Dadong District, Shenyang 110042, Liaoning Province, P R China;
3. National Clinical Research Center for Digestive Diseases, Xi'an, 110001 China.
Received 2017-7-25; Accepted 2017-10-10; Published 2017-11-12
Abstract
Noncoding RNAs play critical roles in regulating protein-coding genes and comprise two major classes: long noncoding RNAs (lncRNAs) and microRNAs (miRNAs). LncRNAs regulate gene expression at transcriptional, post-transcriptional, and epigenetic levels via multiple action modes. LncRNAs can also function as endogenous competitive RNAs for miRNAs and indirectly regulate gene expression post-transcriptionally. By binding to the 3'-untranslated regions (3'-UTR) of target genes, miRNAs post-transcriptionally regulate gene expression. Herein, we conducted a review of post-transcriptional regulation by lncRNAs and miRNAs of genes associated with biological behaviors of gastric cancer.
Keywords: ncRNAs, lncRNAs, miRNAs, gastric cancer, post-transcriptional regulation.
Introduction
In humans, noncoding RNAs (ncRNAs) cannot be translated into protein and can be divided into 2 major categories according to their lengths: long noncoding RNAs (lncRNAs) and small noncoding RNAs (sncRNAs) [1]. LncRNAs exceed 200 nucleotides (nt) in length [2], whereas sncRNAs are less than 200 nt in length and include microRNAs of approximately 21 to 24 nt and Piwi-interacting RNAs [3]. It has been suggested that ncRNAs can regulate gene expression at transcriptional, post-transcriptional, and epigenetic levels, thereby participating in physiological and pathological processes [2, 4].
Post-transcriptional control refers to the regulation of gene expression after RNA transcription. It is a primary mechanism by which ncRNAs regulate gene expression, which is extensively involved in biology, evolution, and pathology [5-7]. LncRNAs mainly form RNA-RNA duplexes with mRNAs, occupy the binding sites of mRNAs and transacting factors [8], and influence mRNAs degradation and stability, thus regulating gene expression post-transcriptionally. LncRNAs can also act as translational regulators to modulate protein expression, degradation, transportation, distribution, and modification. Post-transcriptional regulation by miRNAs is mainly mediated by specific binding to the 3ʹ-UTR of target genes and leads to cytoplasmic degradation and translational inhibition. Aberrant control by lncRNAs and miRNAs is closely related to various human diseases, including cancer [9, 10], and contributes to tumor cell proliferation and apoptosis, invasion and metastasis, induction of tumor angiogenesis, and drug resistance [11]. Currently, mechanisms of post-transcriptional regulation by lncRNAs and miRNAs in cancer have become a hot topic.
Gastric cancer is one of the five most lethal malignant tumors in China, following lung and bronchial cancer [12]. A variety of oncogenes and tumor suppressor genes are involved in multistage and multistep of gastric carcinogenesis. In recent years, lncRNAs and miRNAs have been found to modulate biological behaviors of tumor cells and influence prognosis of patients. Identification of post-transcriptional regulation in gastric cancer can help to elucidate the molecular mechanisms involved in genesis and development of gastric cancer, thus providing new clues for clinical diagnosis and therapy.
We conducted a review of mechanisms involved in post-transcriptional regulation of gene expression related to cell proliferation, apoptosis, the epithelial-mesenchymal transition (EMT), metastasis, angiogenesis, and drug resistance. These mechanisms can be mediated by lncRNAs, miRNAs, and their interactions, causing a series of alterations in gastric cancer. The aims of this review were to summarize the mechanisms of post-transcriptional control of gene expression by which lncRNAs and miRNAs affect biological behaviors in gastric cancer, providing a theoretical basis for the identification of biomarkers for risk assessment and prognosis in gastric cancer as well as therapeutic targets.
Post-transcriptional regulation by lncRNAs in the expression of genes related to biological behaviors of gastric cancer
Most lncRNAs are transcribed by RNA polymerase II (Pol II) and processed with a 5ʹ cap and polyadenylation, similar to mRNA transcripts [13, 14]. The major differences between lncRNAs and mRNAs are that lncRNAs have little protein-coding capacity [15], contain fewer exons, and have weaker splicing and polyadenylation potentials [16]. Recent proteomic analysis has suggested that a putative lncRNA may encode short proteins or “micropeptides” [17]. Many lncRNAs function as molecular signals [18], decoys [19], guide molecules [20], or scaffolds [21]; these activities may be mediated by the expression of related genes, precipitating subcellular localization of lncRNAs to the nucleus or cytoplasm [22]. LncRNAs influence post-transcriptional regulation of genes associated with gastric cancer mainly by altering the process of mRNA degradation and stability and by regulating the expression, degradation, transportation, distribution, and modification of proteins. Therefore, lncRNAs are involved in proliferation, apoptosis, the EMT, metastasis, and drug resistance and contribute to a series of changes in biological behaviors of gastric cancer (Table 1, Figure 1, 3).
Post-transcriptional regulation by lncRNAs on the expression of genes related to proliferation and apoptosis in gastric cancer
Cell proliferation and apoptosis, when functioning appropriately, facilitate maintenance of a steady state, contributing to adaptability to the microenvironment of the human body. If the proliferative function overwhelms that of cellular apoptosis, tumorigenesis may occur. GHET1 (gastric carcinoma highly expressed transcript 1), an intergenic RNA, is located on chromosome 7 and has a length of 1913 nt. High expression of GHET1 is strongly related to poor prognosis in cancer [23]. Yang et al. demonstrated an association between GHET1 and IGF2BP1 (insulin-like growth factor binding protein 1) in gastric carcinoma (GC) cells by RNA immunoprecipitation (RIP) and RNA pull-down. Moreover, GHET1 enhances the interaction between IGF2BP and c-Myc mRNA. Consequently, GHET1 increases the stability and expression of c-Myc mRNA, enhances the numbers of EdU (ethyl deoxyuridine)-positive nuclei, and promotes proliferation of GC cells as well as colony formation.
The lncRNA GAS5 (growth arrest-specific transcript 5) is located on chromosome 1q25.1 and has a length of 650 nt [24]. It has been reported that GAS5, as a tumor suppressor, is down-regulated in gastric cancer, breast cancer, prostate cancer, and other malignant tumors [25-27]. Liu et al. determined that GAS5 was associated with the transcriptional activator YBX1 protein (human Y-box binding protein 1) and that GAS5 depletion promoted protein turnover of YBX1, inhibited the expression of the cell cycle regulator p21, decreased the percentage of cells arrested in G1 phase, and accelerated the cell cycle [28].
Linc-POU3F3 (long intergenic noncoding RNA POU3F3) is positioned on chromosome 2q12.1 and has a length of 2874 nt [29]. Expression of linc-POU3F3 is significantly increased in esophageal carcinoma and colorectal cancer and is strongly related to tumor size [29-31]. By using RIP and western blot, Xiong et al. found that up-regulation of linc-POU3F3 could directly combine with TGF-beta, caused the up-regulation of TGF-beta and phosphorylation of SMAD2 and SMAD3, activated the TGF-beta signaling pathway, ultimately enhanced proliferation of GC cells [31].
TINCR (terminal differentiation-induced ncRNA) is a noncoding RNA located on human chromosome 19p13.3 that has an approximate length of 3700 nt and regulates human epidermal differentiation [32]. Xu et al. demonstrated that highly expressed TINCR could mediate stability and degradation of KLF2 mRNA by acting with TINCR-STAU1, resulting in decreased expression of CDKN1A/P21 and CDKN2B/P15, accelerated cell growth, redistribution of G0/G1 and S phases of the cell cycles well as promotion of cell proliferation, and suppressed apoptosis of GC cells [33].
Post-transcriptional regulation by lncRNAs on the expression of genes related to biological behaviors of gastric cancer.
| LncRNA names | Target genes | Potential mechanisms | Biological behaviors involved | Clinicopathological features |
|---|---|---|---|---|
| 1 Post-transcriptional regulation of lncRNAs on the expression of genes related to proliferation and apoptosis of gastric cancer | ||||
| GHET1 | c-Myc | Up-regulated GHET1 enhances the interaction between IGF2BP and c-Myc mRNA. Consequently, GHET1 increases the stability and expression of c-Myc mRNA in GC cells. | promote proliferation and colony formation | up-regulation of GHET1 is associated with poor prognosis |
| GAS5 | YBX1 protein | Over-expressed GAS5 is associated with the transcriptional activator YBX1 protein and GAS5 depletion promotes protein turnover of YBX1. | GAS5 depletion decrease the percentage of cells arrested in G1 phase | low-GAS5 is related to larger tumor size, greater invasion depth, more lymph nodes and higher tumor stage |
| Linc-POU3F3 | TGF-beta protein | By using RIP and western blot, Xiong et al. found that up-regulation of linc-POU3F3 could directly combine to TGF-beta, causing the up-regulation of TGF-beta and phosphorylation of SMAD2 and SMAD3, activated the TGF-beta signaling pathway. | enhance proliferation | high expression of linc-POU3F3 is strongly associated with tumor size |
| lncRNA TINCR | KLF2 | TINCR could mediate stability and degradation of KLF2 mRNA by acting with TINCR-STAU1. | promote proliferation and suppress apoptosis | highly expressed TINCR is related to advanced TNM stage and deeper invasion depth |
| MALAT1 | SF2/ASF protein | Up-regulated MALAT1 induces SF2/ASF nuclear distribution and expression. | enhance proliferation, decrease the percentage of cells arrested in G0/G1 phase | elevated expression of MALAT1 is strongly associated with poor prognosis |
| 2 Post-transcriptional regulation by lncRNAs in the expression of genes related to the EMT and metastasis of gastric cancer | ||||
| linc00261 | Slug protein | Over-expressed linc00261 may promote degradation of Slug protein by facilitating the interaction between Slug and GSK3β. | inhibit the EMT and invasion | low-linc00261 is associated with invasion depth, lymphatic metastasis and TNM stage |
| E-cadherin, N-cadherin, Vimentin and FN1 protein | Up-regulation of linc00261 inhibits the EMT by downregulating N-cadherin, Vimentin, and FN1 proteins. | prevent migration, invasion and metastasis | ||
| PVT1 | FOXM1 protein | PVT1 interacts with FOXM1 and promotes the protein expression of FOXM1 in GC cells. | enhance growth and metastasis | up-regulation of PVT1 is associated with invasion depth, lymphatic metastasis and tumor stage |
| lncRNA SNHG5 | MTA2 protein | Over-expressed SNHG5 prevents translocation of MTA2 protein from the cytoplasm to the nucleus by interacting with MTA2. | suppress growth, colony formation, invasion and metastasis | reduced expression of SNHG5 is related to TNM stage and tumor embolus |
| WT1-AS | ERK protein | WT1-AS overexpression in GC cells reduces the phosphorylation level of ERK protein without affecting the expression of ERK mRNA. | inhibit proliferation, clonal formation, migration and invasion, elevate the ratio of cells in G1/G0 phase, and reduce the percentage of cells in S and G2/M phases | significant down-regulation of WT1-AS is correlated with tumor size and clinical stage |
| lncRNA KRT7-AS | KRT7 | KRT7-AS upregulates the mRNA and protein expression of KRT7 and prevents KRT7 mRNA from being degraded by forming an RNA-RNA duplex. | enhance growth, proliferation, migration, increase cell percentages in the S+G1/M stage | unknown |
| 3 Post-transcriptional regulation by lncRNAs in the expression of genes related to multidrug resistance and angiogenesis in gastric cancer | ||||
| PVT1 | MDR1, MRP1 | Up-regulation of PVT1 in GC elevates the mRNA and protein expression of MDR1 and MRP1. | inhibit apoptosis, induce cisplantin resistance | up-regulation of PVT1 is associated with invasion depth, lymphatic metastasis, regional lymph nodes and tumor stage |
| ANRIL | MDR, MRP1 | ANRIL is up-regulated in cisplatin- and 5-FU-resistant GC tissues and cells, and reduces the mRNA and protein of MDR1 and MRP1. | induce proliferation, migration, invasion, promote cisplantin- and 5-FU-resistance, inhibit apoptosis, | up-regulation of ANRIL is strongly linked to higher TNM stage and tumor size |
| MALAT1 | VM-related protein | MALAT1 promotes the expression of VM-related proteins and associated signaling pathways. | promote migration, invasion, metastasis, VM formation and angiogenesis | highly expressed MALAT1 is positively correlated with VM and endothelial vessel (EV) density |
Post-transcriptional regulation by lncRNAs on the expression of genes related to biological behaviors of gastric cancer. LncRNAs influence post-transcriptional regulation of genes associated with biological behaviors in gastric cancer by altering the process of mRNA degradation and stability or by regulating the expression, degradation, transportation, distribution, and modification of proteins.
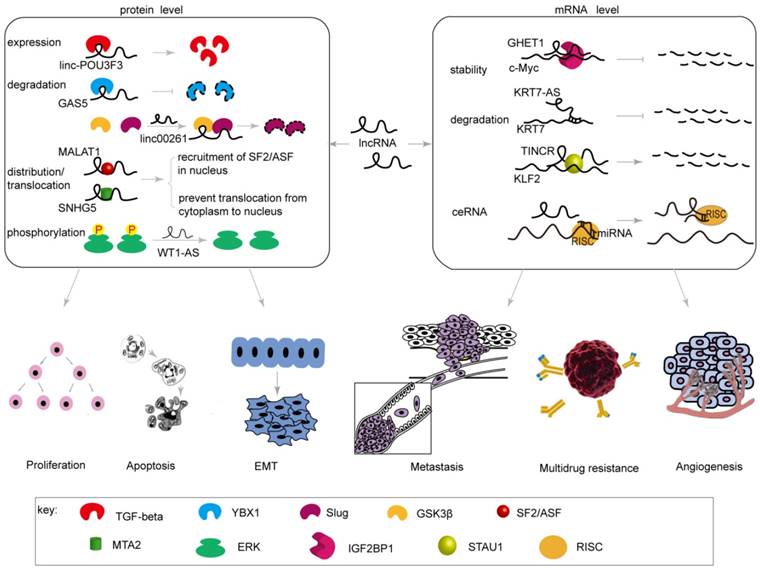
MALAT1 (lung adenocarcinoma metastasis-related transcript 1) is located on chromosome 11q13 and has a length of approximately 6.7 kb [34, 35]. MALAT1 is primarily localized to nuclear splice spots and modulates distribution and phosphorylation of SR (serine/arginine) splicing factors [36, 37]. Up-regulated MALAT1 is strongly linked to poor prognosis in GC [38]. Wang et al. [39] found that the expression of MALAT1 and SF2/ASF protein were significant higher in GC cells. Depletion of MALAT1 may prohibit the distribution of SF2/ASF in the nucleus, thereby inhibiting cell cycle progression and cell proliferation. These authors noted that MALAT1 might affect cell proliferation and cell cycle progression by mediating nuclear distribution of SF2/ASF and its expression in gastric cancer.
Post-transcriptional regulation by lncRNAs in the expression of genes related to the EMT and metastasis of gastric cancer
The EMT (Epithelial-Mesenchymal Transition) has significant effects on tumor invasion and metastasis. In the EMT, epithelial cells obtain migratory abilities and morphologically transform into mesenchymal cells, leading to cell invasion and migration [40]. Slug, a key regulator of the EMT, is highly expressed during this process and is notably associated with malignant phenotypes in GC [41]. Investigators have demonstrated that phosphorylated Slug protein—mediated by GSK3β—proceeds through ubiquitination and proteasome degradation in breast cancer and non-small cell lung cancer [42, 43]. Yu et al. [41] reported that linc00261 might promote degradation of Slug protein as well as inhibition of the EMT and of GC cell invasion by facilitating the interaction between Slug and GSK3β. The vivo data in an animal model indicated that overexpression of linc00216 alleviated GC cell metastasis of the lung. Fan et al. [44] found that up-regulation of linc00261 inhibited the EMT by decreasing N-cadherin, Vimentin, and FN1 proteins and played vital roles in preventing migration, invasion, and metastasis of GC cells.
The lncRNA PVT1 is an intergenic noncoding RNA positioned on chromosome 8q24, close to MYC gene [45]. Up-regulation of PVT1 is found in various malignant tumors [46, 47] and is associated with invasion depth, lymphatic metastasis, regional lymph nodes, and more advanced malignant tumor stage [48, 49]. Xu et al. [50] found that PVT1 interacted with FOXM1 and promoted expression of FOXM1 protein in GC cells. FOXM1 activates transcription of PVT1 by directly binding to its promoter, forming a positive feedback loop of PVT1-FOXM1 that enhances growth and metastasis of GC cells [50]. In addition, PVT1 overexpression significantly facilitates lung metastases in nude mice [50].
The lncRNA SNHG5 (small nucleolar RNA host gene 5) is positioned on chromosome 6q14.3 and is down regulated in many malignant tumors [19]. Expression of SNHG5 is correlated with TNM (Tumor Node Metastasis) stage and tumor embolus [51]. Zhao et al. [51] demonstrated that SNHG5 prevented translocation of MTA2 protein from the cytoplasm to the nucleus by interacting with MTA2 in the cytosol, activated MTA2-mediated signal pathways, such as KAI-1 and E-cadherin, suppressed growth of GC cells, colony formation, invasion, and metastasis. Moreover, growth of SNHG5-overexpressing cells is inhibited substantially in subcutaneous xenografts, resulting in fewer lung metastatic nodules [51].
The lncRNA WT1-AS is an antisense transcript of WT1 (Wilms' tumor gene) [52] that is positioned on chromosome 11p13. Significant down-regulation of WT1-AS is correlated with tumor size and clinical stage in GC tissues [53]. WT1-AS undergoes aberrant splicing in acute myeloid leukemia, suggesting a vital role of this lncRNA in malignant tumors [54]. Du et al. [53] found that WT1-AS overexpression in GC cells reduced the phosphorylation level of ERK protein without affecting the mRNA expression of ERK. WT1-AS may regulate ERK phosphorylation, thereby remarkably inhibiting cell proliferation, clonal formation, migration, and invasion in GC by elevating the percentage of cells in the G1/G0 phase and reducing the ratio of cells in the S and G2/M phases.
The lncRNA KRT7-AS (KRT7 antisense RNA 1) is an antisense transcript that is positioned on chromosome 12q13. Huang et al. [55] demonstrated that the lncRNAs KRT7-AS was remarkable up-regulated and possessed the same expression pattern with KRT7 in GC tissues and cells. KRT7-AS upregulates the mRNA and protein expression of KRT7 and prevents KRT7 mRNA from being degraded by forming an RNA-RNA duplex. This enhances cell growth, proliferation, and migration and increases cell percentages in the S+G1/M stage. In addition, researchers have found that the secondary structure of the RNA-RNA duplex close to the poly(A) tail protects KRT7 mRNA from being degraded by the ribonucleolytic RNA exosome or by miRNAs [56]. KRT7-AS may facilitate transportation from nuclear to cytoplasm of KRT7 mRNA and its interaction with ribosomes.
Post-transcriptional regulation by lncRNAs in the expression of genes related to multidrug resistance (MDR) and angiogenesis in gastric cancer
Multidrug resistance comprises a series of pathological processes involving gene expression or suppression and distribution and utilization of chemotherapy drugs in tumor cells [57, 58]. Several studies have suggested that many lncRNAs participate in multidrug resistance, including ANRIL [59], AK022798 [60], MRUL [61], and CASC9 [62]. Zhang et al. [63] found that up-regulation of PVT1 in GC elevated the mRNA and protein expression of MDR1 and MRP1—which are involved in cisplatin resistance—thereby inhibiting cell apoptosis. Lan et al. [59] observed that ANRIL was up-regulated in cisplatin- and 5-FU-resistant GC tissues and cells; down-regulation of ANRIL in GC cells led to inhibition of cell proliferation, migration, and invasion; promotion of apoptosis; reduction of mRNA and protein of MDR1 and MRP1; and reversal of chemotherapy resistance in GC cells.
Vasculogenic mimicry (VM)—a recently described concept in tumor angiogenesis—provides blood nutrition for sustainable growth of tumor cells [64, 65]. Li et al. [66] found that MALAT1, highly expressed in GC tissues, was positively correlated with VM and endothelial vessel (EV) density and promoted migration, invasion, and metastasis of GC cells. MALAT1 affects VM formation and angiogenesis by mediating the expression of VM-related proteins and associated signaling pathways.
Post-transcriptional regulation by miRNAs in the expression of genes related to biological behaviors of gastric cancer
MiRNAs are sncRNAs with lengths of approximately 22 nt that regulate target genes at the post-transcriptional level. MiRNAs silence or inhibit target genes through 5ʹ seed sequences of 2 to 8 nt that interact with the 3ʹ-UTR of mRNAs completely or incompletely. More than 50% of miRNAs are positioned near tumor-related areas or brittle loci in the genome [67], suggesting that miRNAs participate in carcinogenesis. MiRNAs are involved in proliferation, apoptosis, the EMT, metastasis, and drug resistance by affecting post-transcriptional regulation of mRNAs, which resulting in a series of changes related to biological behaviors in gastric cancer (Table 2, Figure 2, 3).
Post-transcriptional regulation by miRNAs in the expression of genes related to proliferation and apoptosis in gastric cancer
MiR-24 is up-regulated in GC [68], and its expression is closely related to cancer behaviors [69]. Bioinformatics analyses predicted that miR-24 might be the upstream regulator of BCL2L11 [68], an important mediator of cell apoptosis [70, 71]. By means of the luciferase assay, Zhang et al. [68] confirmed that miR-24 directly binded to the 3'-UTR of BCL2L11 mRNA and inhibited mRNA expression of BCL2L11 post-transcriptionally, thus promoted cell proliferation and migration, inhibited apoptosis in GC.
MiR-1207-5p and miR-1266 are remarkable lower in GC tissues. Up-regulation of miR-1266 is correlated with a longer survival time [72, 73]. Chen et al. [72] screened 14 down-regulated miRNAs that potentially regulated hTERT (human telomerase reverse transcriptase) using a combination of an miRNA array and bioinformatics analysis in GC tissues. These authors found that miR-1207-5p and miR-1226 repressed hTERT expression at the post-transcriptional level by directly combining with its 3ʹ-UTR, suppressing the G1-to-S transition. Moreover, miR-1207-5p- and miR-1226-mediated inhibition of cell proliferation prohibit growth of transplanted SGC-7901 cells in xenografts in nude mice.
Post-transcriptional regulation by miRNAs in the expression of genes related to biological behaviors of gastric cancer
| MiRNA names | Target genes | Potential mechanisms | Biological behaviors involved | Clinicopathological features |
|---|---|---|---|---|
| 1 Post-transcriptional regulation by miRNAs in the expression of genes related to proliferation and apoptosis in gastric cancer | ||||
| miR-24-3p | BCL2L11 | MiR-24-3p directly binds to the 3ʹ-UTR of BCL2L11 mRNA and inhibits mRNA expression of BCL2L11 post-transcriptionally. | enhance proliferation, migration, inhibit apoptosis | highly expressed miR-24-3p is correlated with lymph node and liver metastasis |
| miR-1207-5p, miR-1266 | hTERT | MiR-1207-5p and miR-1226 repress hTERT expression at the post-transcriptional level by directly combining with its 3ʹ-UTR. | inhibit proliferation and invasion, suppress the G1-to-S transition | higher expressed miR-1266 is correlated with a longer survival time |
| miR-7 | RELA, FOS, IKKε | When over-expressed, miR-7 targets the 3ʹ-UTR of RELA and FOS and inhibits their protein expression. MiR-7 also mediates RELA indirectly by suppressing the protein level of IKKɛ; in turn, IKKɛ/RELA inhibits miR-7, forming a miR-7/IKKɛ/RELA feedback loop. | inhibit proliferation, colony formation and enhance apoptosis, induce cells to arrest in the G0/G1 phase | decreased miR-7 is correlated with tumor size, grade of differentiation and TNM stage |
| miR-338-3p | P-Rex2a | MiR-338-3p targets the 3ʹ-UTR of P-Rex2a, thus directly inhibiting its translation. | repress proliferation and apoptosis, and reduce cell cycle progression from G1 to S | down-regulated miR-338-3p is associated with TNM stage and invasion depth |
| 2 Post-transcriptional regulation by miRNAs in the expression of genes related to the EMT and metastasis in gastric cancer | ||||
| miR-181a-5p | RASSF6 | MiR-181a-5p negatively controls RASSF6 mRNA and protein by binding to the 3ʹ-UTR of its mRNA. | enhance proliferation, invasion, EMT and peritoneal metastasis, increase the percentage of cells in the S phase | overexpression of miR-181a-5p is correlated with TNM stage, UICC stage, and invasion into vessels and nerves |
| miR-149 | IL-6 | Over-expressed miR-149 targets the 3ʹ-UTR of IL-6 mRNA to negatively regulate IL-6 expression. | inhibit EMT | decreased miR-149 is closely related to tumor differentiation, lymph node metastasis and TNM stage |
| miR-3978 | LGMN | LGMN (which encodes legumain) may be a potential target of miR-3978, miR-3978 interacts with the 3ʹ-UTR of LGMN and suppresses legumain protein. | inhibit proliferation, migration, invasion and peritoneal metastasis | down-expressed miR-3978 is linked to peritoneal metastasis |
| miR-217 | EZH2 | MiR-217 could reduce luciferase activity of Wt-EZH2-3'-UTR and protein expression of EZH2 by binding to the 3ʹ-UTR of EZH2 mRNA. | inhibit proliferation, migration and invasion, restrain liver and lung metastasis | lower expressed miR-217 is strongly correlated with poor differentiation, metastasis, advanced TNM stage and large tumor size |
| 3 Post-transcriptional regulation by miRNAs in the expression of genes related to multidrug resistance and angiogenesis in gastric cancer | ||||
| miR-29c | catenin-δ | MiR-29c is up-regulated by certain chemotherapeutic drugs in GC cells, which prohibits its recently described target catenin-δ. | inhibit migration, invasion, liver and lung metastasis, mediate drug resistance | down-regulated miR-29c is correlated with widespread venous invasion and advanced TNM stage |
| miR-939 | SLC34A2 | MiR-939 combines with the 3ʹ-UTR of SLC34A2, decreases expression of SLC34A2 mRNA and protein. | suppress proliferation, migration and invasion, induced apoptosis and promoted 5-fluorouracil-induced sensitivity | miR-939 is correlated with chemotherapy response |
| miR-125a | VEGF-A | MiR-125a inhibits tumor angiogenesis by binding to the 3ʹ-UTR of VEGF-A mRNA, thus suppresses mRNA and protein expression of VEGF-A. | inhibit HUVECs proliferation, migration and angiogenesis | miR-125a is negatively related to microvessel density |
| miR-126 | VEGF-A | Lenti-miR-126 reduces expression of VEGF-A in GC cells. | inhibit tumorigenesis and angiogenesis of xenografts in nude mice | miR-126 is negatively related to microvessel density |
Post-transcriptional regulation by miRNAs in the expression of genes related to biological behaviors of gastric cancer. MiRNAs, produced or epigenetically mediated by lncRNAs, silence target genes through interacting with the 3ʹ-UTR, resulting in a series of changes related to biological behaviors in gastric cancer.
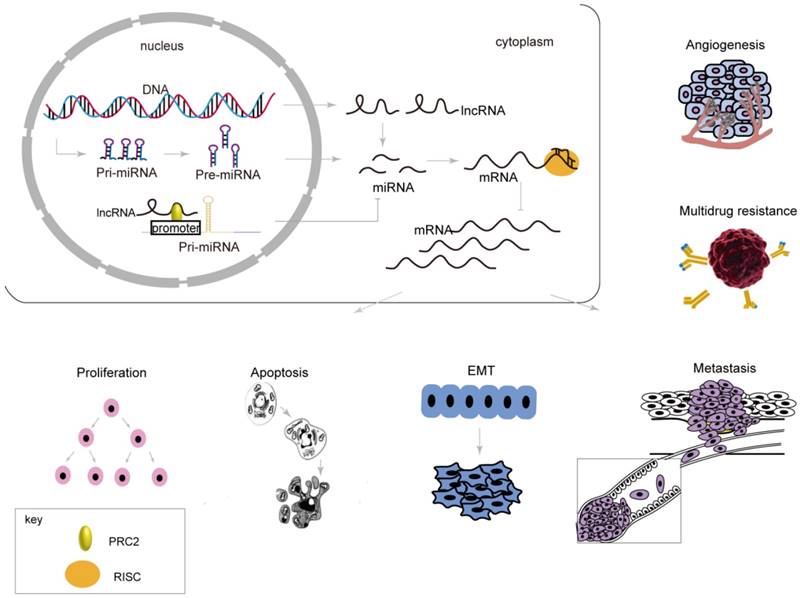
MiR-7 plays a tumor suppressor role, inhibiting cell proliferation and other biological behaviors; miR-7 is down-regulated in numerous tumors [74]. Zhao et al. [75] determined that miR-7 had a lower expression in GC tissues and was negatively correlated with expression of the target genes RELA and FOS. When over-expressed, miR-7 targets the 3ʹ-UTR of RELA and FOS, inhibits their protein expression, represses cell proliferation, induces cells to arrest in the G0/G1 phase, and prohibits growth of GC cells in xenografts in nude mice. MiR-7 also mediates RELA indirectly by suppressing the protein level of IKKɛ; in turn, IKKɛ/RELA inhibits miR-7, forming a miR-7/IKKɛ/RELA feedback loop that regulates proliferation, colony formation, and apoptosis of GC.
MiR-338-3p is down-regulated in hepatocellular carcinoma and GC tissues [76, 77]. It was found that miR-338-3p served as a tumor suppressor by decreasing cell proliferation, and inducing cell cycle progression from G1 to S and cell apoptosis [77]. Guo et al. [77] also determined that miR-338-3p targeted the 3ʹ-UTR of P-Rex2a, thus directly inhibiting its translation. Mechanistically, miR-338-3p activates PTEN and suppresses phosphorylation of AKT by silencing P-Rex2a; in turn, this represses cell proliferation, induces cell cycle arrest in G1-S, and promotes apoptosis of GCs.
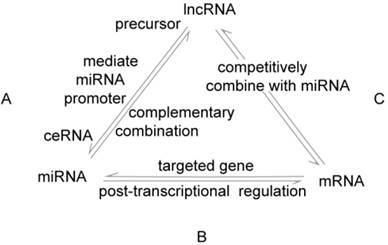
The interactions between lncRNAs, miRNAs and mRNAs. A. There are several links between lncRNAs and miRNAs in GC: lncRNAs produce miRNAs through intracellular splicing; lncRNAs silence miRNAs by epigenetically mediation; lncRNAs regulate the expression of transcripts by combining competitively with miRNAs, called ceRNAs mechanism. B. MiRNAs affect post-transcriptional regulation by interacting with the 3ʹ-UTR of mRNAs. C. LncRNAs compete with mRNAs to combine with miRNAs.
Post-transcriptional regulation by miRNAs in the expression of genes related to the EMT and metastasis in gastric cancer
MiR-181a-5p is up-regulated in breast cancer and acute lymphoblastic leukemia [78, 79]. Overexpression of miR-181a-5p affects TNM stage, UICC stage, and invasion into vessels and nerves [80]. In an analysis of how miR-181a-5p regulates RASSF6, Mi et al. [80] found that miR-181a-5p negatively controlled RASSF6 mRNA and protein by combining with the 3ʹ-UTR. Ectopic expression of miR-181a-5p facilitates cell proliferation, invasion, and the EMT, and elevates the ratio of cells in the S phase and peritoneal metastasis in vitro and in vivo; these effects can be partially reversed by overexpression of RASSF6.
MiR-149 is methylated—and thusobviously decreased—in gastric cancer and is closely related to tumor differentiation, lymph node metastasis, and TNM staging [81, 82]. Down-regulation of miR-149 is related to cell differentiation, lymph node metastasis, and more advanced TNM stage [81, 82]. Li et al. [82] found that miR-149 prevented fibroblast activation and inhibited carcinogenesis of cancer-associated fibroblasts (CAFs) by targeting the 3ʹ-UTR of IL-6 mRNA to negatively regulate IL-6expression. Suppression of miR-149 in GC promotes IL-6 expression, indicating that CAFs enhance the EMT and stem-like characteristics of GC cells according to the miR-149/IL-6 axis, which promotes GC.
MiR-3978 is substantially decreased in gastric cancer, especially in peritoneal metastasis [83], indicating that down-regulation of miR-3978 might be predictive for poor prognosis in GC. By using a murine model, Zhang et al. [83] found that LGMN (which encodes legumain) might be a potential target of miR-3978 because miR-3978 inhibited GC cell proliferation, migration, and invasion and restrained peritoneal metastasis of gastric cancer by interacting with the 3ʹ-UTR of LGMN and suppressing legumain protein. Legumain is dramatically increased in metastatic gastric cancer, suggesting that this factor is also indicative for poor prognosis [84, 85].
MiR-217 is significantly decreased in pancreatic ductal adenocarcinoma (PDAC) and clear cell renal cell carcinoma (ccRCC), exhibiting anticancer characteristics [86, 87]. Chen et al. [88] found that miR-217 was suppressed in GC tissues and cells and was strongly correlated with poor differentiation, metastasis, advanced TNM stage, and large tumor size. MiR-217 inhibits proliferation, migration, and invasion of GC cells and restrains liver and lung metastasis in xenografts in nude mice; in contrary, EZH2 promotes cell proliferation and invasion. Bioinformatics analysis showed that miR-217 could reduce luciferase activity of Wt-EZH2-3'-UTR and protein expression of EZH2 by binding to the 3ʹ-UTR of EZH2 mRNA, which revealed the regulatory mechanism of miR-217/EZH2 in proliferation, migration, and invasion in GC.
Post-transcriptional regulation by miRNAs in the expression of genes related to MDR and angiogenesis in gastric cancer
The miR-29 family contains three members (miR-29a, miR-29b, and miR-29c) and is known for inhibiting cell proliferation and invasion in gastric cancer [89]. Wang et al. [90] found that down-regulated miR-29c was correlated with more widespread venous invasion and more aggressive TNM stage in gastric cancer. MiR-29c is elevated by cisplatin and docetaxel treatment in GC cells and inhibits migration, invasion, and liver and lung metastases in GC cells in nude mice. MiR-29c is up-regulated by certain chemotherapeutic drugs in GC cells, which prohibits its recently described target catenin-δ, resulting in suppressed proliferation, invasion, and metastasis of GC cells and reversal of drug resistance in GC cells [90].
MiR-939 is deregulated in gastric cancer tissues [91]. Zhang et al. [92] found that overexpression of miR-939 in GC cells sharply suppressed cell proliferation, migration, and invasion; induced apoptosis; and promoted 5-fluorouracil-induced sensitivity in GC cells. Subsequent studies demonstrated that miR-939 expression was associated with reduced cell migration, improved chemosensitivity, and decreased expression of SLC342 mRNA and protein; specifically, miR-939 combined with the 3ʹ-UTR of SLC34A2, and blocked the SLC34A2/Raf/MEK/ERK pathway.
Both miR-125a and miR-126 regulate tumor angiogenesis by mediating expression of VEGF-A (vascular endothelial growth factor A) [93, 94]. Dai et al. [94] demonstrated that miR-125a was significantly decreased in GC and was inversely related to microvessel density (MVD) and VEGF-A expression. Down-regulation of miR-125a facilitates VEGF-A secretion and promotes Akt phosphorylation, thereby promoting cell proliferation, migration, and angiogenesis of human umbilical vein endothelial cells (HUVECs). MiR-125 also inhibits tumor angiogenesis by binding to the 3ʹ-UTR of VEGF-A mRNA, thus suppressing mRNA and protein expression of VEGF-A. Chen et al. [93] showed that transfected lenti-miR-126 substantially inhibited tumorigenesis and angiogenesis of xenografts in nude mice by reducing expression of VEGF-A in GC cells.
Post-transcriptional regulation associated with interactions between lncRNAs and miRNAs of the expression of genes related to biological behaviors in gastric cancer
An understanding of the associations between lncRNAs and miRNAs can lend insight into the mechanisms that modulate GC. There are several relationships between lncRNAs and miRNAs in GC: (1) lncRNAs produce miRNAs through intracellular splicing; (2) lncRNAs silence miRNAs by epigenetically mediating the promoters of miRNAs, thus enhancing target genes indirectly; (3) lncRNAs act as endogenous “miRNA sponges” that combine competitively with miRNAs via miRNA response elements (MREs) and regulate the expression of transcripts, a process called the ceRNAs (competing endogenous RNAs) mechanism. LncRNAs and miRNAs are involved cooperatively in proliferation, apoptosis, the EMT, metastasis, and drug resistance by affecting post-transcriptional regulation of mRNAs to produce a series of changes in biological behaviors associated with gastric cancer (Table 3, Figure 1, 2, 3).
Post-transcriptional regulation of interactions between lncRNAs and miRNAs affecting the expression of genes related to proliferation and apoptosis in gastric cancer
H19, located on chromosome 11p15, is thought to have carcinogenic effects on malignant tumors, such as gastric cancer [95-97]. H19 gives rise to miR-675, a vital medium in H19-induced carcinogenesis [98-100]. H19 enhances cell proliferation and inhibits apoptosis by producing miR-675, which suppresses mRNA and protein expression of RUNX1 (tumor-suppressor runt domain transcription factor 1) [101]. H19/miR-675 axis accelerates the activation of the AKT/mTOR signaling pathway by deregulating miR-675 target, RUNX1, and promotes proliferation, colony formation, and migration of GC cells [102]. Zhou et al. [103] noted that H19 binded to miR-141 at a specific sequence and attenuated the negative effect of miR-141 on its target gene ZEB1, thereby promoting proliferation and migration of GC cells.
The lncRNA ANRIL, located on chromosome 9p21, has been identified in familial melanoma [104] where it is mainly expressed in the nucleus. Up-regulation of ANRIL is strongly linked to higher TNM stage and tumor size [105]. Zhang et al. determined that ANRIL inhibited miR-99a/miR-449a by recruiting PRC2 to combine with and inducing methylation of the promoters of miR-99a/miR-449a. In this way, ANRIL indirectly enhances the expression of mTOR and CDK6. Down-regulated miR-449a activates ANRIL indirectly, generating a positive feedback loop that promotes proliferation, cell cycle progression, and diminishes apoptosis in GC cells.
SNHG5 has been demonstrated to lower in GC. To illuminate the potential effect, Zhao et al. [106] found that SNHG5 decreased expression of miR-32 post-transcriptionally, whereas miR-32 inhibited SNHG5 in an Ago-2-dependent manner. It was suggested that miR-32 inhibited KLF4 in a similar way, while overexpression of SNHG5 activated KLF4 mRNA via its 3ʹ-UTR as a decoy for miR-32. This miR-32 mimic also attenuates SNHG5-mediated inhibition of cell proliferation. This study suggested an essential role of SNHG5/miR-32/KLF4 in promoting proliferation, migration, and invasion of GC cells.
The lncRNA BC032469 is highly expressed in GC and is correlated with tumor survival time and expression of human telomerase reverse transcriptase (hTERT), BC032469 is negatively correlated with miR-1207-5p [107]. Lu et al. [107] indicated that BC032469 inhibits miR-1207-5p by directly interacting with the complementary sequence of miR-1207-5p. Furthermore, overexpression of BC032469 substantially enhances expression of hTERT protein and cell proliferation, which are lowered by miR-1207. This study suggested that BC032469 modulates GC cell proliferation, colony formation, and the G1-S transformation. Hence, this lncRNA might act as a ceRNA to downregulate hTERT via miR-1207-5p.
The lncRNA FER1L4 is obviously decreased in gastric cancer [108], and FER1L4 and PTEN are targeted by miR-106a-5p [109]. Down-regulated FER1L4 liberates miR-106a-5p, thus diminishing mRNA and protein expression of PTEN and regulating GC cell proliferation as well as cell cycle progression from G0/G1 to S [109]. Shao et al. [110] constructed a regulatory network of RMRP-miR-206-Cyclin D2 and determined that RMRP promoted cell growth and proliferation and regulated the G0/G1 to S transition by functioning as a ceRNA of miR-206 and regulating Cyclin D2.
Post-transcriptional regulation of interactions between lncRNAs and miRNAs in the expression of genes related to the EMT and metastasis in gastric cancer
Down-regulated E-cadherin and expression of mesenchymal hallmarks such as vimentin are key markers of the EMT transition [111, 112]. HOX transcript antisense RNA (HOTAIR), located on chromosome 12q13, is the first identified lncRNA to exhibit trans-transcriptional regulation. Elevated expression of HOTAIR is closely related to lymph node metastasis and vascular invasion [113]. HOTAIR silences miR-34a which is mediated by histone modification of the miR-34a promoter. This enhances the targets of miR-34a (C-Met and Snail), and induces the EMT among epithelial cells in GC. Another study indicated that HOTAIR interacted competitively with miR-331-3p as a ceRNA to upregulate HER2 expression, which was negatively mediated by miR-331-3p post-transcriptionally; this ultimately promoted growth, migration, and invasion of gastric cancer cells and inhibited apoptosis [114].
Post-transcriptional regulation associated with interactions between lncRNAs and miRNAs of the expression of genes related to biological behaviors in gastric cancer
| LncRNA names | MiRNA names | Target genes | Potential mechanisms | Biological behaviors involved | Clinicopathological features |
|---|---|---|---|---|---|
| 1 Post-transcriptional regulation of interactions between lncRNAs and miRNAs affecting the expression of genes related to proliferation and apoptosis in gastric cancer | |||||
| H19 | miR-675 | RUNX1 | H19 can produce miR-675, which suppresses the mRNA and protein expression of RUNX1. | enhance proliferation, colony formation and migration and inhibit apoptosis | over-expressed H19 is linked to TNM stage |
| miR-141 | ZEB1 | H19 binds to miR-141 at a specific sequence and attenuates the negative effect of miR-141 on its target gene ZEB1. | promote proliferation and migration | ||
| ANRIL | miR-99a /miR-449a | mTOR, CDK6 | ANRIL inhibits miR-99a/miR-449a by recruiting PRC2 to combine with and inducing methylation of the promoters of miR-99a/miR-449a. | contribute to proliferation, reduce cells to arrest in the G0-G1 phase and apoptosis | up-regulation of ANRIL is strongly linked to higher TNM stage and tumor size |
| SNHG5 | miR-32 | KLF4 | MiR-32 inhibits KLF4, whereas overexpression of SNHG5 activates KLF4 mRNA via its 3ʹ-UTR as a decoy for miR-32. | inhibit proliferation, migration and invasion | reduced expression of SNHG5 is related to TNM stage and tumor embolus |
| lncRNA BC032469 | miR-1207-5p | hTERT | BC032469 inhibits miR-1207-5p by directly interacted with the complementary sequence of miR-1207-5p. Furthermore, overexpression of BC032469 substantially enhances expression of hTERT protein, which is lowered by upregulation of miR-1207-5p. | promote proliferation, colony formation and G1-S transformation | up-regulated lncRNA BC032469 is linked to greater tumor volume, poor tumor differentiation and shorter survival time |
| FER1L4 | miR-106a-5p | PTEN | FER1L4 and PTEN are targeted by miR-106a-5p. Down-regulated FER1L4 liberates miR-106a-5p, thus diminishing mRNA and protein expression of PTEN. | inhibit proliferation, cell cycle G0/G1 to S transition | low-FER1L4 is related to tumor size, TNM stage, histologic grade, invasion depth, lymphatic and distant metastasis, and vessel or nerve invasion |
| 2 Post-transcriptional regulation of interactions between lncRNAs and miRNAs in the expression of genes related to the EMT and metastasis in gastric cancer | |||||
| HOTAIR | miR-34a | C-Met, Snail | HOTAIR silences miR-34a by recruiting the PRC2 complex to the miR-34a promoter and mediating histone modification of the promoter. This enhances the targets of miR-34a (C-Met and Snail). | induce EMT | up-regulated HOTAIR is related to greater tumor volume, vascular invasion, lymph node metastasis and shorter overall survival |
| miR-331-3p | HER2 | HOTAIR interacts competitively with miR-331-3p as a ceRNA to upregulate HER2 expression, which is negatively mediated by miR-331-3p post-transcriptionally. | promote growth, migration and invasion, inhibit apoptosis | ||
| H19 | miR-675 | CALN1 | H19 and miR-675 exert similar functions: down-regulation of the miR-675 target gene CALN1. | promote invasion and metastasis | over-expressed H19 is linked to TNM stage |
| CCAT1 | miR-490 | hnRNPA1 | MiR-490 targets CCAT1, whereas CCAT1 decreases expression of miR-490. MiR-490 decreases translation of its target mRNA hnRNPA1 by combining with its 3ʹ-UTR, and miR-490 inhibitors enhance hnRNPA1, which can be reversed by CCAT1 siRNA. | enhance migration | Overexpression of CCAT1 is highly associated with TNM stage |
| Lnc XIST | miR-101 | EZH2 | XIST is negatively associated with miR-101 in GC, and miR-101 down-regulates its target EZH2. XIST knockdown decreases EZH2, and miR-101 inhibitor increases EZH2. | promote proliferation, apoptosis, migration | highly expressed Lnc XIST is correlated with tumor size, lymph node metastasis, and clinical stage |
| miR-497 | MACC1 | lncRNA XIST promotes expression of the target gene of miR-497—MACC1— by decreasing miR-497 as a ceRNA. | promote proliferation, invasion, accelerate cell cycle G1/S transition | ||
| 3 Post-transcriptional regulation of interactions between lncRNAs and miRNAs in the expression of genes related to multidrug resistance and angiogenesis in gastric cancer | |||||
| HOTAIR | miR-126 | VEGFA, PIK3R2 | HOTAIR targets miR-126 and promotes the expression of the miR-126-target genes VEGFA and PIK3R2. | accelerate cisplatin resistance | up-regulated HOTAIR is related to greater tumor volume, vascular invasion and lymph node metastasis, advanced pathological stage and shorter overall survival |
| UCA1 | miR-27b | BCL-2 | Knockdown of UCA1 upregulates miR-27b in MDR gastric cancer cells. Down-regulation of UCA1 or overexpression of miR-27b decreases the expression of the anti-apoptotic protein BCL-2, enhances cleaved caspase-3. | inhibit apoptosis, increase the IC50 values of the chemotherapeutic drugs adriamycin (ADR), cisplatin (DDP) and 5-fluorouracil (5-FU) | highly expressed UCA1 is related to larger tumor size, poor differentiation, and lymphatic/venous invasion |
H19 enhances the EMT and metastasis of GC in several ways. Li et al. [96] identified three candidate RNA-binding proteins as H19 co-expressing ribonucleoproteins (RNPs) using GO (Gene Ontology) and KEGG (the Kyoto Encyclopedia of Genes and Genomes). It was demonstrated that H19 upregulated the expression of ISM1 protein by combining with it directly. These researchers also demonstrated that H19 and miR-675 exert similar functions: down-regulation of the miR-675 target gene CALN1 and promotion of invasion and metastasis of GC cells. Others have noted that H19 inhibits metastasis and expression of EMT markers in hepatocellular carcinoma [115]. The function H19 exerts on carcinogenesis may be related to its target protein ISM1, which promotes endothelial cell growth or apoptosis depending on its physiological state [116].
The lncRNA CCAT1 (colon cancer-related transcript 1) is transcribed upstream of the c-Myc gene [117] and is identified by Nissan et al. [118], occurring primarily in colon cancer. CCAT1 is localized mainly in the nucleus. Overexpression of CCAT1 is highly associated with TNM stage in GC [119]. Zhou et al. found thatCCAT1 siRNA prevented gastric cancer cells from migration, which could be reversed by miR-490 inhibitors. In addition, miR-490 decreases translation of its target mRNA hnRNPA1 by combining with its 3ʹ-UTR and miR-490 inhibitors enhance hnRNPA1, which can be reversed by CCAT1 siRNA.
The lncRNA XIST (X-inactive specific transcript) plays crucial roles in cell proliferation and chromosome maintenance [120]. XIST is highly expressed in gastric cancer tissues, and its expression is strongly correlated with tumor size, lymph node metastasis, and clinical stage [121, 122]. XIST is negatively associated with miR-101 in GC, and miR-101downregulates its target EZH2 (enhancer of zest homolog 2) [121]; while XIST knockdown decreases EZH2, and miR-101 inhibitor increases EZH2. Ma et al. [122] suggested that the lncRNA XIST promoted expression of the target gene of miR-497—MACC1—by decreasing miR-497 as a ceRNA, thus promoting GC cell proliferation and invasion as well as accelerating the G1/S transition.
Post-transcriptional regulation of interactions between lncRNAs and miRNAs in the expression of genes related to MDR and angiogenesis in gastric cancer
HOTAIR is notably elevated in cisplatin-resistant cells and tissues in GC [123]. Moreover, HOTAIR accelerates cisplatin resistance in GC cells by targeting miR-126 and promotes the expression of the miR-126-target genes VEGFA and PIK3R2 [123]. Endothelial cells specifically expressing miR-126 inhibits VEGFA and PIK3R2 expression—mediated by the VEGF/PI3K/AKT signaling activity—and resists angiogenesis [124]. UCA1 is significant higher in gastric cancer [125, 126], and knockdown of UCA1 upregulates miR-27b in MDR gastric cancer cells. Down-regulation of UCA1 or overexpression of miR-27b, sensitizes GC cells to adriamycin (ADR), cisplatin (DDP), and 5-fluorouracil (5-FU) chemotherapeutic agents, suppresses the expression of the anti-apoptotic protein BCL-2 and enhances cleaved caspase-3, inducing apoptosis of GC cells. These effects suggest that the UCA1-miR-27b axis participate in regulation of chemosensitivity in GC cells.
Conclusions and Future Perspectives
We described the interactions effects between the two important categories of ncRNAs, lncRNAs and miRNAs on post-transcriptional regulation of gene expression related to biological behaviors of gastric cancer. In GC cells, lncRNAs can regulate genes associated with protein translation, cell proliferation, apoptosis, the EMT, metastasis, and drug resistance. MiRNAs silencing of target gene expression involves interactions with the 3ʹ-UTR. Interactions between lncRNAs and miRNAs are responsible for post-transcriptional regulation. The most important mechanism is that lncRNAs act as ceRNAs to attenuate the negative function that miRNAs exert on target genes. In this way, lncRNAs and miRNAs can participate in carcinogenesis.
With the accumulating research in the life sciences, our knowledge of ncRNAs has expanded. We can obtain lncRNAs expression profiles and screen specific lncRNAs in tissues from the current databases, or by gene chip and sequencing to explore new lncRNAs. There are a variety of lncRNAs in the genome, which regulate gene expression and epigenetic through multiple modes. We should focus on the further classification of lncRNAs, such as gene structure, genomic mapping, cellular localization, regulation methods, involved biological behaviors of cancerous cell and other aspects to identify the functions of lncRNAs.
The secondary structure of lncRNAs may be closely related to their functions [127]; however, examinations of secondary structure are particularly challenging. Additional research is warranted to summarize whether changes in secondary structure of lncRNAs can affect interactions with other molecules, such as proteins, and thus influence carcinogenesis. Such knowledge could further elucidate the mechanism of post-transcriptional regulation by lncRNAs and miRNAs in cancer, which may shed a new insight for molecular therapy and clinical diagnosis in cancer.
Presently, many lncRNAs have dysregulated expression in cancer tissues. It remains unclear whether ncRNAs can turn into valid targets for molecular therapies in cancer in the context of interactions with mRNAs, and proteins. Bioinformatics and functional analyses have indicated that lncRNAs and miRNAs modulate gene expression post-transcriptionally via interactions with mRNAs, proteins and mutual effects. The effects are to influence proliferation, apoptosis, the EMT, metastasis, multidrug resistance, and angiogenesis, suggesting a potential value of ncRNAs for the treatment of GC.
Abbreviations
5-FU: 5-fluorouracil; ADR: adriamycin; bHLH: helix-loop-helix; CAFs: cancer-associated fibroblasts; CCAT1: colon cancer-related transcript 1; ccRCC: clear cell renal cell carcinoma; ceRNA: competing endogenous RNA; DDP: cisplatin; EdU: ethyl deoxyuridine; EMT: epithelial-mesenchymal transition; ER/PR: estrogen receptor /progesterone receptor; EV: endothelial vessel; EZH2: enhancer of zest homolog 2; HOTAIR: HOX transcript antisense RNA; GAS5: growth arrest-specific transcript 5; GHET1: gastric carcinoma highly expressed transcript 1; GO: Gene Ontology; hTERT: human telomerase reverse transcriptase; HUVECs: human umbilical vein endothelial cells; IGF2BP1: insulin-like growth factor binding protein 1; IKKɛ: IκB kinase; KAI-1: CD82 molecule; KEGG: Kyoto Encyclopedia of Genes and Genomes; KRT7-AS: KRT7 antisense RNA 1; Linc-POU3F3: long intergenic noncoding RNA POU3F3; MALAT1: lung adenocarcinoma metastasis-related transcript 1; MDR: multidrug resistance; MREs: miRNA response elements; MVD: microvessel density; NcRD: nucleosome remodeling and histone deacetylase; NEAT2: nuclear-enriched transcript 2; nrRNA: nuclear-retained regulator; PCA3: prostate cancer associated 3; PDAC: pancreatic ductal adenocarcinoma; Pol II: polymerase II; RIP: RNA immunoprecipitation; RNPs: ribonucleoproteins; RUNX1: tumor-suppressor runt domain transcription factor 1; SNHG5: small nucleolar RNA host gene 5; SR: serine/arginine; TINCR: terminal differentiation-induced ncRNA; TNM: Tumor Node Metastasis; VEGF-A: vascular endothelial growth factor A; VM: Vasculogenic mimicry; WT1: Wilms' tumor gene; XIST: X-inactive specific transcript; YBX1: human Y-box binding protein 1.
Acknowledgements
This work was supported by grants from the National Science and Technology Support Program (2015BAI13B07) and the National Natural Science Foundation (81772987).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42-54
2. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-74
3. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205
4. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-31
5. Covarrubias AA, Reyes JL. Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 2010;33:481-9
6. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407
7. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26-46
8. Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R. et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756-69
9. Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976-96
10. Jiang C, Chen X, Alattar M, Wei J, Liu H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015;22:291-301
11. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-19
12. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
13. Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59
14. Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996-1009
15. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci. 2016;41:761-72
16. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-27
17. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR. et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595-606
18. Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390-403
19. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8
20. Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-14
21. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D. et al. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299-310
22. Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R. et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75
23. Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K. et al. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. Febs j. 2014;281:802-13
24. Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787-93
25. Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195-208
26. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613-23
27. Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP. et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319
28. Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G. et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159
29. Shan TD, Xu JH, Yu T, Li JY, Zhao LN, Ouyang H. et al. Knockdown of linc-POU3F3 suppresses the proliferation, apoptosis, and migration resistance of colorectal cancer. Oncotarget. 2016;7:961-75
30. Li W, Zheng J, Deng J, You Y, Wu H, Li N. et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714-26.e5
31. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 promotes cell proliferation in gastric cancer via increasing T-reg distribution. Am J Transl Res. 2015;7:2262-9
32. Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231-5
33. Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM. et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648-61
34. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-41
35. Gutschner T, Hammerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl). 2013;91:791-801
36. Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919-32
37. Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat Immunol. 2011;12:853-60
38. Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q. et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209-18
39. Wang J, Su L, Chen X, Li P, Cai Q, Yu B. et al. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557-64
40. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
41. Yu Y, Li L, Zheng Z, Chen S, Chen E, Hu Y. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J Cell Mol Med. 2017;21:955-67
42. Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109:16654-9
43. Kao SH, Wang WL, Chen CY, Chang YL, Wu YY, Wang YT. et al. GSK3beta controls epithelial-mesenchymal transition and tumor metastasis by CHIP-mediated degradation of Slug. Oncogene. 2014;33:3172-82
44. Fan Y, Wang YF, Su HF, Fang N, Zou C, Li WF. et al. Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial-mesenchymal transition. J Hematol Oncol. 2016;9:57
45. Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208
46. Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J. et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745-55
47. Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H. et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br J Cancer. 2014;110:164-71
48. Ding J, Li D, Gong M, Wang J, Huang X, Wu T. et al. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625-30
49. Yuan CL, Li H, Zhu L, Liu Z, Zhou J, Shu Y. Aberrant expression of long noncoding RNA PVT1 and its diagnostic and prognostic significance in patients with gastric cancer. Neoplasma. 2016;63:442-9
50. Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q. et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res. 2017;23:2071-80
51. Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T. et al. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35:5770-80
52. Moorwood K, Charles AK, Salpekar A, Wallace JI, Brown KW, Malik K. Antisense WT1 transcription parallels sense mRNA and protein expression in fetal kidney and can elevate protein levels in vitro. J Pathol. 1998;185:352-9
53. Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J. et al. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta. 2016;1862:12-9
54. Dallosso AR, Hancock AL, Malik S, Salpekar A, King-Underwood L, Pritchard-Jones K. et al. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. Rna. 2007;13:2287-99
55. Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z. et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927-36
56. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454-7
57. Noel EE, Yeste-Velasco M, Mao X, Perry J, Kudahetti SC, Li NF. et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am J Pathol. 2010;176:2607-15
58. Sun Y, Zheng S, Torossian A, Speirs CK, Schleicher S, Giacalone NJ. et al. Role of insulin-like growth factor-1 signaling pathway in cisplatin-resistant lung cancer cells. Int J Radiat Oncol Biol Phys. 2012;82:e563-72
59. Lan WG, Xu DH, Xu C, Ding CL, Ning FL, Zhou YL. et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. 2016;36:263-70
60. Hang Q, Sun R, Jiang C, Li Y. Notch 1 promotes cisplatin-resistant gastric cancer formation by upregulating lncRNA AK022798 expression. Anticancer Drugs. 2015;26:632-40
61. Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182-93
62. Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H. et al. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393-8
63. Zhang XW, Bu P, Liu L, Zhang XZ, Li J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem Biophys Res Commun. 2015;462:227-32
64. Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe'er J. et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739-52
65. Zhang S, Zhang D, Sun B. Vasculogenic mimicry: current status and future prospects. Cancer Lett. 2007;254:157-64
66. Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N. et al. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31-44
67. Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J Clin Invest. 2007;117:2059-66
68. Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L. et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell. 2016;7:141-51
69. Cui S, Liao X, Ye C, Yin X, Liu M, Hong Y. et al. ING5 suppresses breast cancer progression and is regulated by miR-24. Mol Cancer. 2017;16:89
70. Concannon CG, Tuffy LP, Weisova P, Bonner HP, Davila D, Bonner C. et al. AMP kinase-mediated activation of the BH3-only protein Bim couples energy depletion to stress-induced apoptosis. J Cell Biol. 2010;189:83-94
71. Kilbride SM, Farrelly AM, Bonner C, Ward MW, Nyhan KC, Concannon CG. et al. AMP-activated protein kinase mediates apoptosis in response to bioenergetic stress through activation of the pro-apoptotic Bcl-2 homology domain-3-only protein BMF. J Biol Chem. 2010;285:36199-206
72. Chen L, Lu MH, Zhang D, Hao NB, Fan YH, Wu YY. et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014;5:e1034
73. Wu G, Liu A, Zhu J, Lei F, Wu S, Zhang X. et al. MiR-1207 overexpression promotes cancer stem cell-like traits in ovarian cancer by activating the Wnt/beta-catenin signaling pathway. Oncotarget. 2015;6:28882-94
74. Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J. et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312-7
75. Zhao XD, Lu YY, Guo H, Xie HH, He LJ, Shen GF. et al. MicroRNA-7/NF-kappaB signaling regulatory feedback circuit regulates gastric carcinogenesis. J Cell Biol. 2015;210:613-27
76. Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ. et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786-94
77. Guo B, Liu L, Yao J, Ma R, Chang D, Li Z. et al. miR-338-3p suppresses gastric cancer progression through a PTEN-AKT axis by targeting P-REX2a. Mol Cancer Res. 2014;12:313-21
78. Liu K, Xie F, Gao A, Zhang R, Zhang L, Xiao Z. et al. SOX2 regulates multiple malignant processes of breast cancer development through the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol Cancer. 2017;16:62
79. Lyu X, Li J, Yun X, Huang R, Deng X, Wang Y. et al. miR-181a-5p, an inducer of Wnt-signaling, facilitates cell proliferation in acute lymphoblastic leukemia. Oncol Rep. 2017;37:1469-76
80. Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K. et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11-22
81. Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, Yang J. et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693
82. Li P, Shan JX, Chen XH, Zhang D, Su LP, Huang XY. et al. Epigenetic silencing of microRNA-149 in cancer-associated fibroblasts mediates prostaglandin E2/interleukin-6 signaling in the tumor microenvironment. Cell Res. 2015;25:588-603
83. Zhang Y, Wu YY, Jiang JN, Liu XS, Ji FJ, Fang XD. MiRNA-3978 regulates peritoneal gastric cancer metastasis by targeting legumain. Oncotarget. 2016;7:83223-30
84. Guo P, Zhu Z, Sun Z, Wang Z, Zheng X, Xu H. Expression of legumain correlates with prognosis and metastasis in gastric carcinoma. PLoS One. 2013;8:e73090
85. Li N, Liu Q, Su Q, Wei C, Lan B, Wang J. et al. Effects of legumain as a potential prognostic factor on gastric cancers. Med Oncol. 2013;30:621
86. Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726-33
87. Li H, Zhao J, Zhang JW, Huang QY, Huang JZ, Chi LS. et al. MicroRNA-217, down-regulated in clear cell renal cell carcinoma and associated with lower survival, suppresses cell proliferation and migration. Neoplasma. 2013;60:511-5
88. Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng ZL, He MM. et al. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868-79
89. Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C. et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497-506
90. Wang Y, Liu C, Luo M, Zhang Z, Gong J, Li J. et al. Chemotherapy-Induced miRNA-29c/Catenin-delta Signaling Suppresses Metastasis in Gastric Cancer. Cancer Res. 2015;75:1332-44
91. Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB. et al. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657-67
92. Zhang JX, Xu Y, Gao Y, Chen C, Zheng ZS, Yun M. et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16:18
93. Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N. et al. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget. 2014;5:11873-85
94. Dai J, Wang J, Yang L, Xiao Y, Ruan Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int J Oncol. 2015;47:1801-10
95. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-21
96. Li H, Yu B, Li J, Su L, Yan M, Zhu Z. et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-29
97. Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J. et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9:e86295
98. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna. 2007;13:313-6
99. Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ. et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-8
100. Gao WL, Liu M, Yang Y, Yang H, Liao Q, Bai Y. et al. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol. 2012;9:1002-10
101. Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-22
102. Liu G, Xiang T, Wu QF, Wang WX. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells. Oncol Res. 2016;23:99-107
103. Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cell Physiol Biochem. 2015;36:1440-52
104. Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963-9
105. Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L. et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276-92
106. Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y. et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. Faseb j. 2017;31:893-903
107. Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY. et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524-34
108. Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613-7
109. Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P. et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445
110. Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X. et al. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812-24
111. Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319-26
112. Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M. et al. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391-9
113. Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH. et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802
114. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92
115. Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS. et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577-86
116. Zhang Y, Chen M, Venugopal S, Zhou Y, Xiang W, Li YH. et al. Isthmin exerts pro-survival and death-promoting effect on endothelial cells through alphavbeta5 integrin depending on its physical state. Cell Death Dis. 2011;2:e153
117. Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513-31
118. Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M. et al. Colon cancer associated transcript-1: a novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130:1598-606
119. Zhou B, Wang Y, Jiang J, Jiang H, Song J, Han T. et al. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201-10
120. Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973
121. Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS. et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142
122. Ma L, Zhou Y, Luo X, Gao H, Deng X, Jiang Y. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget. 2017;8:4125-35
123. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016
124. Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T. et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157-64
125. Fang Q, Chen X, Zhi X. Long Non-Coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) Increases Multi-Drug Resistance of Gastric Cancer via Downregulating miR-27b. Med Sci Monit. 2016;22:3506-13
126. Shang C, Guo Y, Zhang J, Huang B. Silence of long noncoding RNA UCA1 inhibits malignant proliferation and chemotherapy resistance to adriamycin in gastric cancer. Cancer Chemother Pharmacol. 2016;77:1061-7
127. Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034-51
Author contact
![]() Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, North Nanjing Street 155#, Heping District, Shenyang 110001, China Telephone: +86-024-83282153; fax: +86-024-83282383, Email: yuanyuanedu.cn
Corresponding author: Dr. Yuan Yuan, Tumor Etiology and Screening Department of Cancer Institute and General Surgery, North Nanjing Street 155#, Heping District, Shenyang 110001, China Telephone: +86-024-83282153; fax: +86-024-83282383, Email: yuanyuanedu.cn

 Global reach, higher impact
Global reach, higher impact