3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(2):321-330. doi:10.7150/jca.22218 This issue Cite
Research Paper
Positive Expression of SMYD2 is Associated with Poor Prognosis in Patients with Primary Hepatocellular Carcinoma
1. Department of Center Clinical Pharmacology and Pharmacy, The Third Xiangya Hospital, Central South University, Changsha, China;
2. Department of General Surgery, The Second Xiangya Hospital, Central South University, Changsha, China;
3. Department of Hepatobiliary Surgery, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, China;
4. Department of Obstetrics and Gynecology, Taizhou Hospital of Zhe Jiang Province, Taizhou, China;
5. Center for Experimental Medicine Research, The Third Xiangya Hospital, Central South University, Changsha, China.
Received 2017-8-3; Accepted 2017-10-24; Published 2018-1-1
Abstract
Purpose: SET and MYND domain-containing protein2 (SMYD2), a histone lysine methyltransferases, is a candidate human oncogene in multiple tumors. However, the expression dynamics of SMYD2 in hepatocellular carcinoma (HCC) and its clinical/prognostic significance are unclear.
Methods: The SMYD2 expression profile was examined by quantitative real-time polymerase chain reaction (qRT-PCR), and immunohistochemistry (IHC) in HCC tissues and matched adjacent non-tumorous tissues. SMYD2 was silenced in HCC cell lines to determine its role in tumor proliferation and cell cycle progression, and the possible mechanism. Spearman's rank correlation, Kaplan-Meier plots and Cox proportional hazards regression model were used to analyze the data.
Results: The SMYD2 expression in HCC tissues were significantly up-regulated at both mRNA and protein levels as compared with the matched adjacent non-tumorous tissues. By IHC, positive expression of SMYD2 was examined in 122/163 (74.85%) of HCC and in 10/59 (16.95%) of tumor-adjacent tissues. Positive expression of SMYD2 was correlated with tumor size, vascular invasion, differentiation and TNM stage (P < 0.05). In univariate survival analysis, a significant association between positive expression of SMYD2 and shortened patients' survival was found (P < 0.05). Importantly, SMYD2 expression together with vascular invasion (P < 0.05) provided significant independent prognostic parameters in multivariate analysis. Functionally, SMYD2 silenced markedly inhibited cell proliferation and cell cycle progression in SMMC-7721 cell.
Conclusions: Our findings provide evidences that positive expression of SMYD2 in HCC may be important in the acquisition of an aggressive phenotype, and it is an independent biomarker for poor prognosis of patients with HCC.
Keywords: SMYD2, hepatocellular carcinoma, immunohistochemistry, prognosis.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignant tumor that accounts for 80% of all liver cancer cases worldwide [1]. HCC is particularly problematic in China, where the incidence of HCC is much higher than that in other Asian countries due to endemic Hepatitis B virus infection [2,3]. At present, liver transplantation and surgical liver resection are the curative treatments for early-stage HCC. Cellular and molecular mechanisms that contribute to the progression of liver cancer from an acute or a chronic form to metastatic disease are not fully understood [1]. Therefore, the identification of new possible targets for preventing the initiation and progression of HCC is urgent and must be improved.
SET and MYND domain-containing protein 2 (SMYD2), a histone lysine methyltransferases, was first identified as one of the SMYD family members, containing a SET domain and a MYND domain [4]. SMYD2 methylates H3K36 and functions as a transcriptional regulator in cooperation with the Sin3A and HDAC1 histone deacetylase complex [4,5]. It was initially identified expression in muscle tissues and might be associated with muscle cells differentiation [6]. Recently, emerging evidence suggests that SMYD2 inhibits tumor suppressor proteins p53 [7], Rb [5,8], PTEN [9] and enhances the poly (ADP-ribose) activity of the oncogenic protein PARP1 in cancer cells [10]. Moreover, SMYD2 also plays promoting roles in the development and progression of various tumors. The mRNA and protein expression levels of SMYD2 have been reported to be overexpressed in bladder carcinoma [5], oesophageal squamous cell carcinoma (ESCC) [11], paediatric acute lymphoblastic leukaemia [12], gastric cancer [13] and HPV-unrelated head and neck squamous cell carcinoma(HNSCC) [14]. Moreover, SMYD2 expression has been found to be associated with the poor prognosis of ESCC, paediatric acute lymphoblastic leukaemia, gastric cancer and HPV-unrelated head and neck squamous cell carcinoma(HNSCC) [11-14]. However, to date, there has been no report on the clinical and prognostic significance of SMYD2 in patients with HCC. These findings prompted us to investigate the effects of SMYD2 overexpression and activation in HCC.
In the present study, quantitative real-time polymerase chain reaction (qRT-PCR), and immunohistochemistry (IHC) were utilized to examine the expression dynamics of SMYD2 in a cohort of HCC and matched adjacent non-tumorous tissues. We found that the expression level of SMYD2 in human HCC was significantly elevated, and the increased expression level of SMYD2 was associated with unfavorable clinical features and the poor prognosis of the HCC patients. Our in vitro studies demonstrated that SMYD2 promoted the tumor growth of HCC by facilitating cell proliferation and cell cycle progression. Our results indicated that SMYD2 is a promising biomarker of HCC, and can potentially serve as a therapeutic target of HCC.
Materials and Methods
Clinical samples and data
Clinical specimens derived from 163 patients, including 143 males and 20 females who were diagnosed with primary HCC, were examined in the present study. All patients underwent surgical resection at the Department of Hepatobiliary Surgery of Xiangya Hospital (Changsha, China) during January 2003 to December 2012. All patients did not receive any chemotherapy or embolization during the perioperative period. The clinicopathological characteristics are summarized in Table 1. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained for the use of these clinical materials for research purposes. The tumor specimens were obtained from the paraffin blocks of 163 primary HCCs. We also obtained 59 samples, in paraffin blocks, tumor-adjacent hepatocellular tissue from the same HCC patients. Tumor grade and stage were defined according to the criteria of the World Health Organization and the sixth edition of the TNM classification of the International Union Against Cancer (UICC, 2002). Patient follow-up was terminated on Jannuary, 2012. Median follow-up was 18 months (range, 1-93). HCC-related death [15] was defined if there was a computed tomography scan increase of >25% in the sum of recurrent tumor index lesions' cross-sectional areas or new onset of, or increase in, either recurrence, vascular invasion, or metastasis within the last 6 months of life, irrespective of immediate cause of death. Overall survival (OS) was defined as the time interval between the date of resection and the date of death or the last observation.
Immunohistochemistry
Paraffin-embedded samples were cut into 4-μm thick slices. The slides were dried overnight at 37°C, dewaxed in xylene, rehydrated with graded alcohol, and immersed in 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity. Antigen retrieval was carried out in a microwave oven with 10 mM citrate buffer (pH 6.0) for 20 min. The slides were incubated with 10% normal goat serum at room temperature for 30 min to reduce nonspecific reactions. Subsequently, the slides were incubated overnight at 4°C with rabbit polyclonal antibody against SMYD2 (1:50; Proteintech Group, Chicago, USA), overnight at 4°C. After rinsing four times with 0.01 mol/L phosphate-buffered saline (PBS; pH 7.4) for 10 min, primary antibody was detected using a secondary antibody (Maxim, FuZhou, China) for 1 h at room temperature and stained with 3,3-diaminobenzidine (DAB) after washing in PBS again. Finally, the sections were counterstained with Maxim's hematoxylin, dehydrated, and mounted.
Two independent pathologists blinded to the clinicopathological information performed the analysis of IHC for SMYD2. Immunopositive staining was evaluated in randomly selected five areas of the tissue section. For SMYD2 protein expression, sections were scored as positive if epithelial cells showed immunostaining in the nucleus/cytoplasm when observed. The tissue sections were scored based on the percentage of SMYD2 expressing cells in the total population (proportion score 0-10% = 0, 1-25% = 1, 26-50% = 2, 51-75% = 3 and 76-100% = 4) and scoring SMYD2 staining intensity as negative = 0, mild = 1, moderate = 2, intense = 3 using high-power (×400) microscopy. Finally, expression of SMYD2 was graded based on the sum of the intensity score and the proportion score: positive expression (intensity multiply proportion scores >6) or negative expression (intensity multiply proportion scores ≤6).
Correlation of SMYD2 expression in tissue with patients' clinicopathological variables in 163 cases of HCC
| Variables | All cases (N=163) | SMYD2 expression (%) | ||
|---|---|---|---|---|
| Positive expression (N=122) | Negative expression (N=41) | p-valuea | ||
| Age(years) | 0.88 | |||
| ≤50b | 97 | 73(75.25) | 24(24.74) | |
| >50 | 66 | 49(74.24) | 17(25.76) | |
| Gender | 0.07* | |||
| Male | 143 | 104(72.73) | 39(27.23) | |
| Female | 20 | 18(90.00) | 2(10.00) | |
| AFP(ug/L) | 0.81 | |||
| ≤20 | 61 | 45(73.77) | 16(26.23) | |
| >20 | 102 | 77(75.49) | 25(24.51) | |
| HBV | 0.38 | |||
| Negative | 22 | 7(31.82) | 15(68.18) | |
| Positive | 141 | 105(74.47) | 36(25.53) | |
| HCV | 0.99* | |||
| Negative | 157 | 117(74.52) | 40(25.48) | |
| Positive | 6 | 5(83.33) | 1(16.67) | |
| Cirrhosis | 0.68 | |||
| No | 75 | 55(73.33) | 20(26.67) | |
| Yes | 88 | 67(76.14) | 21(23.86) | |
| Tumor size(cm) | 0.04 | |||
| ≤5 | 60 | 57(95.0) | 3(5.00) | |
| >5 | 103 | 65(63.11) | 38(36.89) | |
| Tumor encapsulation | 0.45 | |||
| None | 75 | 54(72.00) | 21(28.00) | |
| Complete | 88 | 68(77.23) | 20(22.73) | |
| Tumor number | 0.51* | |||
| Single | 149 | 110(73.83) | 39(26.17) | |
| Multiple | 14 | 12(85.71) | 2(14.29) | |
| Vascular invasion | 0.01* | |||
| No | 123 | 86(69.92) | 37(30.08) | |
| Yes | 40 | 36(90.00) | 4(10.00) | |
| Distant metastasis | >0.99* | |||
| Negative | 158 | 118(74.68) | 40(25.32) | |
| Positive | 5 | 4(80.00) | 1(20.00) | |
| Differentiation | <0.01 | |||
| Poor-moderate | 127 | 105(82.68) | 22(17.32) | |
| Well | 36 | 19(52.78) | 17(47.22) | |
| TNM stage | 0.04 | |||
| Ⅰ | 99 | 68(68.69) | 31(31.31) | |
| Ⅱ-Ⅲ-Ⅳ | 64 | 54(84.38) | 10(15.63) | |
aChi-square test. bmean age. *Fisher exact test.
HCC cell lines and cell cultures
The human HCC cell lines MHCC-97H, MHCC-97L and HepG2 were obtained from the Jennio Biological Technology (Guangzhou, China) and SMMC-7721 was obtained from the KeyGen Biological Technology (Nanjing, China). MHCC-97H, MHCC-97L and SMMC-7721 Cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. All cells were grown in a humidified incubator at 37°C with 5% CO2.
Paired tumor and adjacent tissues
Ten pairs of HCC tissues and matched adjacent non-tumorous tissues were selected from the Department of Hepatobiliary Surgery of Xiangya Hospital, and then frozen and stored in -80℃ refrigerator until used to compare the expression levels of SMYD2 mRNA.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from the 10 pairs of HCC tissue and matched adjacent non-tumorous tissues using TRIZOL reagent (OMEGA, USA). RNA was reverse-transcribed using ReverTra Ace Qpcr RT Master Mix (TOYOBO, Osaka, Japan) according to the manufacturer's instructions. The SMYD2 sense primer was 5'-ATCTCCTGTACCCAACGGAAG-3', and the antisense primer was 5'-CACCTTGGCCTTATCCTTGTCC-3'. For the β-actin gene, the sense primer was 5'-TGGCACCCAGCACAATGAA-3', and the antisense primer was 5'-CTAAGTCATAGTCCGCCTAGAAGCA-3'. qRT-PCR was done using SYBR Green PCR master mix (TOYOBO, Osaka, Japan) in a total volume of 20 μl on the ABI 7500 thermocycler (Thermo Fisher Scientific, Inc., USA) as follows: 95°C for 1 min, 40 cycles of 95°C for 15s, and 60°C for 20s. A dissociation procedure was performed to generate a melting curve for confirmation of amplification specificity. β-actin was used as the reference gene. The relative levels of gene expression were represented as △Ct=Ctgene-Ctreference, and the fold change of gene expression was calculated by the 2-△△Ct Method. Experiments were repeated in triplicate.
Western blotting
Equal amount of whole-cell lysates were resolved with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Bio-Rad, USA). This was followed by incubation with primary rabbit polyclonal antibody against human SMYD2 (1:1500) (Abcam, Cambridge, MA), cyclin D1 (1:10000) (Abcam, Cambridge, MA). The immunoreactive proteins were detected with enhanced chemiluminescent detection reagents (Millipore Corporation, USA) according to the manufacturer's instructions. The membranes were stripped and reblotted with a rabbit monoclonal anti-GAPDH antibody (Bioworld Technology, Minneapolis, USA) as a loading control.
Small interfering RNA transfection
siRNA oligonucleotide duplexes and siNegative control (siNC) were synthesized by GenePharma, Suzhou. Co., Itd. (Suzhou, China). The siRNA sequences are described in Table 2. SiRNA duplexes (final concentration, 50nM) were transfected into HCC cell lines with an HVJ Envelope (HVJ-E) Vector Kit GenomONE-Neo (Ishikawa Sangyo KaishaLtd., Osaka, Japan) by modified methods described in the manufacturer's instructions and by Tsuchiya et al. [16]. Briefly, HVJ-E (10ul) was mixed with 5ul of siRNA solution and 3ul of Reagent B. After centrifugation, the pellet was resuspended in 27ul of buffer, followed by the addition of 8ul of Reagent C. The siRNA-HVJ-E mixture was combined with HCC cells and centrifuged at 12,000 rpm for 20 min at 4℃. The cells were resuspended in 1 ml of culture media and incubated for 72h.
siRNA Sequences.
| siRNA Name | Sequence | ||||
|---|---|---|---|---|---|
| siNegative control | Sense | UUCCCGAACGUGUCACGU | |||
| Antisense | ACGUGACACGUUCGGAGAA | ||||
| siSMYD2 no.1 | Sense | GAUUUGAUUCAGAGUGACA | |||
| Antisense | UGUCACUCUGAAUCAAAUC | ||||
| siSMYD2 no.2 | Sense | GGUUAAGAGAUUCUUAUUU | |||
| Antisense | AAAUAAGAAUCUCUUAACC | ||||
EdU assay
EdU assays was used to assess cell proliferation. Briefly, cells were cultured in 96-well plates and transfected with siSMYD2 or siNC. Cell proliferation was evaluated on 24h, 48h and 72h. Then, cells were incubated with 50 μl of EdU (Ribobio Co., LTD, Guangzhou, China) for additional 2 h at 37 °C. Cells were fixed with 4% formaldehyde for 30 min, incubated with glycine (2 mg/ml) for 5 min and treated with 0.5% Triton X-100 for 10 min to permeabilize cells. After being washed with phosphate-buffered saline, cells were incubated with Apollo reaction cocktail for 30 min and treated twice with 0.5% Triton X-100. DNA was stained with Hoechst 33342 stain for 30 min and visualized/calculated with Operetta (Perkin Elmer, USA). Six groups of confluent cells were randomly selected from each sample image.
Flow cytometry
For cell cycle analysis, cells were collected at the indicated time points. Cells (1 × 106) were washed with PBS and fixed with cold 70% ethanol at 4°C overnight. Then, cells were treated with RNase and stained with propidium iodide (PI, Wellbio, China). The DNA content of the cells was quantified using a flow cytometer (BD FACSCalibur, USA). In total, 10,000 nuclei were examined in the flow cytometer, and DNA histograms were analyzed by ModFit software (Verity Software House, USA).
Statistical analysis
Data were analyzed using SPSS18.0 software (SPSS Inc.). Significant associations between SMYD2 expression and clinicopathological parameters were assessed using a χ2 test. Survival curves were plotted by Kaplan-Meier analysis and compared by the log-rank test. Cox regression analysis was carried out to assess the significance of variables for survival. Data were expressed as mean ± SD, and the t-test was used to determine the significance of differences between two groups. All tests carried out were two-sided. P < 0.05 was considered statistically significant.
Results
qRT-PCR and Western blotting analysis of SMYD2 expression in HCC tissues and cell lines
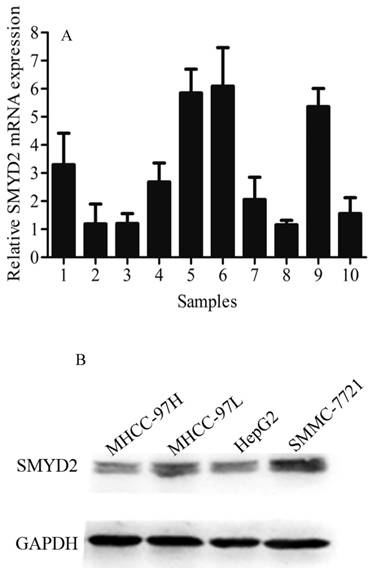
SMYD2 exhibited a markedly higher expression in HCC tumor tissues than in matched adjacent non-tumorous tissues, as determined by qRT-PCR (Figure 1A). SMMC-7721, HepG2, MHCC-97H, and MHCC-97L cells were detected by Western blot analysis to determine the SMYD2 expression in different liver cancer cell lines. The results exhibit as following (in descending order of their SMYD2 levels): SMMC-7721, MHCC-97L, HepG2 and MHCC-97H. Almost positive results were obtained in different liver cancer cell lines (Figure 1B).
IHC analysis of SMYD2 expression in paraffin-embedded of HCC tissues
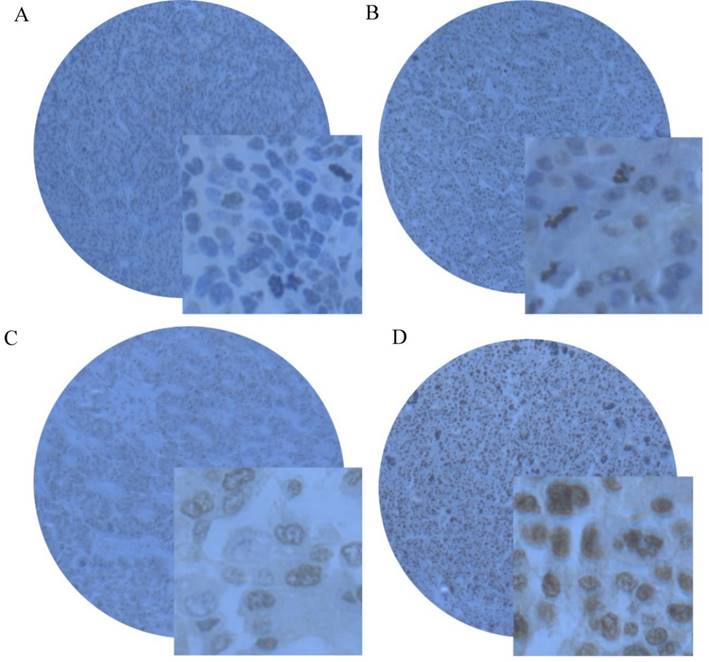
The expression of SMYD2 protein was determined by IHC in paraffin-embedded of HCC tissues containing 163 cases of HCC tissues and 59 specimens of matched adjacent non-tumorous tissues. Using the criteria described earlier, positive expression of SMYD2 was detected in 74.85% (122/163) of HCC tissues, while only 16.95% (10/59) of matched adjacent non-tumorous tissues had positive staining (Figure 2A-2D).
Association of SMYD2 expression with HCC clinicopathological features
The association between SMYD2 expression in HCC detected by IHC and several known clinicopathological features were studied further. SMYD2 levels were inversely correlated with tumor size, vascular invasion, differentiation and TNM stage (P < 0.05, Table 1). There was no significant association between SMYD2 expression and other clinicopathological features, such as patient gender, age, AFP level, and Hepatitis B Virus (HBV), Hepatitis C Virus (HCV), cirrhosis, tumor encapsulation, tumor number and distant metastasis (P > 0.05, Table 1).
Relationship between clinicopathological variables, SMYD2 expression, and HCC patient survival: univariate survival analysis
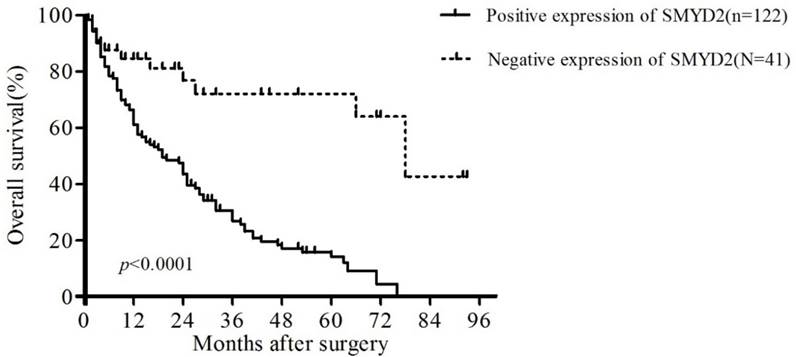
To confirm the representativeness of the HCC in our study, we analyzed the established prognostic predictors of the overall survival (OS) in our cohort. Kaplan-Meier analysis demonstrated a significant impact of well-known clinicopathological prognostic parameters on patient survival, such as vascular invasion, differentiation and TNM stage (P < 0.05, Table 3). Assessment of the overall survival curves showed that patients with positive staining of SMYD2 had significantly reduced OS (P <0.01, Table 3, Figure 3). These data suggest that SMYD2 may be involved in the development and progression of HCC, and can serve as a promising predictor of the prognosis of HCC.
Univariate analysis of factors associated with overall survival of 163 patients with HCC
| OS | |||
|---|---|---|---|
| univariate analysis | |||
| HR | 95%CI | p-value | |
| Age(>50vs.≤50) | 1.15 | 0.77-1.37 | 0.47 |
| Gender (male vs. female) | 1.61 | 0.92-2.81 | 0.10 |
| AFP (≤20 vs.>20) | 0.77 | 0.52-1.14 | 0.19 |
| HBV (positive vs. negative) | 1.03 | 0.59-1.78 | 0.93 |
| HCV (positive vs. negative) | 0.69 | 0.22-2.19 | 0.53 |
| Cirrhosis (yes vs. no) | 1.34 | 0.90-1.99 | 0.15 |
| Tumor size(cm)(>5 vs. ≤5) | 1.19 | 0.80-1.77 | 0.39 |
| Tumor encapsulation (none vs. complete) | 0.74 | 0.49-1.10 | 0.13 |
| Tumor number (multiple vs. single) | 0.95 | 0.51-1.78 | 0.88 |
| Vascular invasion (yes vs. no) | 2.55 | 1.51-4.31 | <0.01 |
| Distant metastasis (positive vs. negative) | 0.56 | 0.21-1.47 | 0.24 |
| Differentiation (poor-moderate vs. well) | 1.74 | 1.13-2.70 | 0.01 |
| TNM stage(Ⅱ-Ⅲ-Ⅳvs.Ⅰ) | 1.62 | 1.08-2.44 | 0.02 |
| SMYD2 expression (positive vs. negative) | 2.89 | 1.87-4.46 | <0.01 |
Independent prognostic factors of HCC: multivariate Cox regression analysis
The expression of SMYD2 as well as other clinical pathological parameters that were significant in univariate analysis (vascular invasion, differentiation, TNM stage and SMYD2 expression), was further examined in multivariate analysis. Positive expression of SMYD2 was found to be an independent prognostic factor for poor overall survival (relative risk: 4.00, 95% confidence interval: 2.03-7.91, P <0.01, Table 4). Of the other parameters, vascular invasion was also demonstrated as independent prognostic factors for overall survival (P < 0.05, Table 4).
Multivariate analysis of prognostic factors on overall survival (Cox regression model)
| OS | |||
|---|---|---|---|
| multivariate analysis | |||
| HR | 95%CI | p-value | |
| Vascular invasion (yes vs. no) | 1.64 | 1.06-2.53 | 0.03 |
| Differentiation (poor-moderate vs. well) | 1.06 | 0.63-1.78 | 0.83 |
| TNM stage(Ⅱ-Ⅲ-Ⅳvs.Ⅰ) | 1.06 | 0.61-1.82 | 0.84 |
| SMYD2 expression (positive vs. negative) | 4.00 | 2.03-7.91 | <0.01 |
Expression of SMYD2 in HCC tumor tissues, matched adjacent non-tumorous tissues and cell lines. (A) qRT-PCR analysis was performed to analyze SMYD2 expression in 10 pairs of HCC tissues and matched adjacent non-tumorous tissues, the data are shown the mean of 2-△△Ct, and the expression of SMYD2 in HCC is significantly higher than that in matched adjacent non-tumorous tissues (P <0.05); (B) Western blot assay was performed to analyze SMYD2 expression in liver cancer cell lines (MHCC-97H, MHCC-97L, HepG2, and SMMC-7721). C: HCC tumor tissues, N: matched adjacent non-tumorous tissues
Representative immunohistochemical staining of SMYD2 in HCC tissues and matched adjacent non-tumorous tissue. (A) Negative staining of SMYD2 in matched adjacent non-tumorous tissue with the magnifying power of 400×. (B) Low. (C) medium and (D) high expression of SMYD2 in HCC tissues.
Kaplan-Meier survival analysis of the correlation between SMYD2 expression in patients with HCC and OS. Patients with positive SMYD2 expression (solid line) exhibited decreased OS.
SMYD2 promotes the growth of HCC cells in vitro
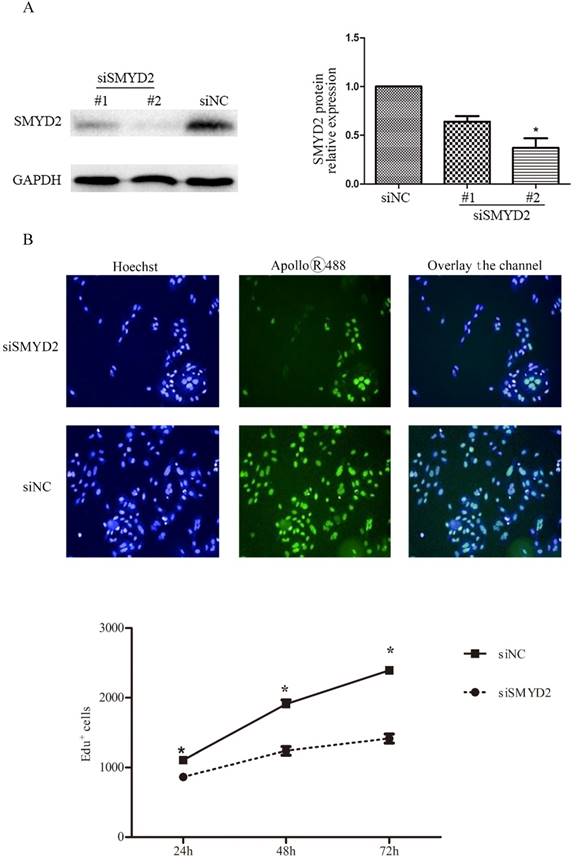
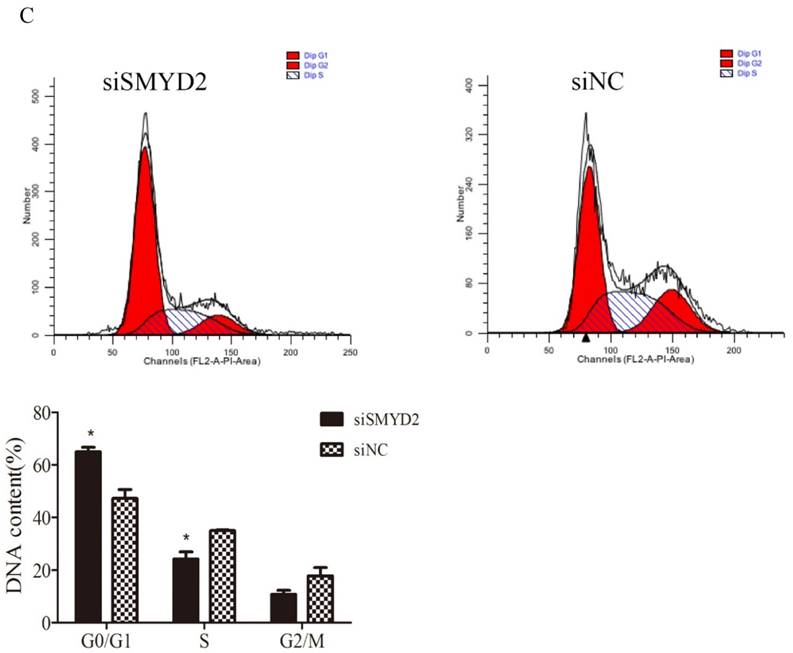
To explore the functional of SMYD2 in HCC, we performed cell proliferation assays using siRNA specific for SMYD2 and investigated whether silence of SMYD2 would suppress proliferation of liver cancer cell lines. We chose the SMMC-7721 cell line for these assays, because it had the highest amount of SMYD2 protein (Figure 1B). Expression of SMYD2 protein in this cell line was efficiently knocked down 72 h after the transient introduction of SMYD2-specific siRNA (siRNA-SMYD2) (Figure 4A) than with the control siRNA (siRNA-NC). Subsequently, the results of the EdU assays showed that the cell proliferation of the SMMC-7721 cells was significantly decreased after SMYD2 knockdown (P <0.05, Figure 4B). In addition, cell cycle assays showed that knockdown of SMYD2 significantly increased the percentage of cells in the G0/G1 phase (P <0.05), and decreased the percentage of cells in the S phase (P <0.05) (Figure 4C). These data suggest that SMYD2 promotes the growth of HCC cells by regulating cell proliferation and cell cycle progression in vitro.
Suppression of SMYD2 expression inhibits the cell proliferation and cell cycle progression in SMMC-7721 cells. (A) SMYD2-specific siRNA significantly inhibited the levels of SMYD2 protein in the SMMC-7721 cells. (B) Cell proliferation as assessed by EdU assay was inhibited after SMYD2 knockdown. (C) Cell cycle assays demonstrated that SMYD2 silience increased the percentage of SMMC-7721 cells in the G0/G1 phase while decreased the percentage of SMMC-7721 cells in the S phase. n=3 independent repeats with similar results. *P <0.05 by t-test.
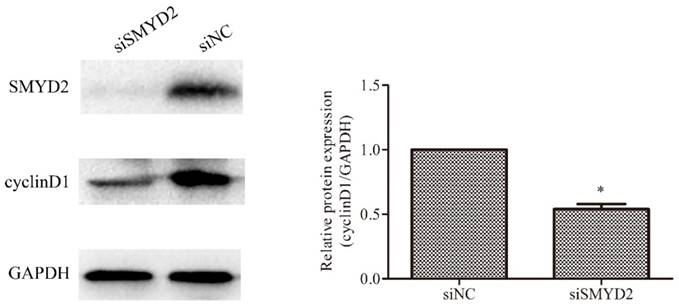
SMYD2 regulates the expression of cyclin D1 in HCC cells. SMYD2 knockdown in SMMC-7721 cells significantly reduced the protein level of cyclin D1. *P <0.05 by t-test.
SMYD2 regulates the expression of cyclin D1 in HCC cells
Cyclin D1 has been found to be overexpressed in HCC [17, 18], and plays an important role in regulating cell cycle progression [19]. Therefore, we ascertained whether SMYD2 regulates the expression of cyclin D1 in HCC cells. The results of the Western blot analyses showed that the expression of cyclin D1 was significantly reduced after downregulation of the expression of SMYD2 (P <0.05, Figure 5). These data indicate that SMYD2 may regulate the expression of cyclin D1 in HCC cells.
Discussion
In spite of significant advances in its diagnosis and treatment, HCC remains one of the most lethal tumors in China [20]. Therefore, it is of great importance to explore HCC-specific biomarkers and effective therapeutic targets, which may promote the early diagnosis and improve the efficacy of molecular-targeted drugs for HCC. Several SMYD family members have been reported to be involved in oncogenesis and tumor progression. For example, Liu et al. reported that overexpression of SMYD3 promoted tumor cell proliferation and was a predictor of poor prognosis in gastric cancer [21], and others have shown that SMYD4 acts as a tumor suppressor in breast cancer [22]. Despite this existing body of knowledge, the role of SMYD2 in the progression of HCC has not been explored.
In the present study, we measured SMYD2 expression in primary HCC samples by qPCR. Similar to previous studies on oesophageal squamous cell carcinoma (ESCC), bladder carcinoma, paediatric acute lymphoblastic leukaemia, gastric cancer and HPV-unrelated head and neck squamous cell carcinoma(HNSCC) [5, 11-14], we found that SMYD2 expression in HCC tumor tissue was significantly increased compared with matched adjacent non-tumorous tissue. Importantly, we found that the positive expression of SMYD2 was associated with poor prognosis and reduced survival periods for HCC patients. Multivariate analysis showed that the positive expression of SMYD2 protein expression could be used as an independent prognostic predictor for HCC patients. Our results indicate that the expression level of SMYD2 protein might provide useful information in the evaluation prognosis and follow up schedule guiding for HCC patients. In addition, Silence of endogenous SMYD2 expression using a specific siRNA suppressed the proliferation and cell cycle progression of SMMC-7721 cells. CyclinD1 was also down-regulated under that condition. Taken together, these findings may suggest that SMYD2 has an oncogenic role in the HCC cell line. Cyclin D1 functions as a well-recognized regulator of cell cycle progression and proliferation, and has been found to be deregulated in many types of human cancers, including HCC [23-25]. In the present study, we confirmed that SMYD2 regulated the expression of cyclin D1 in HCC cells using specific siRNA. SMYD2 as a lysine methyltransferase (KMT) have been validated suppress p53 at Ly370. PTEN protein was shown to be methylated at lysine 313 in breast cell lines [9], and methylated RB at lysine 860 in bladder cancer cell lines [5]. Therefore, SMYD2 might act through different mechanisms in different cancer types.
In summary, the present study confirmed for the first time that SMYD2 is overexpressed in HCC tissues. Patients with positive expression of SMYD2 had adverse clinicopathological features and poorer prognosis. Functionally, SMYD2 can promote the growth of HCC cells by promoting cell proliferation and cell cycle progression. Furthermore, SMYD2 can regulate the expression of cyclin D1 in HCC cells. Therefore, the present study demonstrated that SMYD2 can serve as a promising biomarker for the early diagnosis and prognostic prediction of HCC patients, and can potentially act as an effective therapeutic target of HCC.
Acknowledgements
Shi-Kun Liu designed the study and revised the manuscript critically. Shan-Ru Zuo evaluated the clinical records, carried out the experimental work and drafted the manuscript. Xiao-Cong Zuo contributed for critical revision of statistical analysis and of the manuscript. Wei-Jin Fang, Chun-Jiang Wang, and Yang He contributed for data interpretation and drafted the manuscript. Heng Zou, Pan Chen and Ling-Fei Huang help to collect the samples and participate in the design of the study. Li-Hua Huang and Hong Xiang contributed to carry out the immunohistochemistry assays. This work was supported by Program for New Century Excellent Talents in University (grant number NCET-13-0605, 2013)(to Z.X.C), the New Xiangya Talent Project of the Third Xiangya Hospital of Central South University(grant number 20150218, 2015)(to Z.X.C), and National Natural Science Foundation of China (grant number 81102512) (to Z.X.C).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Aravalli RN, Cressman EN, Steer CJ. Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch Toxicol. 2013;87:227-247
2. Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90
3. Poon RT, Fan ST. Hepatectomy for hepatocellular carcinoma: patient selection and postoperative outcome. Liver Transpl. 2004;10:S39-45
4. Brown MA, Sims RJ 3rd, Gottlieb PD. et al. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer. 2006;5:26
5. Cho HS, Hayami S, Toyokawa G. et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14:476-486
6. Kawamura S, Yoshigai E, Kuhara S. et al. Smyd1 and Smyd2 are expressed in muscle tissue in Xenopus laevis. Cytotechnology. 2008;57:161-168
7. Huang J, Perez-Burgos L, Placek BJ. et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629-632
8. Saddic LA, West LE, Aslanian A. et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285:37733-37740
9. Nakakido M, Deng Z, Suzuki T. et al. Dysregulation of AKT Pathway by SMYD2-Mediated Lysine Methylation on PTEN. Neoplasia. 2015;17:367-373
10. Piao L, Kang D, Suzuki T. et al. The Histone Methyltransferase SMYD2 Methylates PARP1 and Promotes Poly(ADP-ribosyl)ation Activity in Cancer Cells. Neoplasia. 2014;16:257-264
11. Komatsu S, Imoto I, Tsuda H. et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30:1139-1146
12. Sakamoto LH, Andrade RV, Felipe MS. et al. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leukemia Research. 2014;38:496-502
13. Komatsu S, Ichikawa D, Hirajima S. et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. British Journal of Cancer. 2014;112:357-364
14. Ohtomo-Oda R, Komatsu S, Mori T. et al. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Human Pathology. 2016;49:145-155
15. Tao YM, Huang JL, Zeng S. et al. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology. 2013;57:2326-2337
16. Tsuchiya Y, Okuno Y, Hishinuma K. et al. 4-Hydroxy-2-nonenal-modified glyceraldehyde-3-phosphate dehydrogenase is degraded by cathepsin G. Free Radic Biol Med. 2007;43:1604-1615
17. Deane NG, Parker MA, Aramandla R. et al. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 2001;61:5389-5395
18. Joo M, Kang YK, Kim MR. et al. Cyclin D1 overexpression in hepatocellular carcinoma. Liver. 2001;21:89-95
19. Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158-163
20. Shariff MI, Cox IJ, Gomaa AI. et al. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353-367
21. Liu Y, Liu H, Luo X. et al. Overexpression of SMYD3 and matrix metalloproteinase-9 are associated with poor prognosis of patients with gastric cancer. Tumor Biology. 2015;36:4377-4386
22. Hu L, Zhu YT, Qi C. et al. Identification of Smyd4 as a Potential Tumor Suppressor Gene Involved in Breast Cancer Development. Cancer Research. 2009;69:4067-4072
23. Tu K, Liu Z, Yao B. et al. BCL-3 promotes the tumor growth of hepatocellular carcinoma by regulating cell proliferation and the cell cycle through cyclin D1. Oncol Rep. 2016;35:2382-2390
24. Freiburghaus C, Janicke B, Lindmark-Månsson H. et al. Lactoferricin treatment decreases the rate of cell proliferation of a human colon cancer cell line. J Dairy Sci. 2009;92:2477-2484
25. Zwijsen RM, Klompmaker R, Wientjens EB. et al. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol Cell Biol. 1996;16:2554-2560
Author contact
![]() Corresponding author: Shi-Kun Liu, PhD. Department of Center Clinical Pharmacology and Pharmacy, The Third Xiangya Hospital of Central South University, No.172 Tongzipo Road, YueLu District Changsha, Hunan P.R.China, 410013. E-mail: L8618496com
Corresponding author: Shi-Kun Liu, PhD. Department of Center Clinical Pharmacology and Pharmacy, The Third Xiangya Hospital of Central South University, No.172 Tongzipo Road, YueLu District Changsha, Hunan P.R.China, 410013. E-mail: L8618496com

 Global reach, higher impact
Global reach, higher impact