3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(5):778-783. doi:10.7150/jca.22282 This issue Cite
Short Research Communication
FOXG1 Expression Is Elevated in Glioma and Inhibits Glioma Cell Apoptosis
1. Key Laboratory of Molecular Epigenetics of Ministry of Education, Institute of Cytology and Genetics, Northeast Normal University, Changchun, China
2. Department of Neurosurgery, First Hospital of Jilin University, Changchun, China;
3. Department of Neuroscience and Regenerative Medicine and Department of Neurology, Medical College of Georgia, Augusta University, USA.
# Current address: Henan university Joint national laboratory for antibody drug engineering, Henan University, Kaifeng, Henan, China
Received 2017-8-8; Accepted 2017-12-24; Published 2018-2-11
Abstract
FOXG1, a member of forkhead family transcriptional factor, is involved in telencephalon development. Recent studies showed FOXG1 was important for a variety of cellular events in cancer cells. In respect to glioma, FOXG1 has been shown to regulate cell proliferation and cell cycles. However, its impacts on other cellular events were not well studied. Here, we found FOXG1 had high expression in clinical glioma tissues, and its expression positively correlated with glioma malignancy. Moreover, we found FOXG1 played roles in glioma cell apoptosis. The expressions of caspase family members were significantly altered in response to change of FOXG1 expression, indicating a direct regulation of FOXG1 on caspase family members. These data strongly suggest FOXG1 is negative regulator of glioma cell apoptosis.
Keywords: FOXG1, Glioma, Cell proliferation, Cell apoptosis
Introduction
Gliomas, the most common type of brain tumors, are characterized by prominent proliferation and aggressive infiltration [1]. Although surgical resection followed by combined radiotherapy and chemotherapy were extensively applied in glioma treatment, the median survival time of glioma patients were less than 16 months [2]. Therefore, it is imperative to understand the molecular mechanism of gliomagenesis and to discover new drug target.
FOXG1 belongs to the forkhead family of transcriptional regulators [3, 4]. Previous studies on FOXG1 functions were mostly focused on telencephalon development, which showed FOXG1 was a key regulator in both neural progenitor cell differentiation and regional patterning of mammalian forebrain [3, 5, 6]. Recently, some pilot researches also suggested that FOXG1 regulated tumorigenesis. In ovarian cancer, medulloblastoma (MB), glioblastoma (GBM) and neuroepithelium, FOXG1 was shown to be up-regulated and involved in cell proliferation and cell cycles [7-10]. In breast cancer, FOXG1 was shown to have low expression, which was correlated with the poor prognosis [11]. Therefore, FOXG1 could be either a pro- or anti-oncogenic factor in different cancers. In regarding to glioma, FOXG1 was reported to be involved in GBM tumorigenesis via regulating the expressions of cell cycle-related genes [12]. In addition, microarray screening showed FOXG1 may regulate cell apoptosis pathways [13]. However, its roles in glioma cell apoptosis and the underlying mechanisms were still unclear.
Our results showed that FOXG1 is elevated in many human glioma tissues and its expression positively related to glioma grades. Intriguingly, FOXG1 negatively regulated the expression of caspase family proteins and conducted a repressive effect on cell apoptosis. Collectively, these findings suggested that FOXG1 was associated with glioma grades and cell apoptosis.
Materials and Methods
Cells
The U87MG, SHG44 glioblastoma cells and HEK293T cell lines were cultured according to ATCC (American Type Culture Collection) guidelines. All cells were grown in DMEM (Sigma) supplemented with 10% FBS and 1% penicillin and streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Tumor tissues samples
All clinical samples were obtained from the First Clinical Hospital of Jilin University (Changchun, China). Fresh tumor tissues (n=15, No. 1-15, Table S1) and adjacent normal tissues were immediately frozen at -80°C after surgery. The formalin-fixed paraffin-embedded primary glioma samples (n=43, No. 16-58, Table S1) were classified into different grades according to clinical criteria. The informed consents were obtained from all patients while they were staying in hospital. Permissions were obtained from The Regional Ethical Review Board of Jilin University and Northeast Normal University Changchun, China. All experiments were conducted in accordance with approved guidelines, including any relevant details.
Plasmids construction
FOXG1 CDS region was PCR amplified from Flag-FOXG1 construct using LA taq kit and subcloned into the lentiviral expression vector pWPXLd (Addgene plasmid #12258) using BamH1 and EcoR1 restriction enzymes (NEB). The FOXG1 shRNAs were designed using the BLOCK-iT RNA Designer online tool (Thermo Fisher). The shFOXG1 sequences and scramble shRNA sequence were subcloned into the pLL3.7 lentiviral vector (Shanghai CPG Biotech Co. Ltd., Shanghai) using XhoI and HpaI restriction enzymes. The lentiviral packaging and infection protocols were previously described. [14]. The shFOXG1 sequences are: shFOXG11687, 5'- GGACCAGACTGTAAGTGAA -3'; shFOXG13055, 5'- GCCCTTCAGTTCAGGTACA -3'. All constructs were verified by DNA sequencing.
Cell proliferation assay
For cell proliferation assay, 2 × 103 U87MG cells and 1 × 103 SHG44 cells were seeded into each well of a 96-well plate. All other operations were performed according to the manufacture's protocol (CCK8: Dojindo Molecular Technologies, #CK04; CFSE: C34554, invitrogen) [15].
Immunohistochemistry (IHC)
Detailed protocol was described in our previous study [16]. The UltraSensitiveTM SP (Mouse/Rabbit) IHC kit (KIT-9710, MAB, Fuzhou, China) and a peroxidase-based DBA kit (DAB-0031, MAB, Fuzhou, China) were used in this study. Nuclei were counter-stained with hematoxylin. Olympus FV1000 laser scanning confocal microscope (Tokyo, Japan) was used for imaging.
Western blotting
Western blotting was carried out according to the Bio-Rad general protocol (BIO-RAD Bulletin 6376 Rev A). Detailed protocol was previously described [17]. Primary antibodies were used as follow: rabbit anti-FOXG1 (Abcam, Cambridge, UK, ab18259, 1:1000); rabbit anti-caspase-9 (#9502, 1:1000), mouse anti-caspase-8 (#9746, 1:1000), rabbit anti-caspase-3 (#9665,1:1000), rabbit anti-cleaved caspase-3 (#9661, 1:1000), rabbit anti-AKT (#9272, 1:1000), mouse anti-ERK1/2 (#4696, 1:1000) and rabbit anti-c-Myc (#5605, 1:1000) were all from cell signaling (Danvers, MA, USA); Mouse anti-GAPDH was purchased from TransGen Biotech and mouse anti-α-tublin was get from Ray antibody Biotech (China).
Real-time PCR
Total RNA was extracted from cells using TriPure isolation reagents (Cat. No. 11667165001, Roche). The cDNA was synthesized using transcript One-step gDNA removal and cDNA Synthesis SuperMix (AT-311-03, TransGen Biotech, Beijing, China). Real-time PCR was performed using SYBR Premix Ex TaqTM II (Cat No. RR820A, Takara, Dalian, China). The relative expression of each gene was calculated using a comparative CT method. The primers were used as follow: FOXG1 5'- GAGCGACGACGTGTTCATC-3' (forward) and 5'- GCCGTTGTAACTCAAAGTGCTG -3' (reverse).
Apoptosis assay
Annexin V-FITC staining was carried out following the manufacturer's instructions (KeyGEN Biotech, Nanjing, China). More details were described in previous study [16].
Statistical Analysis
All statistical analyses were performed using SPSS 17.0 software. The statistical significance between two groups was determined by Mann-Whitney U test. For CCK8 analysis, the experimental group was compared to the control group at the same time point using Mann-Whitney U test. Kaplan-Meier curve models was applied to analyze the associations between FOXG1 expression and tumor latency. The patients were divided into two groups by the median value of FOXG1 expression. All FOXG1 IOD values were listed in Table S1. All the graphics were performed using GraphPad Prism 5.0. P < 0.05 was considered to be statistically significant.
Results
FOXG1 expression was elevated in gliomas and positively correlated with glioma grades
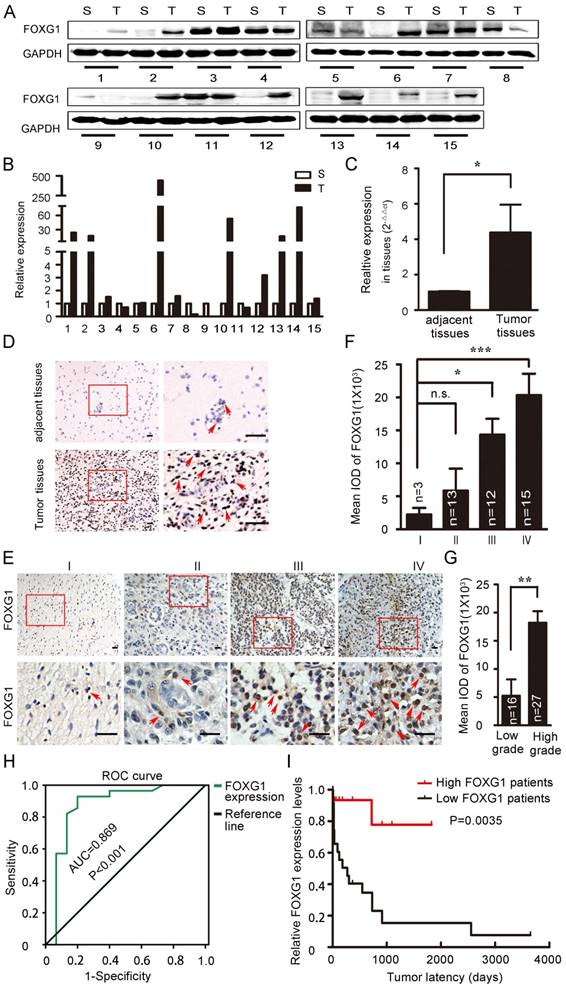
FOXG1 has well-studied roles in neurogenesis and tumor growth [4, 7, 18]. FOXG1 was shown to have high expression in MB or GBM, which was important for tumorigensis [8, 10, 12]. However, the expression of FOXG1 in low grade gliomas and the difference in FOXG1 expression between glioma and its adjacent normal tissues were not well characterized. To explore this, we collected 15 paired primary glioma samples and measured FOXG1 expression by western blotting (figure 1A). FOXG1 expression was found to be elevated in 62.5% (10/16) of glioma tissues compared with each adjacent normal tissue. The overexpression rates ranged from 2 to several hundred folds (figure 1B). To confirm this observation, we further examined FOXG1 expression using real-time PCR analysis. A general of 4-fold up-regulation was detected for FOXG1 in glioma tissues (figure 1C). Immunohistochemistry (IHC) on paired human samples also showed that positive staining of FOXG1 was dramatically increased in glioma samples, and the straining sites were all at the nuclei (figure 1D). These data suggested that the expression of FOXG1 was elevated in clinical glioma samples.
To further study the impact of FOXG1 expression on gliomas, 43 paraffin embedded glioma specimens were collected and applied to IHC (Table S1). Results demonstrated that positive FOXG1 staining was seen in all specimens, and the intensity of FOXG1 staining gradually increased form Grade I to Grade IV (figure 1E, F). There was a significant difference between the expression of FOXG1 in low (I and II) grade and high (III and IV) grade gliomas (figure 1G). Previous reports regarding FOXG1 overexpression were mostly from GBM but not low grade gliomas. Our data showed FOXG1 ubiquitously expressed in all gliomas and its expression level was related to glioma grade. We then constructed the receiver operating characteristics (ROC) curves and calculated the areas under the ROC curve (AUC) [19-21] to assess whether FOXG1 could be a diagnostic marker for glioma. Results showed FOXG1 could be used to distinguish low-grade glioma and high-grade glioma (figure 1H). We also conducted correlation analyses between FOXG1 expression and several known glioma parameters, including ki-67, length of being in hospital, gender. However, negative results were found for all correlation analyses (Table S2), indicating that FOXG1 is independent from such parameters. Nevertheless, the Kaplan-Meier curve [11] analysis showed that patients with higher FOXG1 expression had shorter tumor latency (figure 1I), indicating a faster progress of gliomas. Taken together, these results indicated that FOXG1 correlated with glioma grade in clinic samples and may play a role in gliomagenensis.
FOXG1 negatively regulated glioma cell apoptosis
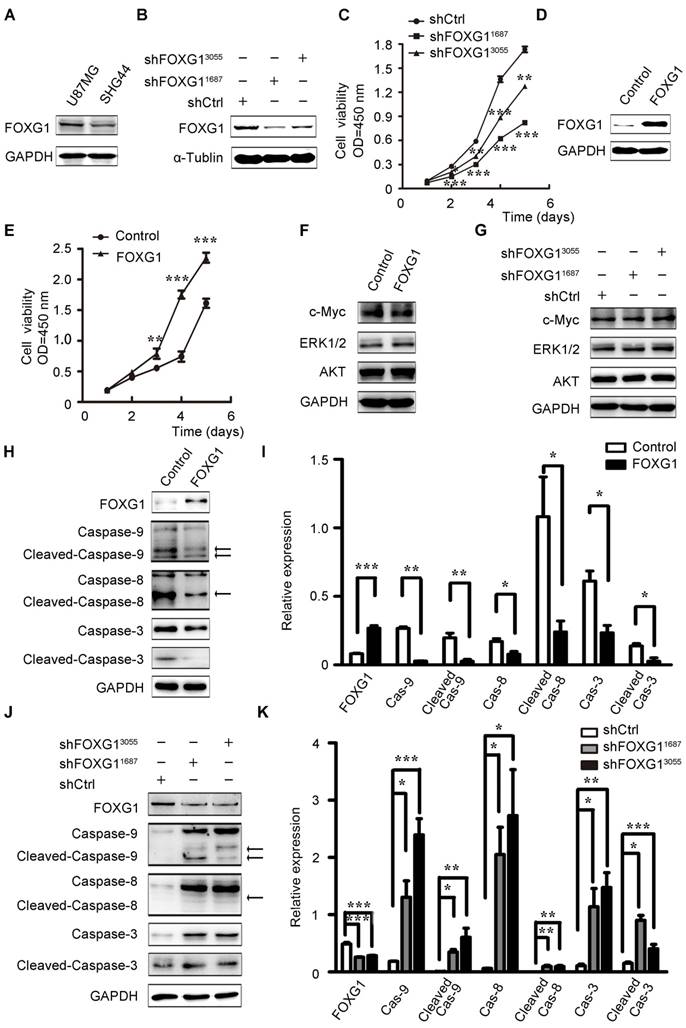
To investigate FOXG1's function in gliomas, we firstly examined FOXG1 expression in two cultured glioma cell lines and found FOXG1 had a higher expression in U87MG cells than SHG44 cells (figure S1A and figure 2A). Therefore, two lentivirus-mediated shRNA and a shCtrl were constructed and infected into U87MG cells. The gene knock down efficiency was assessed by western blotting and qPCR (figure 2B and figure S1B). Then, CCK8 assays were used to measure cell proliferation in FOXG1 knockdown U87MG cells. As expected, both shRNA treated cells had lower proliferation ability than the shCtrl cells (figure 2C). Then, we also constructed a lentivirus mediated expression system to overexpress FOXG1 in SHG44 cells, and the GFP sequence was removed to facilitate the following analyses (figure 2D). Cell proliferation was assessed by both CCK8 (figure 2E) and CFSE assays (figure S1C, D). Both results showed FOXG1 overexpression led to higher proliferation rate. Since FOXG1 is a well-known transcriptional regulator, we thus detected whether FOXG1 regulated the expression of key cell proliferation molecules, such as AKT, ERK1/2 and c-Myc. However, no obvious change was detected of these genes in FOXG1 overexpressing or silencing cells (figure 2F, G).
Then, we wondered whether the increased cell proliferation was a result of decreased apoptosis. Due to the strong GFP signal from the lentivirus-shFOXG1 system, we could not analyze the apoptosis of U87MG cells. We only detected cell apoptosis in FOXG1 overexpressed SHG44 cells. Results showed that overexpressing FOXG1 inhibited cell apoptosis (figure S1E, F). Consistently, FOXG1 overexpression dramatically reduced the expression of caspase-9/8/3 (figure 2H, I) and the cleaved-caspase-9/8/3 (figure 2H, I) in SHG44 cells. These data indicated that cell apoptosis was repressed when overexpressing FOXG1. Correspondently, the expression levels of caspase-9/8/3 (figure 2J, K) and the cleaved-caspase-9/8/3 (figure 2J, K) were elevated in the FOXG1 silencing U87MG cells, suggesting the activation of cell apoptosis. These results together showed a negatively correlation between the expression of FOXG1 and glioma cell apoptosis.
FOXG1 expression elevated in gliomas. (A) Western blotting analyses of paired clinical glioma samples showed that FOXG1 expression was higher in glioma tissues (T) than the adjacent normal tissues (S). GAPDH was used as an internal control. (B) Quantitative analyses of western blotting result using Gel-Pro Analyzer 4 software. (C) Real-time PCR analysis of FOXG1 mRNA levels. (D) Representative images of IHC results. FOXG1 expression was higher in glioma tissues compared with the paired adjacent normal tissues. Boxed areas in the left panel were magnified and shown in the right panel. Scale bar=50 μm. (E) Representative images of IHC showed that FOXG1 expression was gradually increased in glioma samples form Grade I to Grade IV. Boxed areas in the upper panel were magnified and shown in the lower panel. Scale bar = 50 μm. (F) Quantitative analysis of the mean integrated optical density (IOD) of FOXG1 staining using Image-Pro Plus software. (G) IOD of FOXG1 staining in low grade and high grade glioma samples. (H) The receiver operating characteristic (ROC) curves for FOXG1 were constructed to identify whether FOXG1 expression could act as a diagnostic marker. The area under the ROC curve (AUC) is 0.869, indicated that FOXG1 expression could be a diagnostic marker. (I) Kaplan-Meier curves were constructed according to relative FOXG1 expression levels and tumor latency. High FOXG1 patients are marked in red, low FOXG1 patients are marked in black. All data were shown as mean ± SEM. For all statistics analyses in this figure, n.s, p>0.05, *: P < 0.05; **: P < 0.01; ***: P < 0.001.
FOXG1 promoted glioma cell proliferation and inhibited glioma cell apoptosis. (A) Western blotting analysis of FOXG1 expression in glioma cell lines. (B) Western blotting analyses verified the knockdown efficiency of two lentivirus-mediated shRNA systems. The lentivirus-mediated shCtrl was used as control. (C) Cell viability was assessed by CCK8 assay in U87MG cells that were infected with shFOXG1 or shCtrl. (D) Western blotting analysis validated FOXG1 overexpression in SHG44 cells. GAPDH was used as an internal control. (E) CCK8 assay was applied to assess cell viability of infected SHG44 cells. (F and G) Western blotting analyses of the selected oncogenes in FOXG1 overexpressing SHG44 cells and the FOXG1 silencing U87MG cells. GAPDH was used as an internal control. (H and J) Western blotting analysis of the expression of caspase family proteins and the cleaved caspase family proteins in SHG44 cells and U87MG cells. GAPDH was used as an internal control. (I and K) Quantitative analysis of the expression of caspase family proteins and the cleaved caspase family proteins in SHG44 cells and U87MG cells using Gel-Pro Analyzer 4 software. All data were shown as mean ± SEM. For all statistics analyses in this figure *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
Cell proliferation and anti-apoptosis are important tumorigenic processes, which are often targeted in anticancer treatments. Growing evidence has shown that FOXG1 could affect cell proliferation, and thus important for tumorigenesis. In the current study, we found the expression of FOXG1 positively correlated with glioma grades, and FOXG1 could act as a marker to distinguish the low grade and the high grade gliomas. Furthermore, we found FOXG1 promoted glioma cell proliferation and inhibited cell apoptosis. Previous reports on the molecular mechanisms of FOXG1 in gliomas were focused on cell cycles and cell proliferation [9-10, 12]. Though a pilot microarray screening study showed FOXG1 was related to apoptosis in MB [13], the underlying mechanism was still elusive. In FOXG1 silencing and overexpressing glioma cells, the expression of the caspase family proteins, but not the key cell proliferation-related molecules, were significantly changed. Considering the classical functions of FOXG1 [22, 23], cell apoptosis is more likely to be the reason of altered cell proliferation. So far the study of FOXG1 in cell apoptosis was rare. Nevertheless, FOXG1 regulated cell apoptosis was indicated in neurons [24]. In addition, we also found that the expression of cyclin E was altered in FOXG1 silencing or overexpressing cells (data not shown), which is consistent with other reports [12]. Therefore, the effects of FOXG1 on cell proliferation may be the results from multiple cellular events, such as cell apoptosis and cell cycles. Further studies are required to dissect the detailed roles of FOXG1 in promoting tumorigenesis.
In conclusion, our findings identified that FOXG1 may play important roles in glioma cell proliferation through regulating cell apoptosis. Moreover, we found FOXG1 could regulate the expression of caspase family members. These results suggested FOXG1 could be a potential target for glioma treatment.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (31601128, 31671065) and the Science and Technology Development Plan of Jilin Province (20140203002YY, 20140311062YY).
Conflicts of interest
The authors declare no conflicts of interest.
References
1. Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25:867-90
2. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A. et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes & development. 2007;21:2683-710
3. Yao J, Lai E, Stifani S. The winged-helix protein brain factor 1 interacts with groucho and hes proteins to repress transcription. Molecular and cellular biology. 2001;21:1962-72
4. Bourguignon C, Li J, Papalopulu N. XBF-1, a winged helix transcription factor with dual activity, has a role in positioning neurogenesis in Xenopus competent ectoderm. Development. 1998;125:4889-900
5. Huh S, Hatini V, Marcus RC, Li SC, Lai E. Dorsal-ventral patterning defects in the eye of BF-1-deficient mice associated with a restricted loss of shh expression. Developmental biology. 1999;211:53-63
6. Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18:1865-75
7. Adesina AM, Nguyen Y, Guanaratne P, Pulliam J, Lopez-Terrada D, Margolin J. et al. FOXG1 is overexpressed in hepatoblastoma. Human pathology. 2007;38:400-9
8. Adesina AM, Nguyen Y, Mehta V, Takei H, Stangeby P, Crabtree S. et al. FOXG1 dysregulation is a frequent event in medulloblastoma. Journal of neuro-oncology. 2007;85:111-22
9. Chan D, Liu V, To R, Chiu P, Lee W, Yao K. et al. Overexpression of FOXG1 contributes to TGF-β resistance through inhibition of p21WAF1/CIP1 expression in ovarian cancer. British journal of cancer. 2009;101:1433-43
10. Seoane J, Le H-V, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211-23
11. Li JV, Chien CD, Garee JP, Xu J, Wellstein A, Riegel AT. Transcriptional repression of AIB1 by FoxG1 leads to apoptosis in breast cancer cells. Molecular Endocrinology. 2013;27:1113-27
12. Verginelli F, Perin A, Dali R, Fung KH, Lo R, Longatti P. et al. Transcription factors FOXG1 and Groucho/TLE promote glioblastoma growth. Nature communications. 2013:4
13. Adesina AM, Veo BL, Courteau GR, Mehta V, Wu X, Pang K. et al. FOXG1 expression shows correlation with neuronal differentiation in cerebellar development, aggressive phenotype in medulloblastomas, and survival in a xenograft model of medulloblastoma. Human pathology. 2015;46:1859-71
14. Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J. et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nature genetics. 2003;33:401-6
15. Blazquez R, Sanchez-Margallo FM, de la Rosa O, Dalemans W, Álvarez V, Tarazona R. et al. Immunomodulatory potential of human adipose mesenchymal stem cells derived exosomes on in vitro stimulated T cells. Front Immunol. 2014;5:556
16. Wu X, Li Y, Wan X, Kayira TM, Cao R, Ju X. et al. Down-regulation of neogenin accelerated glioma progression through promoter methylation and its overexpression in SHG-44 induced apoptosis. PloS one. 2012;7:e38074
17. Cao R, Chen J, Zhang X, Zhai Y, Qing X, Xing W. et al. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. British journal of cancer. 2014;111:539-50
18. Dou C, Lee J, Liu B, Liu F, Massague J, Xuan S. et al. BF-1 interferes with transforming growth factor β signaling by associating with Smad partners. Molecular and cellular biology. 2000;20:6201-11
19. Nah CW, Ti LK, Liu W, Ng RRG, Shen L, Chew STH. A clinical score to predict acute kidney injury after cardiac surgery in a Southeast-Asian population. Interact Cardiovasc Thorac Surg. 2016;23:757-61
20. Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. Journal of Bone Oncology. 2016;5:74-9
21. Jin C, Shi W, Wang F, Shen X, Qi J, Cong H. et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763-72
22. Ahlgren S, Vogt P, Bronner-Fraser M. Excess FoxG1 causes overgrowth of the neural tube. Developmental Neurobiology. 2003;57:337-49
23. Hanashima C, Shen L, Li SC, Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. Journal of Neuroscience. 2002;22:6526-36
24. Dastidar SG, Landrieu PMZ, D'Mello SR. FoxG1 promotes the survival of postmitotic neurons. J Neurosci. 2011;31:402-13
Author contact
![]() Corresponding author: Xiaoxiao He, E-mail: hexx100edu.cn, Phone: 86-431-85098126, Fax: 86-431-85099768.
Corresponding author: Xiaoxiao He, E-mail: hexx100edu.cn, Phone: 86-431-85098126, Fax: 86-431-85099768.

 Global reach, higher impact
Global reach, higher impact