3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(3):540-547. doi:10.7150/jca.22405 This issue Cite
Research Paper
Identification of Kininogen 1 as a Serum Protein Marker of Colorectal Adenoma in Patients with a Family History of Colorectal Cancer
1. Cancer Institute (Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education), the Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
2. Department of Medical Oncology, the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China.
3. Research Center for Air Pollution and Health, School of Medicine, Zhejiang University, Hangzhou, China
*These authors contributed equally to this work.
Received 2017-8-16; Accepted 2017-12-21; Published 2018-3-8
Abstract
The serum protein markers of colorectal adenoma in patients with a family history of colorectal cancer have been rarely reported. Serum samples from colorectal adenoma patients with or without a family history of colorectal cancer and healthy controls were profiled using Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS). The model to distinguish colorectal adenoma patients with a family history of colorectal cancer from atypical hereditary colorectal families (CRA-H) and sporadic colorectal adenoma patients without a family history of colorectal cancer (CRA-S) was established with 85.0% accuracy. The model distinguishing CRA-H from healthy individuals was established with 90.0% specificity and 86.7% sensitivity. Additionally, five peaks (2202, 5821, 3260, 2480, and 2218) showing differential expression in advanced colorectal adenoma patients with a family history of colorectal cancer were selected. The protein Kininogen 1 (KNG1) was identified in colorectal adenoma patients and validated using Western Blotting. KNG1 may be a biomarker for colorectal adenoma patients with a family history of colorectal cancer.
Keywords: Colorectal adenoma, Family history of CRC, Proteomics
Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-related deaths in many countries. An estimated 1.4 million CRC cases were diagnosed worldwide in 2012(1), and the colorectal cancer rate has also markedly increased during the past 2 decades in China. Most of the newly diagnosed CRC cases were sporadic, but 20% of these individuals were predisposed to CRC because of genetic flaws inherited from their parents(2-4). These cases were recognized as family-aggregated CRC or hereditary CRC. Unfortunately, the predisposing gene mutation has been identified in only a small portion of hereditary CRC(5). Hereditary nonpolyposis colorectal cancer (HNPCC) and family adenomatous polyposis (FAP) are classical hereditary cancer syndromes, but these diseases account for less than 5% of all CRC cases (6, 7). Most of the hereditary CRCs are from atypical cancer families. The deficient genes accounting for the susceptibility of an atypical CRC family are unknown. Moreover, atypical hereditary CRC often observed in a small family with no more than two CRC cases(8). Therefore, it is almost impossible to trace their mutant genes by linkage analysis(9, 10). Recent advances in proteomics study present new techniques for the global measurement of protein markers(11-15). These techniques have been used in combination with bioinformatics approaches to identify potential biomarkers with high sensitivity and specificity for the diagnosis of colorectal cancer (16).
Assuming that individualistic susceptibility to CRC is reflected in the serum proteins of peripheral blood samples, a comparative analysis of the protein profiles in the blood samples of patients with family CRC and those without family CRC might yield consistent differences associated with the features of family history of CRC. To explore this idea, we tentatively explored the serum protein profiles in colorectal adenoma patients with and without a family history of CRC. The objective of the present was to use magnetic bead and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF-MS) combined with Support Vector Machine (SVM) tools to identify the serum protein markers in colorectal adenoma patients that represent hereditary indicators of colorectal cancer families.
Materials and methods
Sample preparation
This study was approved by the Institutional Review Board of Medical Research, The Second Affiliated Hospital, Zhejiang University School of Medicine and all experiments were carried out in accordance with the approved guidelines. A total of 170 serum samples from 145 unrelated patients or healthy controls (HC) were collected between 2011 and 2017, and analyzed at 2017 in the present study. The serum samples were evenly distributed into three groups: colorectal adenoma patients with a family history of colorectal cancer potentially from atypical hereditary colorectal families (CRA-H), sporadic colorectal adenoma patients without a family history of colorectal cancer (CRA-S) and healthy controls (HC). The detailed characteristics of the patients in each group are shown in Table 1. The 50 sera were collected from 25 CRC patients before surgery and after surgery, and 30 sera were collected from patients after colorectal adenomas endoscopic resection for controls (Table 2).
A complete colonoscopy was performed for all subjects. Two experienced pathologists in the Second Affiliated Hospital, Zhejiang University School of Medicine pathologically confirmed all colorectal adenomas. The serum samples were collected prior to colonoscopy in the morning before breakfast. The blood samples were exposed to room temperature for no more than 2 hours and were centrifuged at 3000 rpm for 10 min for the separation of serum samples. The serum samples were immediately stored at -80°C until further use. All authors had access to information that could identify individual participants during or after data collection.
Characteristics of the CRA patients
| CRA-H | CRA-S | ||
|---|---|---|---|
| Age | 40-49 | 5(16.7) | 7(23.3) |
| 50-59 | 15(50.0) | 12(40.0) | |
| 60-74 | 10(33.3) | 11(36.7) | |
| Gender | Male | 15(50.0) | 17(56.7) |
| Female | 15(50.0) | 13(43.3) | |
| Family CRC cases | 1 CRC patient | 19(63.3) | 0 |
| 2 CRC patients | 11(36.7) | 0 | |
| At least 1 CRC patient ≤50 yrs old | 6(20.0) | 0 | |
| Pathological features of adenomas | <1 cm in diameter | 21(70.0) | 21(70.0) |
| ≥1 cm in diameter | 9(30.0) | 9(30.0) | |
| Villous/tuberovillous | 1*(3.33) | 1*(3.33) | |
| High-grade dysplasia | 4*(13.3) | 3*(10.0) |
*All adenomas with villous/tuberovillous structure or high-grade dysplasia were ≥1 cm in diameter.
Control serum samples information.
| Samples | CRC | CRA after resection | HC |
|---|---|---|---|
| Male / Female | 18/7 | 21/9 | 15/15 |
| Average age(y) | 53.44 | 59.35 | 57.53 |
| Stage (I/II/III/IV) | 1/7/14/3 | / | / |
| Total Number | 25 | 30 | 30 |
This study was approved by the Institutional Review Board of Medical Research, The Second Affiliated Hospital, Zhejiang University School of Medicine. Written informed consent was obtained from every patient who donated samples for this study prior to initiating the experiments.
Magnetic bead separation and MALDI-TOF MS detection
Magnetic bead-based weak cation exchange chromatography (MB-WCX) (Bruker Daltonic, Bremen, Germany) was used to separate the peptides in the serum samples. The experiment was conducted using a standard protocol according to the manufacturer's instructions: 5 µl of serum was mixed with 10 µl of binding buffer and 10 µl of MB-WCX beads. The tube was placed into the magnetic separator and the supernatant was subsequently removed. After adding 100 µl of washing buffer to the tube, and the tube was shaken back and forth ten times in the magnetic separator to wash the beads. Subsequently, the supernatant was removed, and the washing step was repeated twice. Next, 5 µl of elution buffer was added to dissolve the beads. The tubes were placed in the magnetic separator, and the clear supernatant was transferred to a fresh tube. Subsequently, 5 µl of stabilizing buffer was added to the tubes. One microliter of the eluted sample was spotted onto a steel target, air-dried, and subsequently 1 μL of the matrix (0.3 mg/ml HCCA, 50% ACN, 2% TFA) was spotted onto the MALDI-TOF steel target, followed by air-drying. The samples were detected using ultrafleXtreme MALDI-TOF/TOF (Bruker Daltonics Corp.) after calibrating the instrument according to the ClinProt standard.
Bioinformatics and biostatistics analysis
The protein profiles were subsequently analyzed using bioinformatics tools Zhejiang University Proteomics Data Analysis System (ZJUPDAS) (11, 15)
We constructed a non-linear SVM classifier to distinguish the different groups. The leave-one-out cross-validation approach was applied to estimate the accuracy of the classifier. This approach leaves one sample out as the test set and retains the remaining samples as the training set. This process continues until each sample has been reserved one time as a test sample.
The capability of each peak in distinguishing multi-groups was estimated based on the p value of the Wilcoxon Rank-Sum Test. The top peaks with the lowest p values were selected for further analysis. All combinations of these peaks were estimated using leave-one-out cross-validation SVM. The peak combinations with the highest accuracy were selected as potential biomarkers, and the SVM model with the highest Youden's index was selected as the final model for detecting familial colorectal adenomas.
Identification of the biomarkers
The sequences of differentially expressed peptides were identified using ultrafleXtreme MALDI-TOF/TOF. After calibration according to a peptide standard (Bruker Daltonic, Germany), we used the reflection mode to detect the isotopic peaks, and subsequently used the lift model to detect the MS/MS fragment. BioTools and mascot software were used to search the database. The peptide mass tolerance was set at 0.1%, the fragment ion mass tolerance was set at 0.5 Da, and the mass type of parent peptide and peptide fragment were set at monoisotopic.
Western blot analysis
To verify the expression of KNG1, the serum samples (10 μl) and the protein lysate from tissues (10 μg) were separated by 15%, sodium dodecyl polyacryamide gel electrophoresis and subsequently transferred to polyvinylidene fluoride membranes. After blocking for 2 h at room temperature with blocking buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.1% TBS/Tween 20 ) containing 5% nonfat milk powder, the membranes were incubated for 2 hours at room temperature with primary antibody KNG1 (1:10,000 Abcam ab124737, Rabbit monoclonal antibody) and β-actin(1:1000, HuaAn Biotechnology M1210-2, for tissues) in blocking buffer. The membranes were washed with TBS/Tween 20, incubated for 1 h at room temperature in secondary antibody (1:10,000, Huabio, HA1001) in blocking buffer, and, finally, washed with TBS/Tween 20. The blots were developed using Immun-Star TMAP Substrate Pack (BioRad, USA) and scanned on an Epson Scan (Agilent Technologies, USA) scanner within the linear range of detection.
Results
The protein markers for distinguishing CRA-H from CRA-S
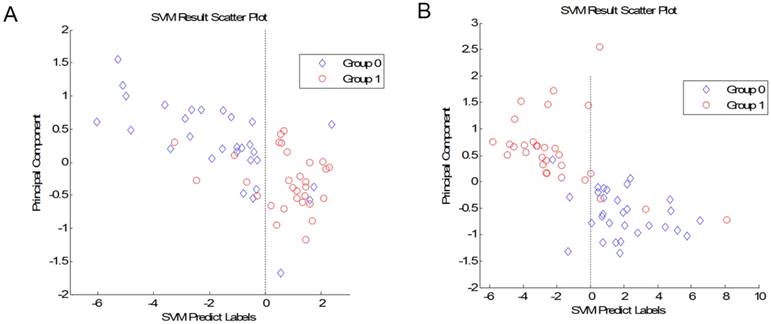
We compared the CRA-H group to the CRA-S group. A total of 12 peaks showed significant differences in the p value of the Wilcoxon rank sum test of less than 0.01. We further established the SVM model to differentiate adenoma patients with a family history from those without a family history. We selected the best combination of 7 peaks to build the model with the highest accuracy. The leave-one-out cross-validation test result of this model showed an accuracy of 86.7% for predicting patients without a family history and 83.3% for predicting patients with a family history (Fig 1A). The 7 peaks were 5776, 6805, 5821, 2072, 2053, 3470 and 1717.
In the acknowledged adenoma-carcinoma sequence, advanced adenomas are more likely to transition to cancer. Thus, the colorectal adenomas were farther differentiated to advanced and non-advanced according to adenoma sizes greater than 1 cm in diameter. Next, we compared 4 groups: Advanced CRA-H, Non-advanced CRA-H, Advanced CRA-S and Non-advanced CRA-S.
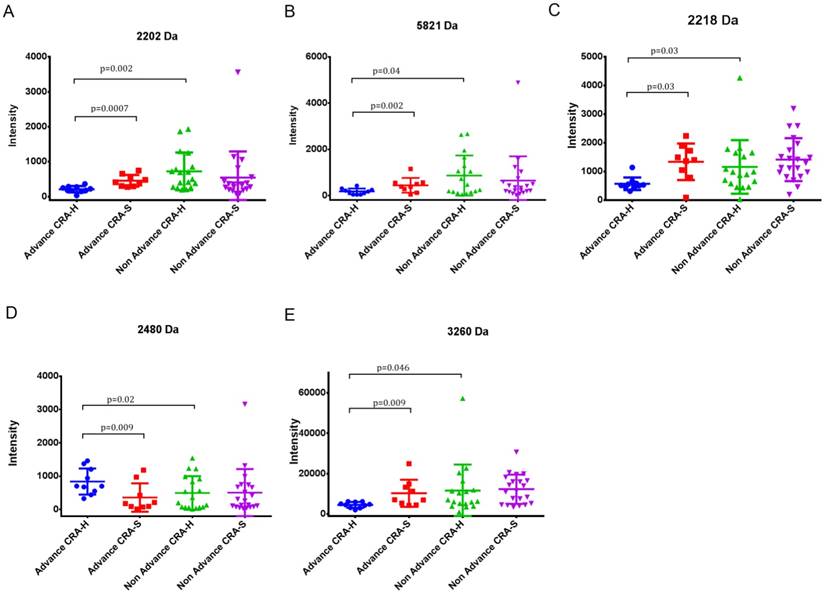
Five peaks (2202, 2218, 2480, 3260 and 5821) showing differences in Advanced CRA-H compared to Non-advanced CRA-H and Advanced CRA-H compared to advanced CRA-S were selected. In contrast, these five peaks showed no differences when comparing Non-advanced CRA-H to Non-advanced CRA-S and advanced CRA-S to Non-advanced CRA-S (Table 3, Fig 2 A, B, C, D and E). These five peaks showed significant differences in advanced colorectal adenoma patients with a family history compared with other groups. Furthermore, the mean intensity of peak 2218 in CRA (544.7±752.9) was significantly up-regulated (p=0.0035) than that in the CRC patients (374.9±767.9), and significantly down-regulated than that in the HCs (1824.6±1383.7; p=7.7E-07; Table 5). Thus, these five peaks may have specificity in Advanced CRA-H.
(A) Plot of the SVM prediction results between CRA-H and CRA-S (Group 1: CRA-S; Group 0: CRA-H). (B) Plot of the SVM prediction results between CRA-H and HC (Group 0: Healthy individuals; Group 1: CRA-H).
Statistics for peaks 2202, 2218, 2480, 3260 and 5821 in Advanced and Non-advanced CRA-H and CRA-S.
| 2202 | 2218 | 2480 | 3260 | 5821 | |
|---|---|---|---|---|---|
| Advanced CRA-H | 209.9±91.0 | 177.6±125.1 | 837.1±392.8 | 4487.5±1449.2 | 571.4±220.0 |
| Non-advanced CRA-H | 722±539.6 | 864.3±867.7 | 493.8±503.9 | 11599±12860.7 | 1158.1±935.6 |
| Advanced CRA-S | 452.7±167.2 | 441.1±320.6 | 358.3±426.9 | 10254.2±6714.5 | 1337.3±636.6 |
| Non-advanced CRA-S | 544.6±746.3 | 644.9±1052.6 | 504.8±707.4 | 12275.7±7239.8 | 1414.1±748.8 |
Comparison of CRA-H patients with healthy individuals
The 30 CRA-H patients were also compared to 30 healthy individuals. We established a model to distinguish CRA-H patients from healthy individuals based on the 8 peaks. This SVM model with peaks 4644, 4660, 4054, 4617, 4678, 1859, 3206, and 1944 had an accuracy of 90.0% for healthy individuals and 86.7% for CRA-H patients (Fig 1B).
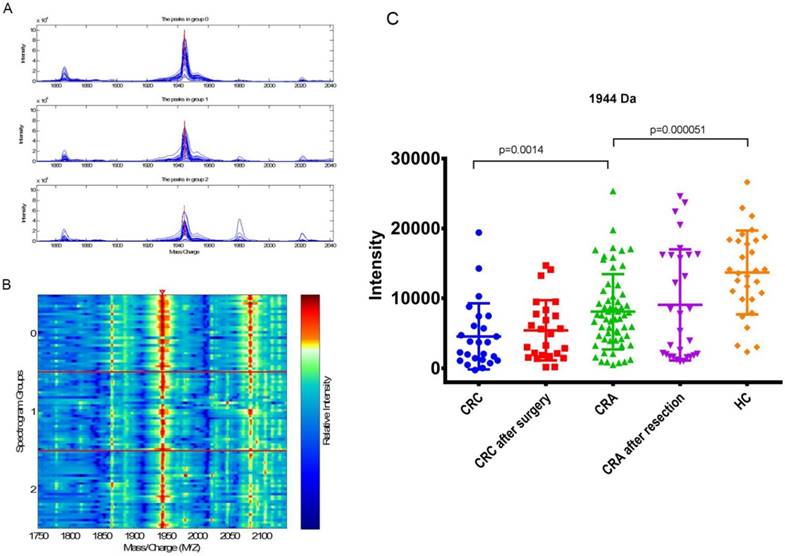
Analyzed the expressions of peak 1944 Da in each group
The mean intensity of peak 1944 in CRC before surgery, CRC after surgery, CRA, CRA after resection and HC was 4556.70±4713.18, 5425.17±4299.569, 8081.98±5381.73, 9058.70±7947.68 and 13698.28±6000.23, respectively.
The expression of peak 1944 in CRA(8081.98±5381.73) was significantly higher (p=0.0014) than that in the CRC patients(4556.70±4713.18), and significantly lower than that in the HCs (13698.28±6000.23; p=5.06E-05; Fig 3 C; Table 5). There were no significant difference for the expression level of peak 1944 between CRC patients before and after surgery (p=0.28), and between CRA patients before and after resection (p=0.92; Table 4). However, the values of peak 1944 were gradually reduced from HC, CRA after resection, CRA, CRC after surgery, and CRC (Fig 3 C). Furthermore, the expression of peak 1944 in CRA-H(6931.62±4150.09) was lower(p=0.049) than that in the CRA-S(9232.36±6242.09).
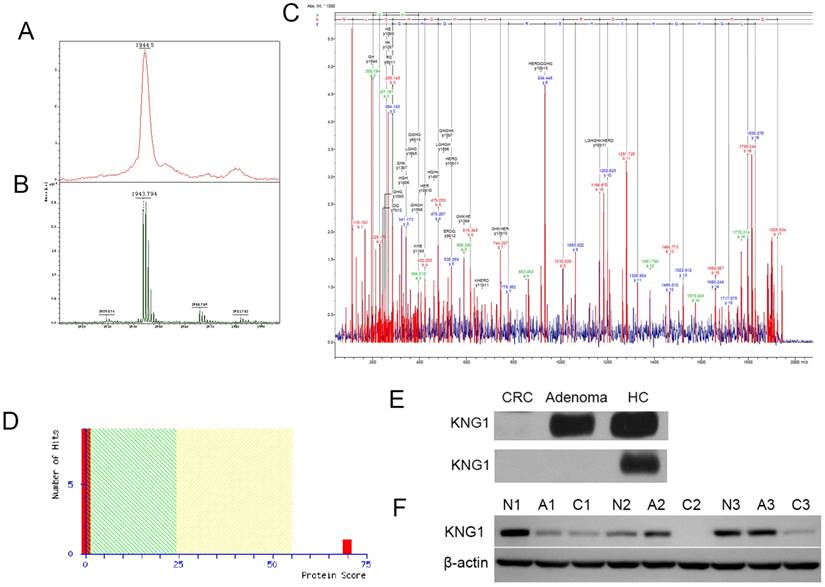
Identification of peak 1944 Da
Peak 1944 showed the low expression of CRA-H compared to HC in the CRA-H distinguishing model identified via MALDI-TOF/TOF (Fig 3 A, B). The peptide sequence was identified as H.NLGHGHKHERDQGHGHQR, and the protein KNG1 had the highest score of 70 according to a Mascot search (Fig 4 A, B, C and D).
Western blot analysis
Western blot experiments were performed to validate the MALDI results. KNG1 was detected in the 6 serum samples (2 colorectal cancer samples, 2 Colorectal adenoma samples, 2 HCs) and 9 tissue samples (3 CRC, 3 colorectal adenoma and 3 adjacent normal paired fresh frozen tissues were surgically collected from 3 CRC patients) using Western blotting. For serum samples, KNG1 was downregulated in Adenoma, and expressed at markedly low levels in CRC in the first group. In another group, KNG1 was expressed at low levels in both Adenoma and CRC but was highly expressed in HC (Fig 4E). For tissue samples, KNG1 was downregulated in CRC than CRA and normal colonic mucosa (Fig 4F). The different expression of KNG1 in sera of CRC, CRA and HC was consistent with which in tissues of CRC, CRA and normal colonic mucosa.
(A) Differential expression of peak 2202 in Advanced CRA-H, Advanced CRA-S, Non-advanced CRA-H and Non-advanced CRA-S groups. (B) Differential expression of peak 5821 in Advanced CRA-H, Advanced CRA-S, Non-advanced CRA-H and Non-advanced CRA-S groups. (C) Differential expression of peak 2218 in Advanced CRA-H, Advanced CRA-S, Non-advanced CRA-H and Non-advanced CRA-S groups. (D) Differential expression of peak 2480 in Advanced CRA-H, Advanced CRA-S, Non-advanced CRA-H and Non-advanced CRA-S groups. (E) Differential expression of peak 3260 in Advanced CRA-H, Advanced CRA-S, Non-advanced CRA-H and Non-advanced CRA-S groups.
Statistics for peaks 2202, 2218, 2480, 3260 and 5821 in HC, CRA and CRC.
| 2202 | 2218 | 2480 | 3260 | 5821 | |
|---|---|---|---|---|---|
| HC | 1399.7±1103.2 | 1824.6±1383.7 | 200±231.3 | 9124.2±7254.6 | 1377.9±959.9 |
| CRA | 483±510.6 | 544.7±752.9 | 503.1±522.4 | 9099±8415.6 | 1059.7±707.5 |
| CRC | 641.5±1093.7 | 374.9±767.9 | 641.5±450.4 | 6027.8±3414.3 | 1078.2±595.8 |
| p value of CRA VS HC | 1.77E-05 | 7.7E-07 | 0.0031 | 0.23 | 0.13 |
| p value of CRA VS CRC | 0.15 | 0.0035 | 0.089 | 0.16 | 0.60 |
Discussion
Colorectal adenoma is recognized as the precursor lesion in most sporadic CRC but not in typical hereditary CRC or HNPCC. Approximately 10% of colorectal adenoma patients have at least one first-degree relative with CRC(17). Genetic predisposition is suspected in these patients. We explored the hereditary factors of colorectal adenomas by comparing the protein profiles in the serum samples obtained from adenoma patients with a CRC family history and individuals without a family history. The results showed that the serum protein profiles of adenoma patients with and without a family history could be distinguished according to their serum protein profiles, indicating that the hereditary factors of colorectal adenoma patients with a family history of this disease might be predicted by serum protein markers.
There are three reasons to explain why we selected colorectal adenoma patients instead of CRC patients with and without a family history for the comparative analysis of their protein profiles. First, the ultimate goal of serum protein marker identification is the early diagnosis of CRC(18). As the precursor lesion of CRC, colorectal adenoma harbors abnormal proteins that might be the driving force for the formation of cancer. Changes in the protein profiles of serum samples from colorectal adenoma patients can reflect such risks. Second, the genetic inheritance of colorectal adenomas other than FAP is still not clear, and the use of typical methods to identify germline mutations in colorectal adenoma is inappropriate(19). Thus, new techniques are needed to uncover the hereditary factors in colorectal adenoma patients with a family history of CRC. Third, the risks of colorectal adenoma in individuals with a positive family history are almost doubled compared with those in individuals without a family history, suggesting that hereditary factors may play an important role in the development of colorectal adenoma(20).
Previous studies have reported that the emergence of colorectal adenoma in a patient may reflect quantitative changes in certain serum proteins. Brown et al observed a progressive increase in serum MIC-1 levels between colorectal adenoma patients and normal individuals(21). Sasaki et al reported significantly higher serum IL-6 levels in colorectal adenoma patients than in controls(22). However, few studies have explored the protein markers that might represent hereditary factors for colorectal adenoma. Numerous studies have focused on germline mutations in the genome of white blood cells for CRC patients. The germline mutations detected in CRC patients are located in the coding region of a gene. The nucleotide changes in this gene cause changes in the amino acid sequence of the protein encoded by the gene. Thus, these proteins are ultimately expressed as abnormal proteins, which might be detectable using high throughout-put techniques. Quaresima et al processed serum samples from FAP patients using a mass spectrometry driven-proteomic strategy (23). These authors successfully identified serum biomarkers specific for two distinct clinical forms of FAP, carpeting FAP and diffuse FAP. These results supported the idea that the serum protein markers for colorectal adenoma are detectable.
Statistics p values for peaks 1944 Da between each group of CRC, CRC after surgery, CRA, CRA after resection and HC.
| p | CRC | CRC after surgery | CRA | CRA after resection | HC |
|---|---|---|---|---|---|
| CRC | 1 | 0.28 | 0.0014 | 0.042 | 1.52E-06 |
| CRC after surgery | 0.28 | 1 | 0.028 | 0.19 | 3.21E-06 |
| CRA | 0.0014 | 0.028 | 1 | 0.92 | 5.06E-05 |
| CRA after resection | 0.042 | 0.19 | 0.92 | 1 | 0.011 |
| HC | 1.52E-06 | 3.21E-06 | 5.06E-05 | 0.011 | 1 |
(A) Mass spectrograph of peak 1944 Da (group 2: CRA-H, group 1: CRA-S, group 0: healthy individuals). (B) Electrophoretogram of peak 1944 Da (the triangle area, Spectrogram Group 2: CRA-H, Spectrogram Group 1: CRA-S, Spectrogram Group 0: HC). (C) Differential expression of peak 1944 Da in CRC, CRC after surgery, CRA, CRA after resection and HC groups.
(A) Detection of peak 1944 in the linear mode of MALDI-TOF-TOF. (B) Detection of peak 1944 in the reflection mode. (C) Detection of a fragment of peak 1944 in the lift mode of MALDI-TOF-TOF and matching the peptide. (D) MASCOT Search sore for KNG1. (E) Western blotting analysis of KNG1 in the serums of CRC, CRA and HC patients. (F) Western blotting analysis of KNG1 in the tissues of CRC(C1, C2, C3), CRA(A1, A2, A3) and adjacent normal(N1, N2, N3). β-actin was used as loading control.
With respect to the comparative analysis of the protein profiles in the present study, colorectal adenoma patients with a family history can be distinguished with an accuracy of 85.0% from those without a family history. It is obvious that Advanced CRA-H is a link in the chain from benign adenoma to malignant altered adenomas and advanced CRC(24). Advanced CRA-H patients are a high-risk group of adenoma patients requiring surveillance. In our study, five peaks (2202, 5821, 3260, 2480, and 2218) were found to have differential expression in Advanced CRA-H. 2480 is expressed in ACRA-H and 2202, 5821, 3260, and 2218 are expressed at low levels in Advanced CRA-H. These five peaks may be specific to Advanced CRA-H.
In the present study, CRA-H and HC can also be distinguished using the SVM model combined with 8 peaks. The peak 1944 showed low regulation in CRA-H than CRA-S. The expression of peak 1944 in CRA was significantly higher than that in the CRC patients, and significantly lower than that in the HCs. The values of peak 1944 were gradually reduced in the occurrence and development of CRC from HC, CRA after resection, CRA, CRC after surgery, to CRC.
Peak 1944 Da was identified using MALDI-TOF-TOF as the protein KNG1. Western blot analysis confirmed that KNG1 was expressed at low levels in CRC and CRA serum.
Kininogen 1 (KNG1) is secreted in human serum. Kininogens are inhibitors of thiol proteases and play important roles in blood coagulation(25). Some groups have reported the detection of KNG1 using mass spectrometry(26-28). However, there are few reports of KNG1 protein expression in tumors sera. Herein, we identified the KNG1 peptide as a potential biomarker for colorectal adenoma patients with a family history, which may also relate to the occurrence and development of CRC.
Abbreviations
MALDI-TOF-MS: matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry; CRA-H: colorectal adenoma patients with a family history of colorectal cancer possibly from atypical hereditary colorectal families; CRA-S: sporadic colorectal adenoma patients without a family history of colorectal cancer; ACRA-H: advance colorectal adenoma patients with a family history of colorectal cancer; NACRA-S: non advance colorectal adenoma patients with a family history of colorectal cancer; HC: healthy controls; CRC: colorectal cancer; HNPCC: hereditary nonpolyposis colorectal cancer; FAP: family adenomatous polyposis; MB: magnetic beads; UDWT: un-decimated discrete wavelet transform: CV: coefficient of variance; KNG1: Kininogens 1; ZJUPDAS: Zhejiang University Proteomics Data Analysis System.
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFC0908200), the Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2014BAI09B07), the National Natural Science Foundation of China (30901731), and the Training Program of the Major Research Plan of the National Natural Science Foundation of China (91229104).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Ivanovich JL, Read TE, Ciske DJ. et al. A practical approach to familial and hereditary colorectal cancer. AM J MED. 1999;107:68-77
3. Jass JR. Familial colorectal cancer: pathology and molecular characteristics. LANCET ONCOL. 2000;1:220-226
4. Kalady MF, Heald B. Diagnostic Approach to Hereditary Colorectal Cancer Syndromes. Clin Colon Rectal Surg. 2015;28:205-214
5. Stoffel EM, Yurgelun MB. Genetic predisposition to colorectal cancer: Implications for treatment and prevention. SEMIN ONCOL. 2016;43:536-542
6. Lynch HT, Lynch JF, Lynch PM. et al. Hereditary colorectal cancer syndromes: molecular genetics, genetic counseling, diagnosis and management. FAM CANCER. 2008;7:27-39
7. Stoffel EM, Boland CR. Genetics and Genetic Testing in Hereditary Colorectal Cancer. GASTROENTEROLOGY. 2015;149:1191-1203
8. Valle L. Recent Discoveries in the Genetics of Familial Colorectal Cancer and Polyposis. Clin Gastroenterol Hepatol. 2017;15:809-819
9. Ballester V, Boardman L. Next Generation Multigene Panel Testing: The Next Step for Identification of Hereditary Colorectal Cancer Syndromes? GASTROENTEROLOGY. 2015;149:526-528
10. Jasperson K, Burt RW. The Genetics of Colorectal Cancer. SURG ONCOL CLIN N AM. 2015;24:683-703
11. Qiu FM, Yu JK, Chen YD. et al. Mining novel biomarkers for prognosis of gastric cancer with serum proteomics. J Exp Clin Cancer Res. 2009:28-126
12. Zheng N, Pan C, Liu W. New Serum Biomarkers for Detection of Endometriosis Using Matrix-assisted Laser Desorption/Ionization Time-of-flight Mass Spectrometry. J INT MED RES. 2011;39:1184-1192
13. Zhai X, Yu J, Lin C. et al. Combining proteomics, serum biomarkers and bioinformatics to discriminate between esophageal squamous cell carcinoma and pre-cancerous lesion. J ZHEJIANG UNIV-SC B. 2012;13:964-971
14. Yu J, Li X, Zhong C. et al. High-throughput proteomics integrated with gene microarray for discovery of colorectal cancer potential biomarkers. ONCOTARGET. 2016;7:75279-75292
15. Wang J, Yu J, Wang L. et al. Application of serum protein fingerprint in diagnosis of papillary thyroid carcinoma. PROTEOMICS. 2006;6:5344-5349
16. Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2007;16:1935-1953
17. Vollmer RT. A Review of Outcomes for Colorectal Adenomas. AM J CLIN PATHOL. 2016;146:567-571
18. Coghlin C, Murray GI. Biomarkers of colorectal cancer: recent advances and future challenges. Proteomics Clin Appl. 2015;9:64-71
19. Buecher B. Colorectal adenomatous polyposis syndromes: Genetic determinism, clinical presentation and recommendations for care. Bull Cancer. 2016;103:199-209
20. Yoshimitsu S, Morita M, Hamachi T. et al. Methionine synthase and thymidylate synthase gene polymorphisms and colorectal adenoma risk: the self defense forces study. Mol Carcinog. 2012;51(Suppl 1):E151-E157
21. Brown DA, Ward RL, Buckhaults P. et al. MIC-1 serum level and genotype: associations with progress and prognosis of colorectal carcinoma. CLIN CANCER RES. 2003;9:2642-2650
22. Sasaki Y, Takeda H, Sato T. et al. Serum Interleukin-6, insulin, and HOMA-IR in male individuals with colorectal adenoma. CLIN CANCER RES. 2012;18:392-399
23. Quaresima B, Crugliano T, Gaspari M. et al. A proteomics approach to identify changes in protein profiles in serum of Familial Adenomatous Polyposis patients. CANCER LETT. 2008;272:40-52
24. Dave S, Hui S, Kroenke K. et al. Is the distal hyperplastic polyp a marker for proximal neoplasia? J GEN INTERN MED. 2003;18:128-137
25. Motta G, Rojkjaer R, Hasan AA. et al. High molecular weight kininogen regulates prekallikrein assembly and activation on endothelial cells: a novel mechanism for contact activation. BLOOD. 1998;91:516-528
26. He X, Wang Y, Zhang W. et al. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. NEOPLASMA. 2014;61:17-26
27. Wen Q, Liu LY, Yang T. et al. Peptidomic Identification of Serum Peptides Diagnosing Preeclampsia. PLOS ONE. 2013;8:e65571
28. Kolialexi A, Anagnostopoulos AK, Papantoniou N. et al. Potential biomarkers for Turner in maternal plasma: possibility for noninvasive prenatal diagnosis. J PROTEOME RES. 2010;9:5164-5170
Author contact
![]() Corresponding author: Dr. Ying Yuan, Department of Medical Oncology, the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou,China,310009, Tel and fax : +086-571-87784501; E-mail: yuanying1999edu.cn
Corresponding author: Dr. Ying Yuan, Department of Medical Oncology, the Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou,China,310009, Tel and fax : +086-571-87784501; E-mail: yuanying1999edu.cn

 Global reach, higher impact
Global reach, higher impact