3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(5):841-850. doi:10.7150/jca.23138 This issue Cite
Research Paper
Ring finger protein 38 promote non-small cell lung cancer progression by endowing cell EMT phenotype
1. Department of Cardiothoracic Surgery, The Second Affiliated Hospital of Nanchang University, Jiangxi Province 330000, P. R. China.
2. Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, Shanghai 200032, P. R. China
3. Department of Cardio-Thoracic Surgery, Kashgar Prefecture Second People's Hospital, Kashgar, Xinjiang 844000, China
† These authors contributed equally to this work.
Received 2017-10-2; Accepted 2017-12-29; Published 2018-2-12
Abstract
Objectives: Ring finger protein 38 (RNF38), as an E3 ubiquitin ligase, plays an essential role in multiple biological processes by controlling cell apoptosis, cell cycle and DNA repair, and resides in chromosome 9 (9p13) which is involvement in cancer pathogenesis including lung cancer. However, its function in tumorigenesis remains unclear. Hence, this study set out to investigate the biological function and clinical implications of RNF38 in non-small cell lung cancer (NSCLC).
Materials and Methods: Immunohistochemistry, quantitative real-time polymerase chain reaction (qRT-PCR) and western blot were used to detect RNF38 protein and mRNA levels in NSCLC and corresponding paratumor tissues. Tissue microarrays (TMA) analysis of 208 NSCLC cases were used to evaluate the relationship between RNF38 expression and clinical implications. Prognostic value was assessed by Kaplan-Meier analysis and log-rank tests. Wound-healing assays, trans-well assays, colony formation assays and CCK8 were used to assess cell migration, invasion and proliferative ability respectively. The analysis of epithelial-to-mesenchymal transition (EMT) phenotype was carried out by immunofluorescence and western blot.
Results: Our data revealed that elevated RNF38 expression were more common in NSCLC tissues than paired normal tissues in both mRNA (2.82 ± 0.29 vs. 1.23 ± 0.13) and protein (2.75 ± 0.09 vs. 1.24 ± 0.02) level. High levels of RNF38 expression were significantly associated with lymph node metastases, higher TNM stages (p=0.011), larger tumor size (p=2.09E-04) and predicted poor prognosis. RNF38 expression was inversely correlated with E-cadherin expression (P= 0.025). Moreover, downregulation of RNF38 impaired the proliferation, metastatic and invasive abilities in NSCLC cells. In addition, aberrant RNF38 expression could modulate the key molecules of EMT.
Conclusions: Our results indicate that elevated expression of RNF38 is significantly associated with the proliferation and metastatic capacity of NSCLC cells, and RNF38 overexpression can serve as a biomarker of NSCLC poor prognosis.
Keywords: RNF38, NSCLC, EMT, Prognosis, Survival
Introduction
Lung cancer is the most commonly diagnosed cancer in males, the third most common cancer in females, and the leading cause of cancer-related death worldwide [1, 2]. Of the estimated 1.8 million new lung cancer cases each year, more than 85% are diagnosed as non-small cell lung cancer (NSCLC) [3, 4]. Despite improvements in surgical technology and the application of targeted drugs, the 5-year survival rate of NSCLC is still lower than 21%, especially for those with advanced stage (metastatic) disease (approximately 2% of cases) [3, 5, 6]. The identification and clarification of new biomarkers that regulate cancer invasion and spread will assist with explorations of reliable prognostic markers and more effective therapeutic targets to improve NSCLC patient outcomes.
Ubiquitination is a process that plays an essential role in protein post-translational modification and participates in almost all cellular processes, including cell proliferation, cell cycle, apoptosis, and DNA repair [7]. Dysregulation of ubiquitin or ubiquitin-like proteins (Ubls) has been reported to be involved in tumor formation and the progression of multiple cancers [7], including lung cancer [8-10], colorectal cancer [11], and hepatocellular carcinoma [12, 13]. Ring finger protein 38 (RNF38) is a functional ubiquitin protein ligase (E3) that resides on chromosome 9 (9p13) [14]. Interestingly, human chromosome 9p12-p13 is deeply involved in many human cancers, including lung cancer [15, 16], ganglioglioma [17], and hepatocellular carcinoma [18]. RNF38 contains a RING-H2 motif (C3H2C2) and a coiled-coil motif at its carboxy terminus, which allows its involvement in both protein-DNA and protein-protein interactions [14]. For example, a previous study showed that RNF38 recognized and ubiquitinated p53 through its E3 ubiquitin ligase activity. RNF38 overexpression can also alter p53 localization [19]. However, few studies have been dedicated to exploring the function of RNF38 to date. Considering the extensive involvement of the function of the tumor suppressor gene p53 in tumors [19], there is an urgent need to reveal the prognostic value and molecular mechanisms of RNF38 in NSCLC.
The present study investigated altered RNF38 expression in NSCLC and its connection with clinical and pathological characteristics. Subsequently, we determined the molecular mechanisms of the involvement of RNF38 in lung cancer progression.
Materials and methods
Oncomine analysis
RNF38 mRNA expression patterns from patients with different cancers were extracted from the Oncomine database tool (www.oncomine.org). The results were filtered to reduce the false discovery rate. Briefly, we chose the cancer vs. normal tissue analysis and clinical specimen in the primary and sample filters, respectively. The threshold we selected was p<0.01.
Patients and specimens
The study population included 208 patients with NSCLC who underwent curative resection at the department of thoracic surgery, Zhongshan Hospital of Fudan University (Shanghai, China) in 2005. The tumors and matched adjacent non-tumor tissues were collected after surgery, frozen at -80°C, and then used for tissue microarray (TMA) construction. The fifteen and eight paired NSCLC specimens used for the qRT-PCR and western blotting analyses, respectively, were collected between 2016 and 2017. The Research Ethics Committee of Zhongshan Hospital provided ethical approval. Informed consent was obtained from all patients to collect and conserve their samples and details. The clinicopathological information was described in our previous studies [20, 21]. The median follow-up period was 43 months (range, 1-66 months) and was completed in July 2010. Overall survival (OS) was defined as the interval between the day of surgery and the date of death or between surgery and the date of the last follow-up visit.
Cell lines
The human NSCLC cell lines 95C, H460, A549 and 95D were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China), and a normal lung epithelial cell line (16HBE) was purchased from Xiangbio Company (Shanghai, China). The 95C and 95D cell lines were cultured in RPMI 1640 medium (HyClone, USA)supplemented with 10% fetal bovine serum (Gibco, USA), penicillin (100 IU/ml), and streptomycin sulfate (100 μg/ml) at 37°C in a thermostatic incubator with 5% CO2. The 16HBE, H460 and A549 cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM, HyClone, USA) with the same supplementation and incubated under the same environment.
Tumor microarray and immunohistochemistry
Detailed information concerning the tissue microarray (TMA) construction was provided in our previous report [22]. Briefly, the TMA included 85 cases of squamous carcinoma, 110 adenocarcinomas and 13 other types of NSCLC. Following dewaxing at 60°C for 2 h and rehydration, the paraffin sections were blocked using 3% H2O2 for 15 min to inactivate endogenous peroxidase activity. Subsequently, the sections were incubated in citric acid epitope retrieval reagent at 100°C for 20 min and allowed to cool to room temperature. To block nonspecific binding sites, the sections were treated with 5% bovine serum albumin (BSA) (YESEN, Shanghai, China) for 1 h. Then, the sections were incubated with primary antibodies overnight at 4°C. The primary antibodies were rabbit anti-human RNF38 (25132-1-AP; 1:100 dilution; Proteintech) and rabbit anti-human E-cadherin (EP700Y, 1:500 dilution, Epitomics). The slides were incubated with a horseradish peroxidase (HRP)-labeled secondary antibody (Gene Tech, Shanghai, China) for 1 h at room temperature. After washing off the residual secondary antibody that had not bound the primary antibody, the sections were stained with diaminobenzidine (DAB, Gene Tech, Shanghai, China) and observed under a microscope.
Immunohistochemistry evaluation
RNF38 expression was evaluated using a combination of both intensity (scores ranging from 0 to 3 representing no, weak, moderate, and strong staining, respectively) and the percentage of positive tumor cells (1, ≤25%; 2, 25%-50%; 3, 50%-75%; and 4, >75%) according to our previously described method [4]. The combined scores ranged from 0 to a maximum of 7. A cutoff of ≤3 was considered the RNF38low group, and higher scores were considered the RNF38high group. The standards were also applied to the evaluation of E-cadherin expression.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from both the tissues and cultured cells using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Two micrograms of total RNA was subjected to reverse transcription using the PrimeScript RT Reagent Kit (RRO47A; Takara, Dalian, China) according to the manufacturer's protocol. Each qPCR reaction was performed in the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA) in duplicate with triplicate samples in a total reaction volume of 10 μl (SYBR Green Real-Time PCR Master Mix; Takara, Japan). The housekeeping gene GADPH was used as an internal standard. The standard curve method was applied for quantification. The GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze relative mRNA expression. The primers were designed as follows: RNF38, forward: 5'- AACACGGAGAGCAGTTCCAC-3' and reverse: 5'- CCTGGCATACGTCTTCAACA-3'; GADPH, forward: 5'-GGTATGACAACGAATTTGGC-3' and reverse 5- GAGCACAGGGTACTTTATTG-3'.
Western blotting
Tissues or cells were collected on ice, lysed using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) and centrifuged for 20 min at 12,000 rpm at 4°C. The supernatants were transferred to a fresh tube, and the protein concentration was measured using a bicinchoninic acid (BCA) kit (Beyotime, Shanghai, China), Protein samples were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking with 5% skim milk for 1 h, the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies were rabbit anti-human RNF38 (25132-1-AP; 1:1000 dilution; Proteintech), E-cadherin (#3195, 1:1000 dilution, Cell Signaling Technology), rabbit anti-human vimentin (ab92547, 1:1000 dilution, Abcam) and mouse anti-β-actin (AA128, 1:1000 dilution, Beyotime, China) or mouse anti-GADPH (AG019, 1:1000 dilution, Beyotime, China); the latter two antibodies were used as internal controls. The next day, the membranes were washed TBST (Tris Buffered saline Tween) three times and incubated with a HRP-conjugated secondary antibody (Beyotime, Shanghai, China) for 1 h at room temperature. Then, the membranes were washed three times and visualized using a chemiluminescent HRP substrate (Millipore; USA). The relative protein expression analysis was conducted with the Image-Pro Plus software version 6.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Proliferation, colony formation and wound healing assays
Cell proliferation was detected using the Cell Counting Kit-8 (CCK8) (Yesen, Shanghai, China) according to the manufacturer's protocol. For the colony formation assay, 500 cells were plated in each well in 12-well plates, and the sizes and numbers of the cell colonies were observed for 10 days. For the wound healing assay, transfected cells were seeded into 6-well plates and incubated at 37°C with 5% CO2. The following day, after the cells in the individual wells reached 90% confluence, the cell monolayers were wounded using a sterile 200-μl pipette tip. The detached cells were rinsed with PBS, and the cells were incubated with DMEM (HyClone, USA) or RPMI 1640 (HyClone, USA) medium containing 10% FBS. Microscope photographs were taken at 0 and 48 h. Three independent experiments were performed.
Transwell assays
The invasive abilities of each cell line were evaluated using 24-well transwell plates (8-μm pore size, Corning, NY, USA). Half a million cells suspended in serum-free medium were plated in the upper chamber with Matrigel (BD Transduction). Then, 0.6 ml of DMEM or RPMI 1640 medium with 30% FBS was added to the lower chamber. After 48 h of incubation, the upper cell layer on the filter was wiped with a cotton swab. The cells on the underside of the transwell were dipped in methanol for 20 min, stained with 0.5% crystal violet, and counted under a microscope.
Lentivirus construction and transfection
The RNF38-control and RNF38-shRNA (short hairpin RNA) expression lentiviruses were designed and provided by Genomeditech (Shanghai, China). The target shRNA sequences were designed as shown in Supplementary Table 1. The lentivirus supernatant was transfected into A549 and 95D cells with 8 μg/ml of polybrene (Genomeditech, Shanghai, China) to establish the stable cell lines A549-shRNA-RNF38 and 95D-shRNA-RNF38. The controls were A549-shRNA-control and 95D-shRNA-control, respectively.
Immunofluorescence
Stable transfected cells were seeded onto cover glasses in 6-well plates. When the cells on each glass reached 20-30% confluence, the cells were fixed with 4.0% paraformaldehyde, permeabilized with 0.2% Triton X-100 (Beyotime, Shanghai, China) for 10 min at room temperature, and washed three times with PBS. Finally, the cells were incubated with rabbit anti-human vimentin (ab92547, 1:100, Abcam) or mouse anti-human E-cadherin (ab1416, 1:100, Abcam) antibodies at 4°C overnight. Then, the cells were washed with PBS and incubated with an immunofluorescent-labelled secondary antibody (Yesen, Shanghai, China) in darkness at room temperature for 2 h, followed by three washes (5-10 min each). The nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI) (Yesen, Shanghai, China) for 10 minutes. The slides were visualized, and photos were taken with a fluorescence microscope (Olympus, Tokyo, Japan).
Statistical analysis
The SPSS 21.0 software (SPSS Inc., Chicago, IL, USA) and Prism software version 5.0 (GraphPad, La Jolla, CA, USA) were applied for the statistical analysis. Statistical differences in clinicopathological parameters between groups were assessed using the χ 2 test or t test. Evaluations of 5-year OS and OS were analyzed using the Kaplan-Meier method (log-rank test to evaluate the differences). Univariate and multivariate analyses and Cox models were used to test whether the quantity and intensity of RNF38 expression possessed a predictive value or an independent prognostic predictive value when added into the base model of traditional factors. The p values were two-sided, and p<0.05 was considered significant.
Results
RNF38 gene expression is increased in several types of cancer
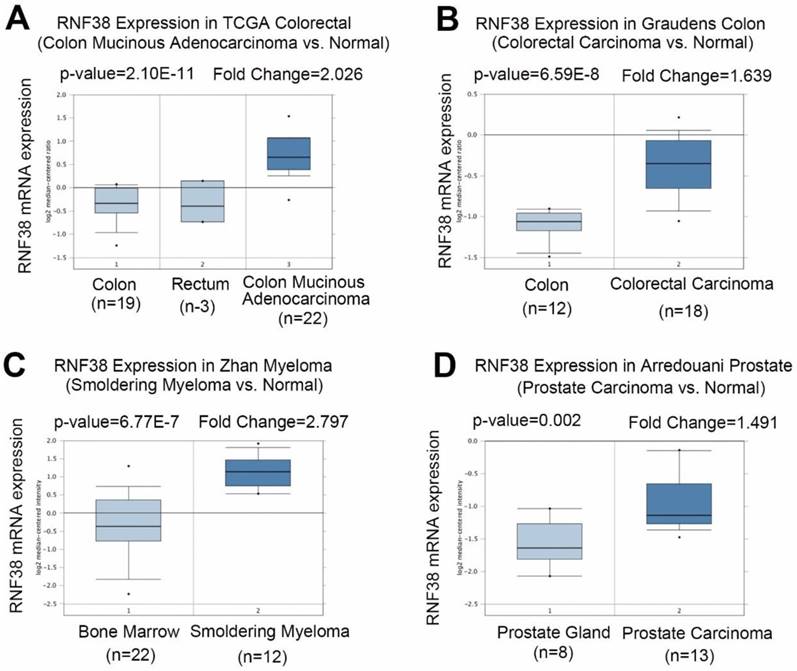
To clarify the role of RNF38 in cancer, first we investigated RNF38 mRNA expression levels using Oncomine, which is a publicly available data source (Fig. 1A). Analysis of colon mucinous adenocarcinoma vs. normal tissue showed that RNF38 was significantly overexpressed in colon mucinous adenocarcinoma (The Cancer Genome Atlas (TCGA) colon data retrieved from Oncomine; p=2.10E-11, fold change=2.026). A similar result was obtained in the study of Graudens, which found significant RNF38 up-regulation in colorectal carcinoma vs. normal tissues (Fig. 1B; p=6.59E-8, fold change=1.639) [23]. The RNF38 transcript levels were also significantly up-regulated in smoldering myeloma vs. normal bone marrow tissues (Fig. 1C; p=6.77E-7, fold change=2.797) [24]. Additionally, a high RNF38 mRNA expression level was observed in prostate carcinoma compared with normal tissues (Fig. 1D; p=0.002, fold change=1.491) [25].
RNF38 is highly expressed in NSCLC
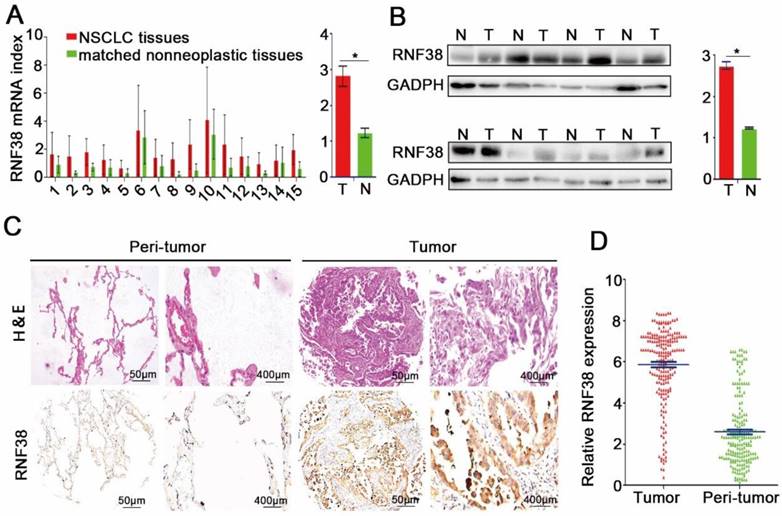
qRT-PCR, western blotting and immunohistochemistry were employed to investigate RNF38 expression in NSCLC. First, we evaluated the mRNA expression levels in 15 NSCLC patients; as shown in Fig. 2A, RNF38 mRNA expression was much higher in the NSCLC tissues than that in the normal lung tissues (2.82 ± 0.29 vs. 1.23 ± 0.13). Moreover, RNF38 protein expression was obviously elevated in 8 paired NSCLC tumors compared with that in the adjacent non-tumor tissues (Fig. 2B; 2.75 ± 0.09 vs. 1.24 ± 0.02). Using immunohistochemistry, we detected RNF38 expression in both the nucleus and cytoplasm. The RNF38 staining intensity was weaker in the nontumorous tissues than that in the NSCLC tissues (Fig. 2C). RNF38 positive staining was detected in 49% (102/208) of the tumor tissues (Fig. 2D). Thus, our results indicated that RNF38 expression was significantly increased in NSCLC.
Correlation between RNF38 expression and the clinicopathological features of NSCLC
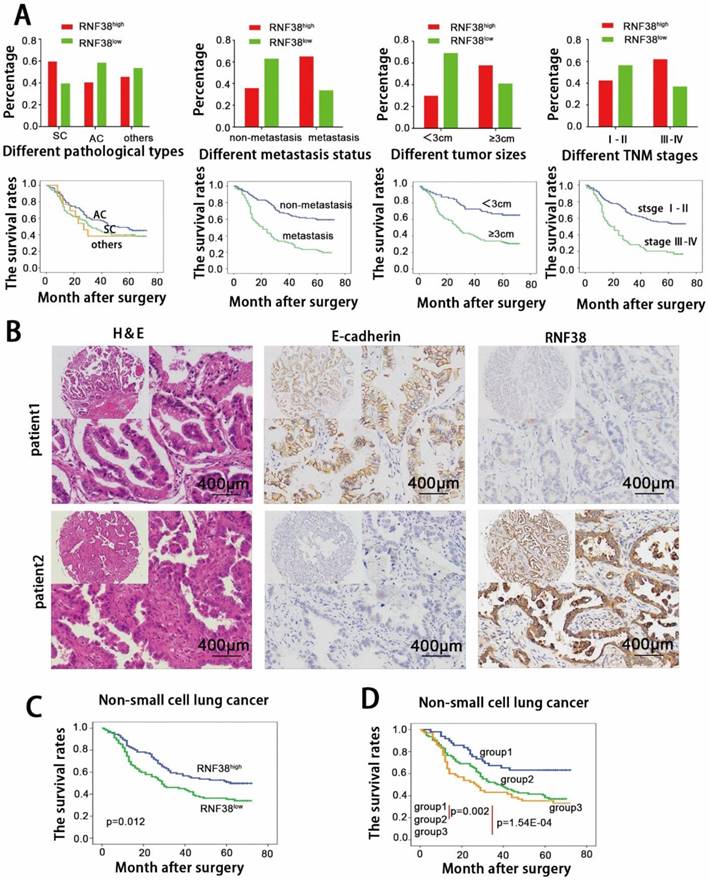
Next, we focused on RNF38 expression and the clinicopathological features of NSCLC using TMAs. Detailed clinical and pathological information was described in our previous study [20, 22]. Based on the RNF38 immunostaining intensity described above, we found no significant association between RNF38 and age, gender, or smoking status. A significant positive association was observed between the RNF38 expression level and the histological type (p=0.030), tumor, node, metastasis (TNM) stage (p=0.011), lymph node metastasis status (p=4.43E-05), and tumor size (p=2.09E-04, Table 1). High RNF38 expression levels were more frequent in squamous cell carcinomas (60%, 51/85) than those in adenocarcinomas (40.9%, 45/110). Elevated RNF38 expression was more frequent in patients with TNM stages III-IV (62.5%, 40/64) and larger tumor sizes (58.3%, 81/139) than that in patients with TNM stages I-II (43%, 62/144) and small tumor sizes (30.4%, 21/69). Additionally, compared to tissues without lymph node invasion (43/118; 36.4%), tissues with lymphatic metastasis (65.6%, 59/90) showed an increased proportion of high RNF38 expression. The OS of the different subtypes is presented in Fig. 3A. Patients with a higher TNM stage, larger size, and lymph node metastasis showed more unfavorable OS.
Increased RNF38 transcript levels in different types of cancer. (A) Analysis RNF38 mRNA level in colon mucinous adenocarcinoma vs. normal (TCGA colon data retrieved from Oncomine). (B) A significant RNF38 upregulation in colorectal carcinoma as compared to normal tissue. (C) RNF38 transcript levels in smoldering myeloma versus normal bone marrow tissues are shown significant upregulation. (D) High level of RNF38 mRNA was observed in prostate carcinoma compared with normal tissue.
RNF38 overexpression predicts an adverse prognosis for NSCLC patients
The 5-year OS rate in the whole population of this study was 42.3%. The univariate analysis revealed that a large tumor size (≥3 cm), high TNM stage and lymphatic metastasis were predictors of poor OS. Other characteristics, including gender and smoking status, had no prognostic impact on OS. We showed that the 5-year OS was significantly poorer in the RNF38high group than that in the RNF38low group (53% vs. 34.3%; p=0.012; Fig. 3C). The multivariate analysis indicated that tumor size (p=0.002), TNM stage (p=0.046) and lymph node metastasis (p=0.002) served as independent predictors of OS (p=0.944; Table 2).
Correlation between RNF38 and clinicopathological characteristics in 208 NSCLCs.
| Variables | No. of patients | RNF38 expression level | ||
|---|---|---|---|---|
| low | high | P | ||
| Age | 0.553 | |||
| <60 | 102 | 52 | 50 | |
| ≥60 | 106 | 54 | 52 | |
| Gender | 0.278 | |||
| Male | 148 | 73 | 75 | |
| Female | 60 | 33 | 27 | |
| Smoking status | 1.000 | |||
| Smokers | 84 | 43 | 41 | |
| Non-smokers | 124 | 63 | 61 | |
| Histological type | 0.030 | |||
| Squamous cell carcinoma | 85 | 34 | 51 | |
| Adenocarcinomas | 110 | 65 | 45 | |
| Othera | 13 | 7 | 6 | |
| Tumor stage | 0.011 | |||
| I-II | 144 | 82 | 62 | |
| III-IV | 64 | 24 | 40 | |
| Lymph node metastasis | 4.43E-05 | |||
| Yes | 90 | 31 | 59 | |
| No | 118 | 75 | 43 | |
| Tumor size | 2.09E-04 | |||
| <3 cm | 69 | 48 | 21 | |
| ≥3 cm | 139 | 58 | 81 | |
| Differentiation | 0.403 | |||
| Well/moderate | 115 | 62 | 53 | |
| Poor | 93 | 44 | 49 | |
NOTE: Bold values are statistically significant (P< 0.05). a Other including adenosquamous carcinoma, large-cell carcinoma, mucoepidermoid carcinoma and carcinosarcoma. * P value was analyzed by squamous cell carcinomas vs. adenocarcinomas.
Univariate and multivariate analysis of factors associated with OS.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | ||
| Gender (male vs. female) | 0.789 | 0.526-1.183 | 0.251 | ||||
| Smoking status (non-smokers vs. smokers) | 0.779 | 0.543-1.118 | 0.175 | ||||
| Tumor size (≥3 cm vs.< 3 cm) | 2.755 | 1.758-4.318 | 9.76E-06 | 2.113 | 1.322-3.377 | 0.002 | |
| Lymph node metastasis (yes vs. no) | 3.042 | 2.103-4.399 | 3.47E-09 | 2.061 | 1.302-3.262 | 0.002 | |
| Tumor stage (III-IV vs. I-II) | 2.771 | 1.922-3.993 | 4.65E-08 | 1.562 | 1.009-2.418 | 0.046 | |
| Differentiation (well/moderate vs. poor) | 1.431 | 1.000-2.049 | 0.050 | 1.193 | 0.826-1.722 | 0.346 | |
| RNF38 level (low vs. high) | 1.579 | 1.101-2.265 | 0.013 | 1.014 | 0.692-1.486 | 0.944 | |
| RNF38/ E-cadherin expression | 1.90E-04 | 0.106 | |||||
| group1 vs. group2 | 1.498 | 1.148-1.954 | 0.003 | 1.282 | 0.974-1.687 | 0.076 | |
| group1 vs. group3 | 1.398 | 1.165-1.677 | 3.16E-04 | 1.102 | 0.900-1.350 | 0.345 | |
Abbreviations and note: OS, overall survival; 95% CI, 95% confidence interval; multivariate analysis, Cox proportional hazards regression model. Variables were adopted for their prognostic significance by univariate analysis with forward stepwise selection (forward, likelihood ratio). Variables were adopted for their prognostic significance by univariate analysis (p < 0.05).
The relationship between RNF38 and E-cadherin expression was appraised based on the scores of the TMA evaluation mentioned above. A significant negative correlation was observed between RNF38 and E-cadherin expression (p=0.025; Supplementary Table 2). The correlation analysis showed that NSCLCs with RNF38high expression tended to express low levels of E-cadherin and vice versa. The representative immunohistochemical staining is shown in Fig. 2B. To evaluate the combined influence of RNF38 and E-cadherin on the prognosis of NSCLC patients, the patients were classified into three subgroups. Group 1 had low RNF38 and preserved E-cadherin expression (n=52), group 2 had RNF38 and E-cadherin levels that were both either high or low (n=88), and group 3 had high RNF38 and impaired E-cadherin expression (n=68). Interestingly, we discovered that the 5-year OS rates in groups 1, 2 and 3 were 65.4%, 36.4% and 33.8%, respectively. Significant differences were found among the 3 groups (group 1 vs. group 2, p=0.002; group 1 vs. group 3, p=1.54E-04, Fig. 4D). The OSs of groups 2 and 3 were significantly lower than the OS of group 1.
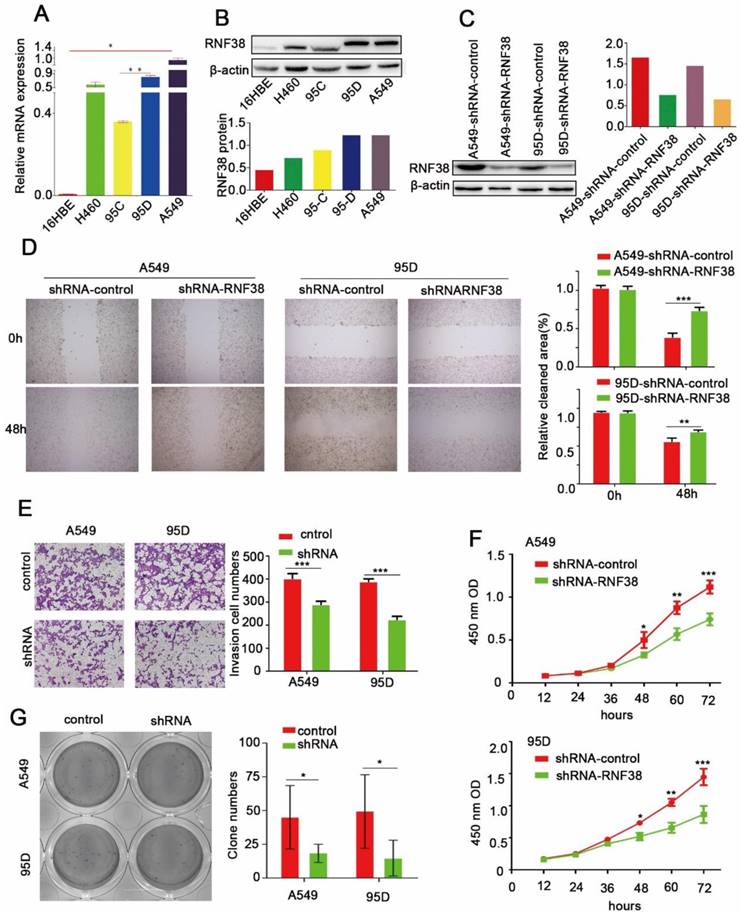
Elevated RNF38 levels are involved in NSCLC cell migration, proliferation and invasion
In this study, RNF38 exhibited a significant relationship with lymph node metastasis (p=4.43E-05) and a large tumor size (p=2.09E-04), which prompted us to investigate the potential biological function of RNF38. Among the four NSCLC cell lines (Fig. 4A and 4B), A549 and 95D possessed the highest RNF38 expression at both the mRNA and protein levels. We silenced RNF38 expression in the A549 and 95D cells using the RNF38-shRNA lentivirus and its control. Western blotting was used to detect the knockdown efficiency of RNF38 (Fig. 4C). Cell motility was dramatically inhibited in the RNF38-silenced NSCLC cells compared with the control group in the scratch test (Fig. 4D). An impaired invasion ability of the 95D-shRNF38 and A549-shRNF38 cells was consistently detected in the transwell Matrigel invasion assay (Fig. 4E). Significantly reduced colony formation was observed in the RNF38-silenced NSCLC cells (Fig. 4G). Cell proliferation was measured with a CCK8 kit and a microplate reader at an OD value of 450 nm. Cells with RNF38 interference showed a reduced proliferative capacity (Fig. 4F). Generally, RNF38 promoted the proliferation, migration and invasion of NSCLC cells.
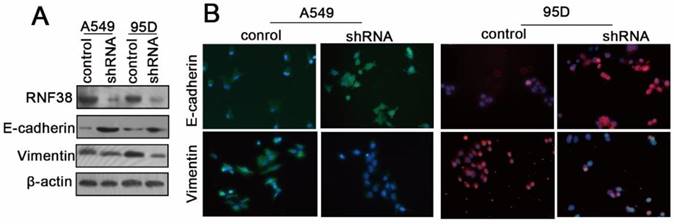
RNF38 interference results in E-cadherin up-regulation and vimentin down-regulation in NSCLC cells
The RNF38 mRNA and protein levels in the normal lung endothelial cell (16HBE) and NSCLC cell lines (H460, 95C, 95D and A549) were also determined using qRT-PCR and western blotting, respectively (Fig. 4A and 4B). To elucidate the mechanisms by which RNF38 was involved in the regulation of cell migration, proliferation and invasion, we evaluated the expression changes of key epithelial-to-mesenchymal transition (EMT) molecules by employing western blotting and an immunocytochemical assay (ICCA). We showed that down-regulation of RNF38 resulted in the up-regulation of E-cadherin and a decreased level of the mesenchymal marker vimentin in the A549 and 95D cells (Fig. 5).
Increased RNF38 expression in NSCLC patients. (A) RNF38 mRNA expression in 15 paired NSCLC tissues (T) and adjacent non-tumor tissues(N). (B)The protein expression of RNF38 in 8 paired NSCLC tissues (T) and adjacent non-tumor tissues (N). *P < 0.05. (C). Representative pictures of RNF38 staining in 208 paired NSCLC tissues matched adjacent normal tissue. (D) Analysis of the integrated optical density (IOD) value of RNF38 immunostaining through IPP6.0 in 208 paired NSCLC tissues matched adjacent normal tissue. *P<0.05, **P<0.01, ***P<0.001.
Discussion
Here, we report that RNF38 protein and mRNA expression was markedly increased in NSCLC tissues compared with adjacent normal lung tissues, which was consistent with Oncomine analysis of colorectal carcinoma [23], smoldering myeloma [24] and prostate carcinoma [25]. A significant positive association between RNF38 expression and clinicopathological features was observed, including the histological type, high TNM stage, lymph node metastasis and a large tumor size. Kaplan-Meier analysis revealed that the RNF38high group patients had significantly shorter OS than the patients in the RNF38low group. Our primary study showed that RNF38 was inversely correlated with E-cadherin expression. The combination of RNF38 and E-cadherin was used to define three subgroups to predict different outcomes for NSCLC patients. Moreover, our results found decreasing proliferation, migration and invasion abilities after RNF38 knockdown. The above results indicate that high RNF38 levels promote NSCLC progression and can serve as a marker of a poor NSCLC prognosis.
Previous studies revealed that the EMT played an important role in promoting cancer development and invasion [26, 27]. The EMT phenomenon is a multistep process in several carcinoma cells during which epithelial cells lose their polarity and markers, such as E-cadherin, and gain mesenchymal markers, such as vimentin [28, 29]. Due to the essential role of the EMT in cancer progression, we evaluated changes in EMT markers after RNF38 knockdown in NSCLC cells. We found up-regulated E-cadherin and decreased vimentin levels after RNF38 silencing. A growing amount of research has provided evidence that Ring finger proteins participate in tumor initiation and progression by inducing tumor suppressor protein degradation, inhibiting apoptosis and improving cell proliferation [7]. For example, overexpression of RNF168 results in DNA damage repair in response to genotoxic treatments and resistance to proteotoxic stress [30]. Importantly, another study revealed that p53 was a substrate of RNF38 and that the nuclear localization of p53 could be altered by RNF38 [19]. To the best of our knowledge, no related studies have directly reported the significance of RNF38 in cancer, and this study is the first to demonstrate a potential oncogenic role of RNF38 in NSCLC. We found that high RNF38 expression in NSCLC tissues was inversely associated with E-cadherin expression. Moreover, interference with RNF38 expression in NSCLC cells impaired their proliferation, migration and invasion abilities, and down-regulation of RNF38 resulted in the retention of E-cadherin and the loss of vimentin. Thus, RNF38 may play a role in the EMT to promote NSCLC progression.
High level of RNF38 is negatively correlated with E-cadherin expression and predict poor prognosis of patients with NSCLC. (A) Analysis of correlation between different RNF38 level and different clinicopathological features of this cohort of NSCLC patients as well as the estimate of their corresponding overall survival. (AC: adenocarcinoma, SC: squamous cell carcinoma, RNF38high: high expression of RNF38, RNF38low: low expression of RNF38). (B) Representative photos of 208 NSCLC patients showing that RNF38 expression was negatively associated with E-cadherin expression. (C) Kaplan-Meier survival curves for 208 NSCLC patients according to RNF38 expression status. (D) Prognostic significance assessed by Kaplan-Meier survival estimates and log-rank tests Kaplan-Meier according to the combination of RNF38 and E-cadherin expression. (Group 1, low RNF38 and preserved E-cadherin; Group 2, RNF38 and E-cadherin both high or both low; Group 3, high RNF38 and impaired E-cadherin.)
RNF38 accelerate NSCLC cell proliferation, migration and invasion. (A, B). RNF38 mRNA and protein levels was confirmed by real‑time qRT‑PCR and western blot respectively in NSCLC cell lines. β‑actin was used as a loading control. (C). RNF38 expression in A549 and 95D cells was reduced distinctly by RNA interference. (D). Wound healing assays were used to evaluate the migration ability of A549 and 95D cells. (E). Transwell assays were used to measure the changes in invasive abilities of shRNF38-transfected A549 and 95D cells. (F) For measurement of cell growth, CCK‑8 assays were used to detect changes in NSCLC cells after down-regulation of RNF38 at different time intervals (from12 to 72 h). (G). Changes in Colony formation activity of NSCLC cells treated with sh-control or sh-RNF38. P values were calculated using Student's t-test All bar graphs depicted quantification of triplicate results with mean ± SD. *P<0.05, **P<0.01, and ***P<0.001
RNF38 promoted NSCLC cell invasion by promoting EMT. (A) The changes of E-cadherin, vimentin, key molecules of EMT, expression levels were determined by Western blot after knockdown of RNF38 in A549 and 95D cells. (B) Representative pictures of Fluorescence staining for E-cadherin and Vimentin in A549 and 95D cells. DAPI-blue stains the nuclei.
Conclusion
In summary, our findings identify RNF38 as a predictor of OS in patients with NSCLC. Combined analysis of RNF38 and E-cadherin may be a more effective risk stratification scheme for the prognosis of NSCLC patients. Moreover, these findings propose that RNF38 may be a potential therapeutic target for NSCLC patients.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported by the National Natural Science Foundation (No. 81560401).
Conflict of interest
None declared.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics. CA Cancer J Clin. 2015;65:87-108
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA Cancer J Clin. 2017;67:7-30
3. Ettinger DS, Wood DE, Akerley W. et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4. J Natl Compr Canc Netw. 2016;14:255-64
4. Liao YF, Wu YB, Long X. et al. High level of BRD4 promotes non-small cell lung cancer progression. Oncotarget. 2016;7:9491-500
5. Miller KD, Siegel RL, Lin CC. et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2016;66:271-89
6. Wang X, Adjei AA. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev. 2015;34:169-71
7. Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776-88
8. Lind H, Zienolddiny S, Ekstrom PO. et al. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718-21
9. Zhao GY, Lin ZW, Lu CL. et al. USP7 overexpression predicts a poor prognosis in lung squamous cell carcinoma and large cell carcinoma. Tumour Biol. 2015;36:1721-9
10. Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319-24
11. Wasch R, Engelbert D. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene. 2005;24:1-10
12. Lee K. Transactivation of peroxisome proliferator-activated receptor alpha by green tea extracts. J Vet Sci. 2004;5:325-30
13. Cai JB, Shi GM, Dong ZR. et al. Ubiquitin-specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor-interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology. 2015;61(5):1603-14
14. Eisenberg I, Hochner H, Levi T. et al. Cloning and characterization of a novel human gene RNF38 encoding a conserved putative protein with a RING finger domain. Biochem Biophys Res Commun. 2002;294:1169-76
15. Kim SK, Ro JY, Kemp BL. et al. Identification of three distinct tumor suppressor loci on the short arm of chromosome 9 in small cell lung cancer. Cancer Res. 1997;57:400-3
16. Girard L, Zochbauer-Muller S, Virmani AK. et al. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894-906
17. Yin XL, Hui AB, Pang JC. et al. Genome-wide survey for chromosomal imbalances in ganglioglioma using comparative genomic hybridization. Cancer Genet Cytogenet. 2002;134:71-6
18. Liew CT, Li HM, Lo KW. et al. Frequent allelic loss on chromosome 9 in hepatocellular carcinoma. Int J Cancer. 1999;81:319-24
19. Sheren JE, Kassenbrock CK. RNF38 encodes a nuclear ubiquitin protein ligase that modifies p53. Biochem Biophys Res Commun. 2013;440:473-8
20. Zhao GY, Ding JY, Lu CL. et al. The overexpression of 14-3-3zeta and Hsp27 promotes non-small cell lung cancer progression. Cancer. 2014;120:652-63
21. Liao Y, Gu J, Wu Y. et al. Low level of 5-Hydroxymethylcytosine predicts poor prognosis in non-small cell lung cancer. Oncol Lett. 2016;11:3753-3760
22. Gu J, Ding JY, Lu CL. et al. Overexpression of CD88 predicts poor prognosis in non-small-cell lung cancer. Lung Cancer. 2013;81:259-65
23. Graudens E, Boulanger V, Mollard C. et al. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19
24. Zhan F, Barlogie B, Arzoumanian V. et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood. 2007;109:1692-700
25. Arredouani MS, Lu B, Bhasin M. et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794-802
26. Thiery JP, Acloque H, Huang RY. et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
27. Ke AW, Shi GM, Zhou J. et al. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology. 2011;140:1629-41 e15
28. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-96
29. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429-37
30. Chroma K, Mistrik M, Moudry P. et al. Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene. 2017;36:2405-2422
Author contact
![]() Corresponding author: Jian-Jun Xu, M.D., Ph.D., Department of Cardiothoracic Surgery, The Second Affiliated Hospital of Nanchang University, NO.1 Mingde Road, Nanchang, Jiangxi, P.R. China, E-mail: jianjxucom or Ji-Jun Li, M.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, E-mail: 13239873691com or Jian-Yong Ding, M.D., Ph.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, NO.180 Fenglin Road, Shanghai, P.R. China, 200032 Tel. & Fax: +86-21-64041990 E-mail: dingjianyongmdcom
Corresponding author: Jian-Jun Xu, M.D., Ph.D., Department of Cardiothoracic Surgery, The Second Affiliated Hospital of Nanchang University, NO.1 Mingde Road, Nanchang, Jiangxi, P.R. China, E-mail: jianjxucom or Ji-Jun Li, M.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, E-mail: 13239873691com or Jian-Yong Ding, M.D., Ph.D., Department of Thoracic Surgery, The Affiliated Zhongshan Hospital of Fudan University, NO.180 Fenglin Road, Shanghai, P.R. China, 200032 Tel. & Fax: +86-21-64041990 E-mail: dingjianyongmdcom

 Global reach, higher impact
Global reach, higher impact