Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(1):21-31. doi:10.7150/jca.21749 This issue Cite
Research Paper
Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profile in Pancreatic Cancer Derived Exosomes Treated Dendritic Cells by Microarray Analysis
Department of General Surgery, Sir Run Run Shaw Hospital, the affiliated hospital of Zhejiang University School of Medicine, Hangzhou, China.
* These authors contributed equally to this work
Received 2017-7-3; Accepted 2017-10-8; Published 2018-1-1
Abstract
Background: Pancreatic cancer is a devastating disease with a low five-year survival rate. Dendritic cells (DCs), which are the most potent antigen-presenting cells in the human body, play a pivotal role in the immune response. However, few studies have investigated the role of pancreatic cancer-derived exosomes (PEXs) in DC-meditated immune escape. The expression profiles of long noncoding RNAs (lncRNAs) and mRNAs of PEX-treated dendritic cells are unknown.
Methods: We used integrated lncRNA and mRNA microarrays to determine the expression profiles of PEX-treated DCs and normal DCs derived from five healthy donors. Gene Ontology (GO), KEGG, and cancer genomics analyses were performed to identify significant functions, pathways, and the associations of differentially expressed mRNAs. A coexpression network was constructed to identify the correlation between differentially expressed lncRNAs and mRNAs and further validated using real-time quantitative PCR in twenty healthy donors. The AnnoLnc program was used to perform an annotation analysis of lncRNAs.
Results: We identified 3,227 and 924 differentially expressed lncRNAs and mRNAs, respectively, in PEX-treated DCs. GO and pathway analysis revealed differentially expressed mRNAs involved in many critical biological processes and molecular functions. Cancer genomics analysis revealed that 36 of the most differentially expressed mRNAs were involved in a pancreatic cancer network and were associated with many critical mutated genes such as TP53, KRAS, SMAD4, and CDKN2A. LncRNAs such as ENST00000560647 and mRNAs such as legumain (lgmn) were differentially expressed in PEX-treated DCs, and the data were validated using RT-qPCR.
Conclusions: To our knowledge, this is the first study to detect the differential expression of lncRNAs and mRNAs associated with PEX-treated DCs. LncRNAs such as ENST00000560647 and mRNAs such as lgmn might play a critical role in immune escape of DCs treated with PEX. Further investigation is required to validate the functions and associations of these RNAs.
Keywords: pancreatic cancer, dendritic cells, exosomes, long noncoding RNAs.
Introduction
Pancreatic cancer is a devastating disease that is difficult to diagnose, which helps to explain the 10% five-year survival rate [1]. Moreover, approximately 53,070 Americans were predicted to have been diagnosed with pancreatic cancer, and approximately 41,780 Americans were predicted to have died of pancreatic cancer in 2016. This lethal disease is the fourth most common cause of cancer-related deaths among Americans [2]. Surgical resection, radiation therapy, chemotherapy, and adoptive immunotherapy are continuously improving; however, patients diagnosed with pancreatic cancer remain extremely vulnerable to relapse and death. New strategies to prevent and cure pancreatic cancer are therefore urgently required. For example, exosomes and nanovesicles secreted by living cells serve as intercellular couriers of mRNA, microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and proteins and have thus garnered considerable interest [3]. Tumor-derived exosomes can deliver tumor antigens to dendritic cells [4] that participate in tumor progression [5, 6] and play an important role in the immune system [7-10].
Dendritic cells (DCs) are the most potent antigen-presenting cells in the human body, because they play a pivotal role in the immune response, which is inhibited in the tumor microenvironment [11]. The regulatory factor X-associated protein (RFXAP), is an important transcription factor for MHC II, and is inhibited by miR-212-3p transferred from pancreatic cancer-derived exosomes (PEXs) to inhibit MHC II expression [12]. Further, PEXs inhibit the expression of toll-like receptor 4 (TLR4) and tumor necrosis factor-alpha (TNF-alpha) via miR-203 and induce immune escape of DCs [13].
Long noncoding RNAs (lncRNAs) (>200 nucleotides) interact with DNA, RNA, or protein molecules to regulate gene expression and affect cellular processes [14, 15]. Tumor-derived exosomes are key players in the communication between cancer cells and their microenvironment through their cargoes, which include proteins, lncRNAs, mRNAs, and miRNAs [16]. However, few studies [12, 13] investigated the role of PEXs in DCS that mediate immune escape. The expression profiles of lncRNAs and mRNAs in DCs treated with PEXs are unknown.
To identify potential differentially expressed lncRNAs and mRNAs in PEX-treated DCs (exo-DCs), we determined their expression profiles in exo-DCs and normal dendritic cells (nDCs) derived from twenty five healthy donors. For this purpose, we employed integrated lncRNA and mRNA microarrays, and differentially expressed lncRNAs and mRNAs were subsequently evaluated using real-time quantitative PCR (RT-qPCR). We conducted bioinformatics analysis to identify the potential functions of differentially expressed lncRNAs and mRNAs.
Materials and Methods
Cell culture and exosome preparations
The human pancreatic cancer cell line PANC-1 was obtained from Chinese Academy of Sciences (Shanghai, China), tested, and authenticated. The cell line was cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 1% antibiotics (Gibco, USA) and 10% fetal bovine serum (Gibco) until 90% confluent. The medium was then replaced with serum-free DMEM for 48 h. The cell-free supernatant was collected by centrifugation at 300 × g for 10 min and 2000 × g for 10 min to remove residual cells and debris and then for 10,000 × g for 30 min to remove microparticles. The supernatant was then passed through a 0.22-µm filter (Millipore, USA) and then centrifuged at 100,000 × g for 70 min to pellet the small vesicles that corresponded to exosomes. The pellet was washed in a large volume of phosphate-buffered saline (PBS), to eliminate contaminating proteins and centrifuged at the same speed [17] using an Optima TLX Ultracentrifuge (Beckman Coulter, USA). The final pellet was resuspended in 100-µl PBS. The size distribution of exosomes was determined using nanoparticle tracking analysis (NTA) Version 2.3 Build 0034. The exosomes were placed on carbon-coated 400-mesh copper grids, stained with 2% uranyl acetate, air-dried, and imaged using transmission electron microscopy.
Western blotting
PEX preparations were lysed in RIPA buffer (Sigma-Aldrich, USA) containing a cocktail of protease inhibitors (Thermo, USA). Proteins were denatured in 2 × SDS buffer at 95°C, separated using 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, USA). After blocking with 5% milk powder for 1 h at room temperature, the membranes were incubated with the antibodies as follows: mouse monoclonal anti-TSG101 (1:500, Abcam, UK), mouse monoclonal anti-CD63 (1:1000, Abcam, UK), mouse monoclonal anti-ALIX (1:1000, Abcam, UK), and mouse monoclonal anti-GAPDH (1:2000, Abcam, UK) as the control. The samples were incubated with a secondary goat anti-mouse antibody (1:5000, Pierce, USA) for 1 h at room temperature. The blots were visualized using a GE Amersham Imager 600.
Generation of DCs and treatment of DCs with PEXs
Peripheral blood mononuclear cells (PBMCs) were isolated from 25 healthy donors (Table S1). Briefly, after Ficoll-Paque density gradient centrifugation (GE, USA), monocytes were sorted from PBMCs using human anti-CD14-coated microbeads (Miltenyi Biotec, German). The resulting CD14-positive monocytes were cultured for up to 5 days in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco) containing 100 U/ml penicillin, 100 µg/ml streptomycin, interleukin-4 (IL-4, 40 ng/ml, PeproTech, USA), and granulocyte-macrophage colony stimulating factor (GM-CSF, 100 ng/ml; PeproTech) to generate the immature DCs [12]. To activate immature DCs, human recombinant TNF-α (rTNF-α; 20 ng/ml; R&D Systems) was added on day 5, and the culture was continued for 2 days to convert these cells into mature DCs (mDCs).
Phenotypic analysis of DCs was performed using flow cytometry. On day 7 of culture, cells were harvested and stained with a monoclonal antibodies against HLA-DR (APC-conjugated anti-HLA-DR; BD Biosciences, USA) and CD83 (PE-conjugated anti-CD83; BD Biosciences) in the dark for 30 min at 4ºC. Cells were then washed twice with PBS. Fluorescence-activated cell sorting (FACS) was performed using a FACSCalibur flow cytometer (BD Biosciences, USA) and CellQuest Software (BD Biosciences). On day 8, DCs were treated with PEXs (20 µg/mL) for 24 h (exo-DCs), and PBS was used as negative control for nDCs.
T cell proliferation
CD4+ and CD8+ T cells were sorted from PBMCs using human anti-CD4-conjugated and human anti-CD8-conjugated microbeads (Miltenyi Biotec, German). To determine the proliferation of T cells, mixed DCs and T cells were continuously cultured for 7 days with 25 U/ml recombinant IL-2 (R&D Systems), which was added on days 3 and 5, [3H]-thymidine was added on day 6, and T cell proliferation was subsequently assessed.
RNA extraction
Total RNAs of exo-DCs and DCs were isolated and purified using an miRNeasy Mini Kit (Qiagen, USA). Each step followed the manufacturer's instructions. RNA quality was assessed using a NanoDrop ND-2000 (Thermo Fisher Scientific, USA), and RNA integrity was assessed using electrophoresis through an agarose gel performed under denaturing conditions.
Microarray processing and analysis
An Agilent lncRNA Gene Expression 4 × 180K Microarray (Design ID: 74348, Agilent, USA) was used to query 77,103 lncRNAs and 18,853 mRNAs. Sample labeling, microarray hybridization, and washing were performed according to the manufacturer's standard protocols. Briefly, total RNA was amplified and labeled using a Low Input Quick Amp WT Labeling Kit (Agilent) following the manufacturer's instructions. Labeled cRNAs were purified using an RNeasy Mini Kit (Qiagen, Germany). Each slide was hybridized with 1.65 µg of Cy3-labeled cRNA using a Gene Expression Hybridization Kit (Agilent) in a Hybridization Oven (Agilent) according to the manufacturer's instructions. After hybridization for 17 h, slides were washed in staining dishes (Thermo Fisher Scientific, USA) using a Gene Expression Wash Buffer Kit (Agilent) following the manufacturer's instructions. Slides were scanned using an Agilent Microarray Scanner (Agilent) with the default settings as follows: dye channel, green; scan resolution = 3 µm; PMT, 100%; 20 bits. Data were extracted using Feature Extraction software 10.7 (Agilent). Raw data were normalized using the Quantile algorithm, GeneSpring Software 12.6.1 (Agilent).
mRNA GO, KEGG, and cancer genomics analyses
Gene Ontology (GO, www.geneontology.org/) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg/) analysis, and cancer genomics analysis, which is based on a TCGA pancreatic adenocarcinoma dataset with 186 samples and 185 patients using the cBioPortal for Cancer Genomics program [18, 19] (http://www.cbioportal.org/), were performed to identify the functions and associated enriched pathways of differentially expressed mRNAs. The mRNA expression z-score threshold was defined as ± 2.0.
Construction of an lncRNA-mRNA coexpression network
According to the normalized signal intensities of differentially expressed lncRNAs and mRNAs, an lncRNA-mRNA coexpression network was built to identify the correlations between lncRNAs and mRNAs. For each lncRNA-mRNA pair, the Pearson correlation was calculated to identify significantly correlated pairs. The Pearson correlation value cutoff = 0.95, P < 0.05.
RT-qPCR
RT-qPCR was performed to validate the expression of significantly altered levels of lncRNAs and mRNAs. Total RNA was isolated and purified using an miRNeasy Mini Kit (Qiagen, USA) following the manufacturer's instructions. RT-qPCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, USA) with the primer pairs listed (Table S2). The relative expression ratios of lncRNAs and mRNAs are presented as fold-changes normalized to those of GAPDH.
LncRNA annotation analysis
LncRNAs annotation analysis was performed using the AnnoLnc program [20], a portal for systematically annotating novel human lncRNAs, by analyzing secondary structure, transcriptional regulation, and GO annotation.
Statistical analysis
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was used to perform statistical analysis. The Student t test was used to evaluate the significance of differences in expression among lncRNAs and mRNAs differentially expressed by exo-DCs and DCs. The threshold value used to designate differentially expressed lncRNAs and mRNAs was a fold change > 2.0 or < 0.5, P < 0.05.
Ethics statement
The Research Ethics Committee of Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University reviewed and approved the protocol. All participants or their guardians gave written consent for the use of subjects' tissue samples and the medical information used for scientific research purposes.
Results
Characterization of PEXs and DCs
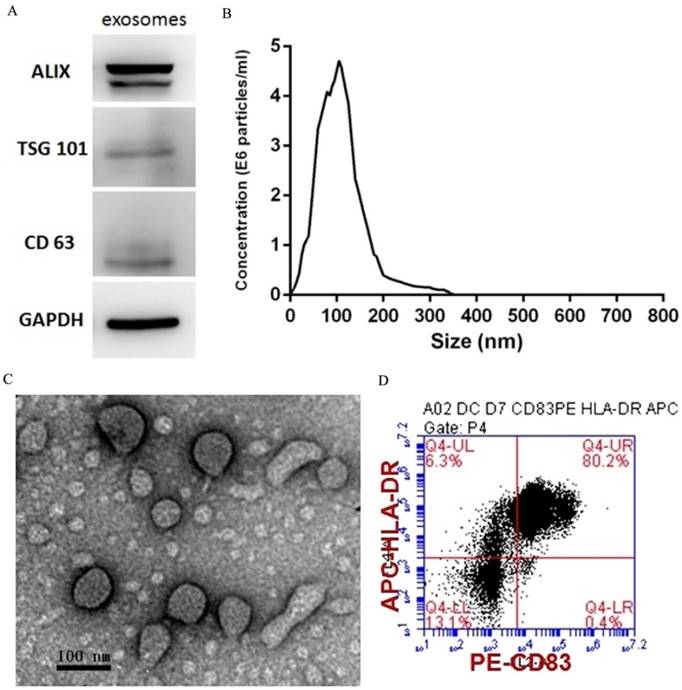
PEXs were detected using western blotting analysis of the expression of the exosome-specific markers ALIX, TSG101, and CD63 (Fig. 1A). The size distribution of the PEXs ranged from 20-200 nm (Fig. 1B). Transmission electron microscopy showed typical vesicular structures, and the size distribution was consistent with that determined using NTA (Fig. 1C). DCs were identified using flow cytometry. The percentages of CD83 and HLA-DR-double positive cells reached 80.2% (Fig. 1D).
Characterization of PEXs and DCs. (A) Western blotting analysis of PEXs detected the presence of the exosomal markers ALIX, TSG101, and CD63. (B) According to nanoparticle tracking analysis, the sizes of the PEXs ranged from 20 nm to 200 nm. (C) Transmission electron microscopy of PEXs. (D) Flow cytometric analysis of DCs.
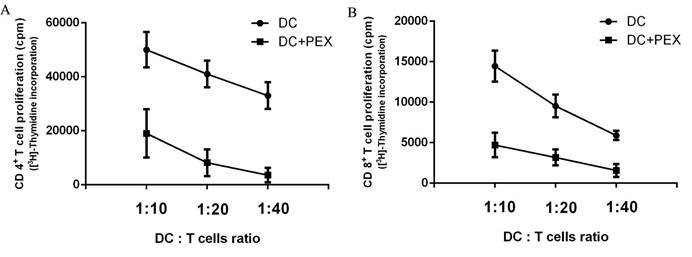
[3H]-Thymidine incorporation by autologous CD4+ and CD8+ T cells. Compared with normal DCs, exo-DCs had a reduced ability to activate autologous (A) CD4+ T cells (P < 0.01) and (B) CD8+ T cells (P < 0.01) at DC/T cell ratios = 1:10, 1:20, and 1:40. The data are expressed as the mean values of quintuplicate assays.
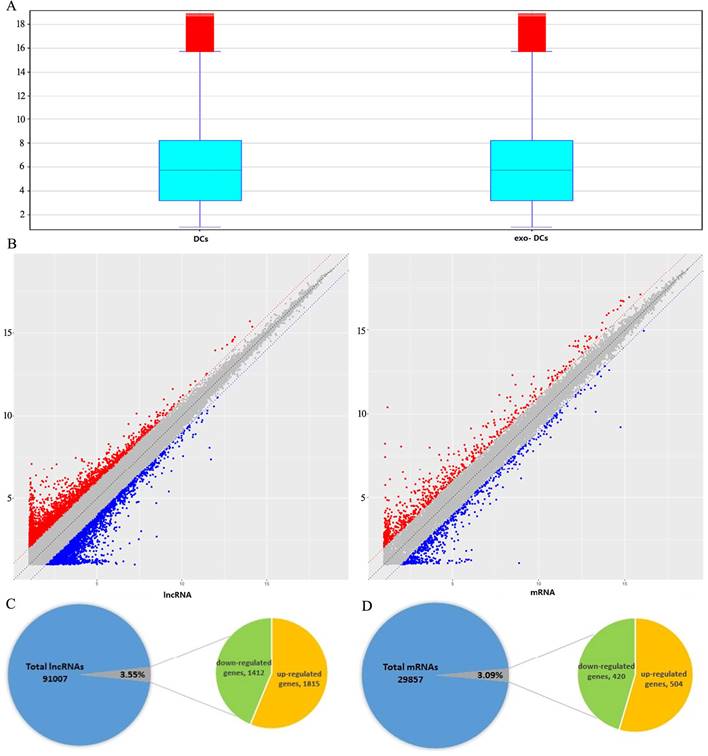
Summary of microarray results of lncRNA and mRNA expression in PEX-treated DCs (exo-DCs). (A) Box plots of the distributions of lncRNAs and mRNAs. (B) Scatter plots of the distributions of lncRNAs and mRNAs. (C) 3,227 of 91,007 (3.55%) differentially expressed lncRNAs, including 1,815 up-regulated lncRNAs and 1,412 down-regulated lncRNAs. (D) 924 of 29,857 (3.09%) differentially expressed mRNAs, including 504 up-regulated mRNAs and 420 down-regulated mRNAs.
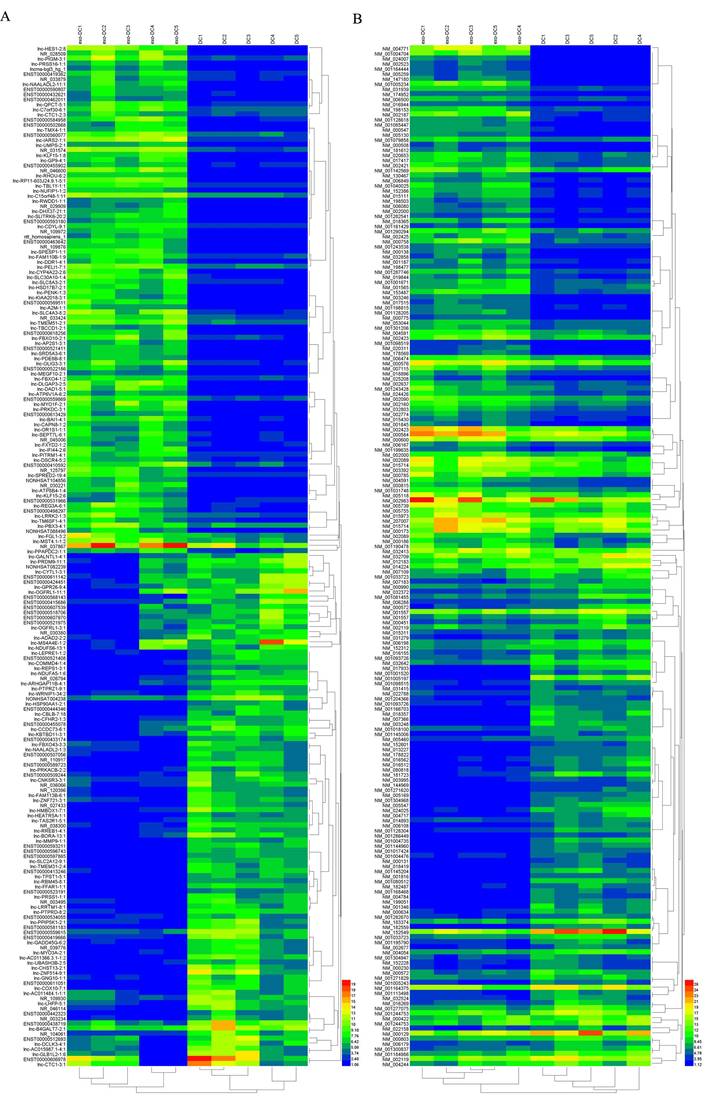
Hierarchical clustering of the differentially expressed lncRNAs and mRNAs in exo-DCs. (A) The top 200 differentially expressed lncRNAs. (B) The top 200 differentially expressed mRNAs.
PEX-treated DCs have reduced ability to activate autologous T cells
The functions of exo-DCs and DCs were evaluated by measuring their capacity to induce the proliferation of autologous CD4+ and CD8+ T cells. Compared with normal DCs, exo-DCs showed significantly less potency to stimulate autologous CD4+ (P < 0.01) and CD8+ (P < 0.01) T cells at DC:T cell ratios = 1:10, 1:20, and 1:40 (Fig. 2).
Expression profiles of LncRNAs and mRNAs
In exo-DCs, we detected 3,227 (3.55%) differentially expressed lncRNAs and 924 (3.09%) mRNAs, including 1,815 up-regulated lncRNAs and 1,412 down-regulated lncRNAs as well as 504 up-regulated mRNAs and 420 down-regulated mRNAs (fold change > ± 2, P < 0.05) (Fig. 3). The lncRNA and mRNA expression profiles differed significantly between exo-DCs compared with DCs (Fig. 4, Fig. S1 and Fig. S2).
GO, KEGG, and cancer genomics analyses
GO analysis revealed differentially expressed mRNAs involved, for example, in the positive regulation of leukocyte chemotaxis, positive regulation of leukocyte migration, cytokine activity, and monocyte chemotaxis (Fig 5). Pathway analysis revealed differentially expressed mRNAs involved, for example, in the TNF and NOD-like receptor signaling pathways, cytokine-cytokine receptor interaction, the Toll-like receptor signaling pathway, and cytosolic DNA-sensing interaction. (Fig. 5). Cancer genomics analysis generated a network that contains 86 nodes, including 36 seed genes (36 most differentially expressed mRNAs) and the 50 most frequently altered neighboring genes total = 2,158 genes) (Fig. 6).
lncRNA-mRNA coexpression network
We constructed an lncRNA-mRNA coexpression network to identify potential interactions between differentially expressed lncRNAs and mRNAs. The results show that the coexpression network comprised 3,867 connections between lncRNAs and mRNAs (Fig. 7).
RT-qPCR validation
To further validate the results of the microarray analysis, the top 10 differentially expressed lncRNAs and mRNAs were analyzed using RT-qPCR in 20 healthy donors (Table S1). LncRNAs ENST00000560647, lnc-G0S2-3:2, lnc-SPINK7-1:4, ENST00000512538, lnc-C1orf137-1:1, lnc-ITPK1-3:1, lnc-HSPA6-2:4, lnc-ASZ1-3:1, lnc-CXCR2-1:4, lnc-ADIPOR2-3:1 and mRNAs WNT5A, MYO1B, CTTNBP2, FCGR2B, LGMN, Sim2, FCGR2A, CTTNBP2, WNT5B, CXCR2 were validated (Fig. 8, Tables S3 and S4). The microarray data correlated well with the RT-qPCR results.
The top 30 enriched GO terms and KEGG pathways.
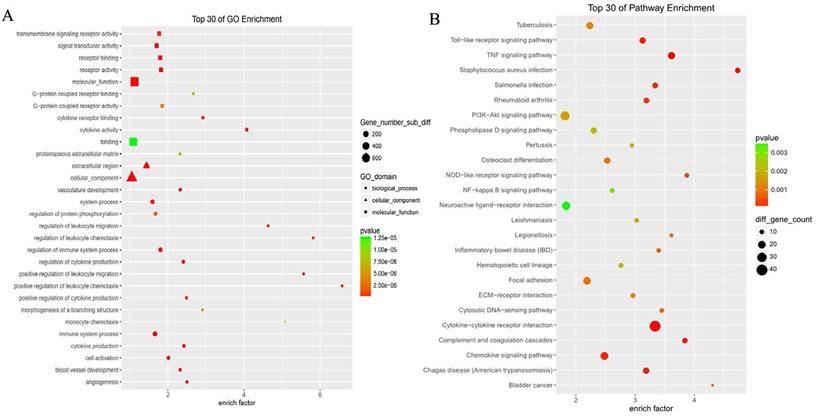
Cancer genomics analysis based on the cBioPortal Cancer Genomics program. The 36 most differentially expressed mRNAs and the 50 most frequently altered neighboring genes (total = 2,158 genes).
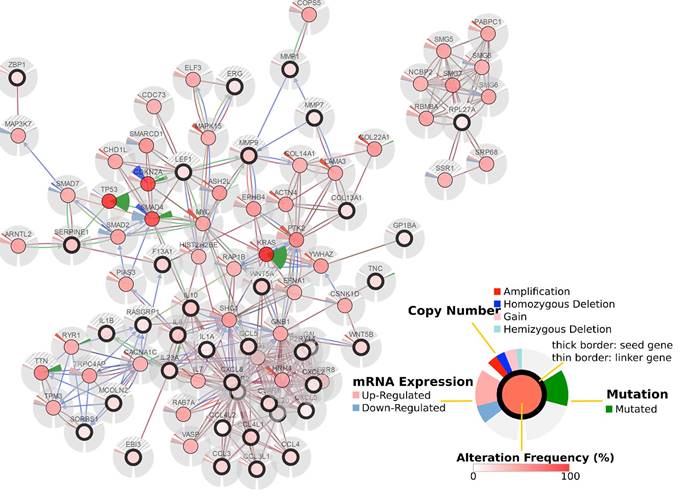
LncRNA-mRNA coexpression network. The blue nodes denote mRNAs and the green triangles denote lncRNAs. The degrees of lncRNAs or mRNAs are indicated by the size of the nodes or triangles.
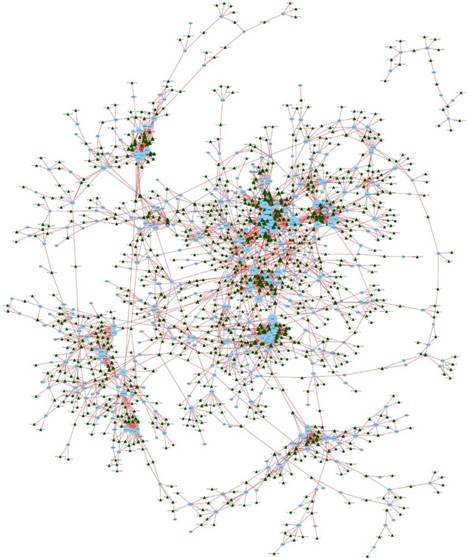
RT-qPCR validation of differentially expressed lncRNAs and mRNAs.
LncRNA ENST00000560647 annotations determined using the AnnoLnc program. (A) Predicted secondary structure of ENST00000560647. (B) The 60 transcription factors predicted to regulate the expression of ENST00000560647. (C) The predicted interaction of five miRNAs.
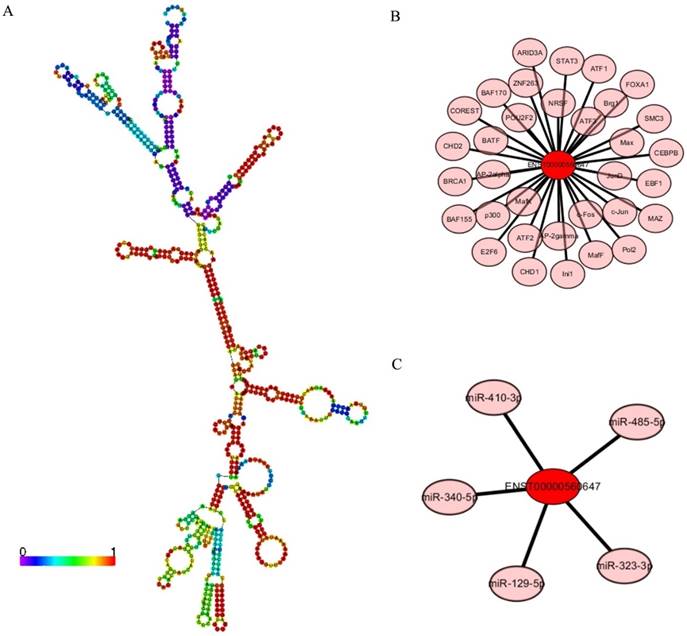
Annotation of lncRNA ENST00000560647
LncRNA ENST00000560647 was up-regulated by approximately 6.33- to 7.16-times in exo-DCs. According to the genes that were coexpressed with this lncRNA, we attempted to determine if they were enriched in functions that might provide hints that would illuminate function. The predicted secondary structure of lncRNA ENST00000560647 (702 bp; chromosome 15, sequence positions 45,742,316 to 45,805,965) is shown in Fig. 9A. Its expression was predicted to be regulated by 60 transcription factors (Fig. 9B) and to interact with 5 miRNAs (Fig. 9C). The table shows the GO annotation of lncRNA ENST00000560647. The GO annotations of biological process and molecular function of the positive coexpression gene sets are shown in Tables 1 and 2.
Biological Processes (Positive Correlation).
| GO Term | Description | P Value |
|---|---|---|
| GO:0018149 | peptide cross-linking | 2.07e-2 |
| GO:0031424 | keratinization | 2.07e-2 |
| GO:2000249 | regulation of actin cytoskeleton reorganization | 2.36e-2 |
| GO:0008544 | epidermis development | 4.02e-2 |
| GO:0043588 | skin development | 4.02e-2 |
| GO:0032956 | regulation of actin cytoskeleton organization | 4.02e-2 |
Molecular Function (Positive Correlation).
| GO Term | Description | P Value |
|---|---|---|
| GO:0030506 | ankyrin binding | 5.45e-3 |
| GO:0070004 | cysteine-type exopeptidase activity | 2.01e-2 |
| GO:0016807 | cysteine-type carboxypeptidase activity | 2.01e-2 |
| GO:0001948 | glycoprotein binding | 2.86e-2 |
| GO:0030984 | kininogen binding | 3.61e-2 |
Discussion
Tumor-derived exosomes are involved in cancer development, invasion, and metastasis [21-24]. Exosomal lncRNAs are potential biomarkers and therapeutic targets for multiple cancers [23, 25-29]. However, few studies [12, 13] investigated the role of PEXs in DC-meditated immune escape. The expression profiles of lncRNAs and mRNAs of exo-DCs are unknown.
In the present study, the expression profiles of lncRNAs and mRNAs revealed 3,227 differentially expressed lncRNAs and 924 differentially expressed mRNAs, including 1,815 up-regulated lncRNAs and 1412 down-regulated lncRNAs as well as 504 up-regulated mRNAs and 420 down-regulated mRNAs in exo-DCs. Functional annotation of mRNAs revealed, for example, differentially expressed mRNAs involved in positive regulation of leukocyte chemotaxis, positive regulation of leukocyte migration, cytokine activity, monocyte chemotaxis. Pathway analysis revealed, for example, differentially expressed mRNAs involved in the TNF and NOD-like receptor signaling pathways, cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway, and cytosolic DNA-sensing interaction. These are critical biological processes and molecular functions. For example, monocyte chemotaxis is a vital biological process of DCs, and PEXs might impair the chemotaxis of DCs and thus contribute to DC-meditated immune escape. Moreover, TNF-α and TLR4 expression decreased in DCs treated with PEXs [13], and pathway analysis of the present study supports these results.
To further explore the relationships to pancreatic cancer among these differentially expressed mRNAs, we performed a cancer genomics analysis, which was based on a TCGA pancreatic adenocarcinoma dataset comprising 186 samples and 185 patients. The results show that the 36 most differentially expressed mRNAs are involved in the pancreatic cancer network and are associated with many critical mutated genes such as TP53, KRAS, SMAD4, and CDKN2A.
To further validate the results of the microarray analysis, the top 10 differentially expressed lncRNAs and mRNAs in 10 healthy donors were analyzed using RT-qPCR. The results indicated that the microarray data correlate well with the RT-qPCR results. Lgmn expression decreased by approximately 3-fold in PEX-treated DCs. Lgmn encodes an enzyme that may be involved in the processing of bacterial peptides and endogenous proteins for MHC class II presentation to the lysosomal/endosomal systems. Further, lgmn plays a critical role in endocytic Toll-like receptor processing and signaling in DCs [30]. This result suggests that PEXs may inhibit the function of DCs by down-regulating the expression of Toll-like receptor and MHC class II proteins. Further research is required to validate this hypothesis.
LncRNA ENST00000560647 was up-regulated by approximately 6.33- to 7.16-times in PEX-treated DCs. However, its function is unknown. Annotation analysis based on the AnnoLnc program, predicted that lncRNA ENST00000560647 (702 bp; chromosome 15, nucleotides 45,742,316 to 45,805,965), is regulated by 60 transcription factors and that it interacts with five miRNAs. These five miRNAs are involved in many cancers, including pancreatic cancer [31-37]. For example, Wang et al. [34] found that miR-323-3p expression in pancreatic cancer tissues and cell lines is significantly decreased. Re-expression of miR-323-3p might provide a new therapeutic target against metastasis in patients with pancreatic cancer. Certain lncRNAs can serve as “sponges” to titrate microRNAs based on the competing endogenous RNA mechanism [38, 39]. Further validation is required to establish the relationship between ENST00000560647 and miR-323-3p in pancreatic cancer.
Conclusions
In summary, to our knowledge, this is the first study to screen and analyze the expression profiles of lncRNAs and mRNAs of exo-DCs and reveals a set of lncRNAs and mRNAs that were differentially expressed. LncRNAs such as ENST00000560647 and mRNAs such as lgmn may play critical roles in immune escape mediated by exo-DCs. Further investigation is required to validate the function and relationships among these lncRNAs and mRNAs.
Supplementary Material
Fig. S1. Hierarchical clustering of the differentially expressed lncRNAs. Fig. S2. Hierarchical clustering of the differentially expressed mRNAs. Table S1. Clinic profiles of 25 healthy donors Table S2. Primers for RT-qPCR. Table S3. RT-qPCR validation of top 10 lncRNAs. Table S4. RT-qPCR validation of top 10 mRNAs.
Acknowledgements
This study was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No.LY17H160015 and the National Natural Science Foundation of China (No.81572323) and Zhejiang Provincial Medical and Health Science and Technology Program (No. 2018KY491). We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Ethics
The Ethics Committee of Sir Run Run Shaw Hospital, Hanghzou, China approved this study.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sirri E, Castro FA, Kieschke J, Jansen L, Emrich K, Gondos A. et al. Recent Trends in Survival of Patients With Pancreatic Cancer in Germany and the United States. Pancreas. 2016;45:908-14
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7-30
3. Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A. et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PloS one. 2016;11:e0147236
4. Yao Y, Wang C, Wei W, Shen C, Deng X, Chen L. et al. Dendritic cells pulsed with leukemia cell-derived exosomes more efficiently induce antileukemic immunities. PloS one. 2014;9:e91463
5. Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem. 2016;74:103-41
6. Xiao D, Barry S, Kmetz D, Egger M, Pan J, Rai SN. et al. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016;376:318-27
7. Hannafon BN, Carpenter KJ, Berry WL, Janknecht R, Dooley WC, Ding WQ. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol Cancer. 2015;14:133
8. Katakowski M, Chopp M. Exosomes as Tools to Suppress Primary Brain Tumor. Cell Mol Neurobiol. 2016;36:343-52
9. Koyama Y, Ito T, Hasegawa A, Eriguchi M, Inaba T, Ushigusa T. et al. Exosomes derived from tumor cells genetically modified to express Mycobacterium tuberculosis antigen: a novel vaccine for cancer therapy. Biotechnol Lett. 2016;38:1857-66
10. Sharma A, Khatun Z, Shiras A. Tumor exosomes: cellular postmen of cancer diagnosis and personalized therapy. Nanomedicine (Lond). 2016;11:421-37
11. Kranz LM, Diken M, Haas H, Kreiter S, Loquai C, Reuter KC. et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396-401
12. Ding G, Zhou L, Qian Y, Fu M, Chen J, Chen J. et al. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget. 2015;6:29877-88
13. Zhou M, Chen J, Zhou L, Chen W, Ding G, Cao L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cellular immunology. 2014;292:65-9
14. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-41
15. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
16. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-26
17. Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology. 2006 Chapter 3: Unit 3.22
18. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1
19. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
20. Hou M, Tang X, Tian F, Shi F, Liu F, Gao G. AnnoLnc: a web server for systematically annotating novel human lncRNAs. BMC genomics. 2016;17:931
21. Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151-68
22. Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Scientific reports. 2016;6:24922
23. Hewson C, Morris KV. Form and Function of Exosome-Associated Long Non-coding RNAs in Cancer. Current topics in microbiology and immunology. 2016;394:41-56
24. Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L. et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2007-12
25. Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F. et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155
26. Gezer U, Ozgur E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell biology international. 2014;38:1076-9
27. Isin M, Uysaler E, Ozgur E, Koseoglu H, Sanli O, Yucel OB. et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Frontiers in genetics. 2015;6:168
28. Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M. et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Medical oncology (Northwood, London, England). 2014;31:148
29. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y. et al. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653-68
30. Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E. et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737-48
31. Zhang YF, Yu Y, Song WZ, Zhang RM, Jin S, Bai JW. et al. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncology reports. 2016;36:480-6
32. Kang M, Ren MP, Zhao L, Li CP, Deng MM. miR-485-5p acts as a negative regulator in gastric cancer progression by targeting flotillin-1. American journal of translational research. 2015;7:2212-22
33. Sun X, Liu Y, Li M, Wang M, Wang Y. Involvement of miR-485-5p in hepatocellular carcinoma progression targeting EMMPRIN. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2015;72:58-65
34. Wang C, Liu P, Wu H, Cui P, Li Y, Liu Y. et al. MicroRNA-323-3p inhibits cell invasion and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7:14912-24
35. Wozniak M, Sztiller-Sikorska M, Czyz M. Diminution of miR-340-5p levels is responsible for increased expression of ABCB5 in melanoma cells under oxygen-deprived conditions. Experimental and molecular pathology. 2015;99:707-16
36. Wu Q, Yang Z, Xia L, Nie Y, Wu K, Shi Y. et al. Methylation of miR-129-5p CpG island modulates multi-drug resistance in gastric cancer by targeting ABC transporters. Oncotarget. 2014;5:11552-63
37. Yu Y, Zhao Y, Sun XH, Ge J, Zhang B, Wang X. et al. Down-regulation of miR-129-5p via the Twist1-Snail feedback loop stimulates the epithelial-mesenchymal transition and is associated with poor prognosis in breast cancer. Oncotarget. 2015;6:34423-36
38. Song X, Cao G, Jing L, Lin S, Wang X, Zhang J. et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. Journal of cellular and molecular medicine. 2014;18:991-1003
39. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358-69
Author contact
![]() Corresponding author: Professor Liping Cao, Department of General Surgery, Sir Run Run Shaw Hospital, the affiliated hospital of Zhejiang University School of Medicine, No. 3, Qingchun Road, Hangzhou, 310000, China E-mail: 11518310edu.cn
Corresponding author: Professor Liping Cao, Department of General Surgery, Sir Run Run Shaw Hospital, the affiliated hospital of Zhejiang University School of Medicine, No. 3, Qingchun Road, Hangzhou, 310000, China E-mail: 11518310edu.cn

 Global reach, higher impact
Global reach, higher impact