Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(8):1500-1505. doi:10.7150/jca.23017 This issue Cite
Research Paper
Shared liver-like transcriptional characteristics in liver metastases and corresponding primary colorectal tumors
1. Fujian Key Laboratory of Medical Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350122, China
2. Department of Systems Biology, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, 150086, China
3. Fujian Key Laboratory of Tumor Microbiology, Fujian Medical University, Fuzhou, 350122, China
* These authors contributed equally to this work.
Received 2017-9-26; Accepted 2018-1-27; Published 2018-4-6
Abstract
Background & Aims: Primary tumors of colorectal carcinoma (CRC) with liver metastasis might gain some liver-specific characteristics to adapt the liver micro-environment. This study aims to reveal potential liver-like transcriptional characteristics associated with the liver metastasis in primary colorectal carcinoma.
Methods: Among the genes up-regulated in normal liver tissues versus normal colorectal tissues, we identified “liver-specific” genes whose expression levels ranked among the bottom 10% (“unexpressed”) of all measured genes in both normal colorectal tissues and primary colorectal tumors without metastasis. These liver-specific genes were investigated for their expressions in both the primary tumors and the corresponding liver metastases of seven primary CRC patients with liver metastasis using microdissected samples.
Results: Among the 3958 genes detected to be up-regulated in normal liver tissues versus normal colorectal tissues, we identified 12 liver-specific genes and found two of them, ANGPTL3 and CFHR5, were unexpressed in microdissected primary colorectal tumors without metastasis but expressed in both microdissected liver metastases and corresponding primary colorectal tumors (Fisher's exact test, P < 0.05). Genes co-expressed with ANGPTL3 and CFHR5 were significantly enriched in metabolism pathways characterizing liver tissues, including “starch and sucrose metabolism” and “drug metabolism-cytochrome P450”.
Conclusions: For primary CRC with liver metastasis, both the liver metastases and corresponding primary colorectal tumors may express some liver-specific genes which may help the tumor cells adapt the liver micro-environment.
Keywords: colorectal cancer, liver metastasis, microdissection, micro-environment, transcriptional characteristics
Introduction
Colorectal cancer (CRC) is one of the most common malignant cancers1,2 and liver is recognized as the most frequent metastasis site of CRCs. About 20~25% of CRC patients are accompanied with liver metastases at the initial diagnosis, 40~50% of patients eventually developed the liver metastasis after resection operations3,4, and more than two thirds of mortalities are attributable to metastases5,6. It has been found that the tumors from the primary and the corresponding metastatic lesions are highly consistent in transcription7,8, somatic mutation9, copy number and other genetic characteristics10 though some minor differences may exist in both genome8,11 and transcriptome7,12. In addition, it has been reported that some genes such as PTEN13 and AURKA14 with similar expression levels in the primary colorectal tumors and the corresponding liver metastases are associated with prognosis. Thus, the National Cancer Institute of the National Institutes of Health recommended that patients with metastasis should be treated based on the characteristics of the primary tumors. In current clinical practices, the same or similar therapeutic regimens for CRC, including FOLFOX, 5-fluorouracil/leucovorinand (5-FU/LV) and capecitabine combined with oxaliplatin (CapeOX) or capecitabine monotherapy15-17, are commonly used to treat the patients with liver metastasis.
However, the underlying molecular mechanism for the tissue-specific metastasis of CRCs remains elusive. Considering that cancer cells may gain metastatic characteristics in primary lesions18,19, we hypothesized that CRC patients with liver metastases may express some “liver-specific” genes in both the primary colorectal tumors and liver metastases in order to adapt to the microenvironment of liver tissues19-21. To the best of our knowledge, there have been no reports on whether the primary colorectal tumors with liver metastases may gain some liver-specific transcriptional characteristics or not. Therefore, it would be of great importance to understand the mechanism of liver-targeted metastasis of CRCs and adjust clinical treatments to patients with liver metastasis on the basis of the liver-like characteristics, rather than on the molecular features of CRC alone22,23.
In this study, we analyzed the gene expression profiles of normal colorectal tissues, normal liver tissues, tumors of primary CRCs without metastasis, tumors of primary hepatocellular carcinoma (HCC) and paired primary colorectal tumors and liver metastases. Among 3958 genes up-regulated in normal liver tissues versus normal colorectal tissues, we identified 12 liver-specific genes which were virtually not expressed in either normal colorectal tissues or tumors of primary CRCs without metastasis but highly expressed in liver tissues. Especially, two of these liver-specific genes, ANGPTL3 and CFHR5, were also expressed in microdissected liver metastases and corresponding primary colorectal tumors. Functional enrichment analysis revealed that the genes whose expression levels were correlated with the expression levels of ANGPTL3 and CFHR5 were significantly enriched in liver-characteristic metabolism pathways. In short, both the liver metastases and corresponding primary colorectal tumors might express some genes which are specifically expressed in liver tissue to help cancer cells adapt the liver micro-environment.
Materials and Methods
Data and preprocessing
The gene expression profiles used in this study were downloaded from the Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/)24, as described in Table 1. The raw mRNA expression datasets (.CEL files) measured by the Affymetrix platform were preprocessed using the Robust Multi-array Average algorithm25. Probe-set IDs were mapped to their Entrez gene IDs with the corresponding platform files. If multiple probes were mapped to a gene, the arithmetic mean of the expression values of those probes was used to represent the expression value of the gene.
As described in Table 1, 114 normal colon samples detected by the GPL570 platform were collected from four datasets, including GSE8671 (32 samples), GSE9254 (19 samples), GSE21510 (25 samples) and GSE37364 (38 samples), respectively. Notably, the dataset GSE41258 detected by the GPL96 platform included 54 normal colon samples, 13 normal liver samples and 323 samples for primary colon tumors, liver metastases, lung metastases, microadenomas, normal lung tissues, ployps and cell lines. In this study, we only used the expression profiles of the 54 normal colon samples and the 13 normal liver samples for the analysis of differentially expressed genes between the two types of tissues. Here, the samples of the microdissected primary colon tumors and the unmatched liver metastases in GSE41258 were not analyzed because we focused on the analysis of the microdissected primary colon tumors and the matched liver metastases in order to eliminate the contamination of the residual liver tissues in samples of liver metastases. And in order to validate that the “liver-specific” genes are not expressed in primary colon tumors without liver metastases, we only analyzed the 103 samples of stages I-III primary CRCs without metastasis from GSE17536, excluding other 74 samples of stage IV patients or patients who experienced recurrence after curative surgery which indicated potential occult metastases26.
Datasets analyzed in the study
| Dataset | Microarray Platform | Tissue Type* | Sample Size |
|---|---|---|---|
| GSE40367 | GPL570 | pCRC# | 7 |
| mCRC# | 7 | ||
| pHCC# | 10 | ||
| normal liver | 5 | ||
| GSE28702 | GPL570 | mCRC | 23 |
| pCRC | 56 | ||
| GSE41258 | GPL96 | normal liver | 13 |
| normal colon | 54 | ||
| GSE21510 | GPL570 | mCRC# | 66 |
| GSE17536 | pCRC | 103 | |
| GSE8671 | normal colon | 32 | |
| GSE9254 | 19 | ||
| GSE21510 | 25 | ||
| GSE37364 | 38 | ||
| GSE45267 | GPL570 | normal liver | 39 |
| pHCC | 48 |
*pCRC, primary colorectal tumor samples; mCRC, colorectal-liver metastasis; and pHCC,
primary hepatocellular carcinoma samples. #represented that the samples were microdissected.
Statistical analysis
Student's t-test was used to detect differentially expressed genes between two groups of samples. Pearson's correlation analysis was used to assess the correlation between the expression levels of genes. Functional enrichment analysis was performed based on the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/)27. And the P values were adjusted using the Benjamini-Hochberg procedure28.
The ranks of expression levels of 12 liver-specific genes in four types of tissues: Colorectal tissues included 114 normal colorectal tissues (red) and 103 Primary CRCs without metastasis (blue); 48 HCCs (green); 39 normal liver tissues (black).
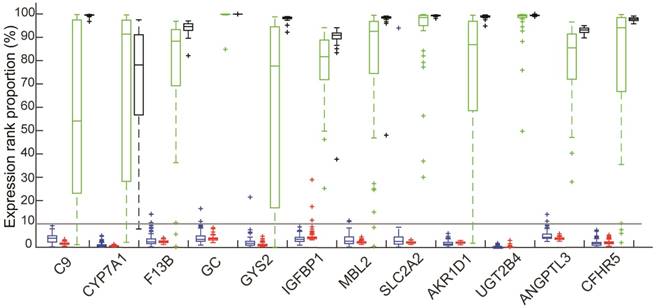
Identification of non-expressed genes in a tissue
For each sample, all genes were ranked according to their expression values. Those genes, which were ranked among the bottom 10% both in more than 95% of tumor tissues and in 95% of normal tissues for the same organ, were defined as the non-expressed genes in this organ, taking into account the background measurement noise and variation in the microarray techniques29,30.
Results
Identification of “liver-specific” genes
Using 13 normal liver samples and 54 normal colorectal samples from GSE41258, we identified 3958 dysregulated genes (DEGs) that were up-regulated in normal liver tissues compared with normal colorectal tissues (Student's t test, FDR < 5%). From these genes, we searched genes that were unexpressed in either normal colorectal tissues or colorectal tumors of primary CRCs without metastasis using a rank-based method. We found 82 genes whose expression values were ranked among the bottom 10% of all the measured 20486 genes in more than 95% of 114 normal colorectal tissue samples collected from 4 datasets (GSE8671, GE9254, GSE21510 and GSE37364) and in more than 95% of 103 tumors of stages I-III primary CRCs without metastasis from GSE17536. Considering the existence of large measurement variations and background noises of high-throughput gene expression profiling techniques29,30, we defined these genes as non-expressed genes in either normal colorectal tissues or tumors of primary CRCs without metastasis.
Among the 82 genes, the mean expression levels of 12 genes, including C9, CYP7A1, F13B, GC, GYS2, IGFBP1, MBL2, SLC2A2, AKR1D1, UGT2B4, ANGPTL3 and CFHR5, were ranked among the top 50% of all the measured genes in all the 39 samples of normal liver tissues and all the 48 tumor tissues of HCCs from GSE45267 and in the 5 samples of microdissected normal liver tissues and 10 microdissected tumor tissues of HCCs from the GSE40367 dataset, as shown in Figure 1. This indicated that these 12 genes were expressed in both the normal liver tissues and HCCs. Therefore, we defined these genes as liver-specific genes (see Figure 1).
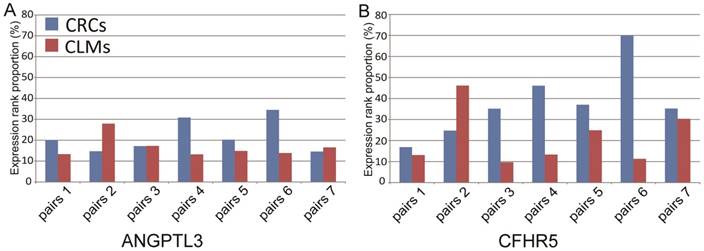
We further analyzed the expression patterns of the 12 liver-specific genes in 7 paired microdissected liver metastases and primary colorectal tumors. The microdissection technique was used to eliminate the possibility that residual liver tissues might contaminate the tumor samples. The result showed that 2 of the 12 genes, ANGPTL3 and CFHR5, were expressed at both the primary and metastatic sites: the expression levels of ANGPTL3 were ranked higher than the bottom 10% within each of paired primary colorectal tumors and metastatic liver tumors; the expression levels of CFHR5 were ranked higher than the bottom 10% in 6 out of the 7 paired samples. And the mean rank percentiles of the expression levels for ANGPTL3 and CFHR5 were 21.71% and 37.87% in primary colorectal tumors, respectively, and 16.70% and 21.23% in liver metastases, respectively (see Figure 2). Furthermore, these 2 genes were unexpressed in all 66 microdissected tumor tissues of stages I-III primary CRCs without metastasis from the GSE21510 dataset, which were unlikely to happen by chance (Fisher's exact test, P <0.05).
The above results suggested that both the liver metastases and corresponding primary colorectal tumors might express some liver tissue-specific genes to adapt the liver micro-environment for CRC patients with liver metastasis.
Functional analysis of ANGPTL3 and CFHR5
The two liver-specific genes, ANGPTL3 and CFHR5, were further analyzed for their biological functions. Using 23 liver metastasis samples of primary CRC patients from GSE28702, we found that the expression levels of 267 genes were significantly correlated with the expression level of ANGPTL3 (FDR< 5%, Pearson's correlation analysis). These 267 genes were significantly enriched in 15 functional pathways including “tyrosine metabolism”, “drug metabolism-cytochrome P450”, “starch and sucrose metabolism” and “drug metabolism-other enzymes” (see Table 2). Similarly, the expressions of 367 genes were found significantly correlated with the expression of CFHR5 and they were significantly enriched in 9 functional pathways characterizing liver tissues, including “tyrosine metabolism” and “drug metabolism-cytochrome P450” besides others (see Table 2). The results indicated that the tumor cells might gain some transcriptional characteristics of liver that might help them adapt the liver microenvironment.
KEGG pathways enriched with the genes significantly co-expressed with ANGPTL3 and CFHR5, respectively.
| KEGG pathway | P-value |
|---|---|
| ANGPTL3-correlated genes | |
| Complement and coagulation cascades | 2.20E-16 |
| Retinol metabolism | 1.11E-16 |
| Drug metabolism-cytochrome P450 | 1.61E-11 |
| Bile secretion | 9.07E-10 |
| Metabolism of xenobiotics by cytochrome P450 | 1.03E-08 |
| Fatty acid degradation | 5.67E-07 |
| Tyrosine metabolism | 3.72E-05 |
| Drug metabolism - other enzymes | 1.06E-04 |
| Glycine, serine and threonine metabolism | 2.46E-04 |
| Steroid hormone biosynthesis | 5.03E-04 |
| Glycolysis / Gluconeogenesis | 9.90E-04 |
| PPAR signaling pathway | 1.19E-03 |
| Starch and sucrose metabolism | 2.46E-03 |
| Fat digestion and absorption | 2.46E-03 |
| Linoleic acid metabolism | 2.54E-03 |
| CFHR5-correlated genes | |
| Complement and coagulation cascades | 2.20E-16 |
| Retinol metabolism | 3.42E-12 |
| Drug metabolism-cytochrome P450 | 6.21E-10 |
| Metabolism of xenobiotics by cytochrome P450 | 2.30E-08 |
| Bile secretion | 3.09E-08 |
| Steroid hormone biosynthesis | 5.42E-06 |
| Fatty acid degradation | 4.56E-05 |
| Drug metabolism - other enzymes | 4.62E-04 |
| Tyrosine metabolism | 1.38E-03 |
The ranks of expression levels of ANGPTL3 and CFHR5 in 7 paired microdissected primary colorectal tumors and liver metastases.
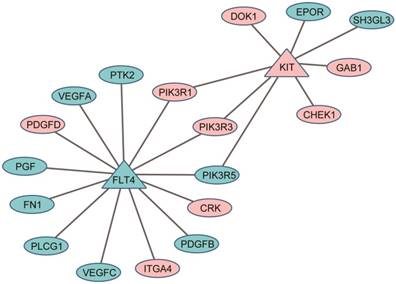
Protein-protein interaction (PPI) links between the DEGs (ellipses) and the target genes of anti-liver cancer drugs (triangles): pink for down-regulated genes and cyan for up-regulated genes.
Discussion
As demonstrated in this study, for primary CRC patients with liver metastasis, both the liver metastases and corresponding primary colorectal tumors shared some transcriptional features of livers. Especially, we identified two liver-specific genes, ANGPTL3 and CFHR5, which were expressed in livers and in the matched liver metastases and corresponding primary colorectal tumors, but not expressed in normal colorectal tissues or in colorectal tumors of the primary CRCs without metastasis. Functional enrichment analysis on those genes significantly correlated with the two genes in the expression patterns found a number of liver-characteristic metabolic pathways31-33. In addition, it has been reported that protein ANGPTL3 is associated with tumor development and invasion by inducing endothelial cell adhesion and migration through stimulating the integrin signaling pathway and promoting angiogenesis34-36. Protein ANGPTL3 could also stimulate mitogen-activated protein kinase (ERK/MAPK) signaling pathway related to cell-cycle G1/S transition37,38 and proliferation39. Protein CFHR5 is a plasma glycoprotein generated in liver and can increase the risk of metastasis40. Taken together, our analysis suggested that primary colorectal tumors may express some liver tissue-specific genes to help CRC cells adapt the liver micro-environment in the metastases to liver.
In clinical practice, the same or similar therapeutic regimens for the treatment of the primary CRC cancer were used to treat those patients with liver metastases15-17. However, as shown here, the metastasized tumors possessed some features of liver including enhanced metabolic efficiency on drugs. The results suggested that when developing therapeutic regimens for the patients with liver metastasis, we should consider the liver-like characteristics of the metastatic cancers, possibly through combining the anti-tumor agents for primary colorectal tumor with the anti-tumor agents of liver cancer (such as sorafenib, an oral multikinase inhibitor for treatment of HCC)15. For example, among 3164 DEGs between 23 liver metastases and 56 non-paired primary colorectal tumors from the GSE28702 dataset (Student's t-test, FDR < 5%), two targeted genes of sorafenib41, KIT42 and FLT443, were significantly up-regulated (KIT) and down-regulated (FLT4) in the liver metastases compared with primary colorectal tumors. Eighteen DEGs have direct protein-protein interaction (PPI) links with KIT and FLT4, indicating that sorafenib might be an effective molecular targeting drug for the treatment of the CRC patients with liver metastasis (see Figure 3).
One limitation of this study is that the size of microdissected samples is not very large. In order to further confirm and elucidate the biological characteristics of the metastatic tumors, more and purer tumor samples, even single cells, should be collected from the primary and metastatic lesions19,44. To gain new insights on the molecular characteristics of the metastases will be of great importance to develop individualized treatment to improve the survival of the CRC patients with liver metastases.
Acknowledgements
This work was supported by the Natural Science Foundation of China (Grant No. 81372213, 81572935, 81602738, 61602119, 61601151 and 21534008), the Joint Scientific and Technology Innovation Fund of Fujian Province (Grant No. 2016Y9044) and the Health and Family Planning Commission of Fujian Province (Grant No. 2017-ZQN-56).
Author Contributions
JC, XS and ZG designed the study and developed the method, YG and XS performed the data analysis, JC and XW drafted the manuscript, ZG, XW, WZ and HL revised the manuscript, JZ and RC interpreted the function annotations and MC and LA drew the figures. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Miron N, Susman S, Balacescu O. et al. Novel cellular and molecular approaches to stratification and treatment of colorectal cancer. J Gastrointestin Liver Dis. Dec. 2012;21(4):413-421
2. Penna C, Nordlinger B. Colorectal metastasis (liver and lung). Surg Clin North Am. Oct. 2002;82(5):1075-1090 x-xi
3. Jiang H, Liu J, Chen Y, Ma C, Li B, Hao T. Up-regulation of mir-10b predicate advanced clinicopathological features and liver metastasis in colorectal cancer. Cancer Med. Oct. 2016;5(10):2932-2941
4. Garden OJ, Rees M, Poston GJ. et al. Guidelines for resection of colorectal cancer liver metastases. Gut. Aug. 2006;55(Suppl 3):iii1-8
5. Kopetz S, Chang GJ, Overman MJ. et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. Aug 01. 2009;27(22):3677-3683
6. Abdalla EK, Adam R, Bilchik AJ, Jaeck D, Vauthey JN, Mahvi D. Improving resectability of hepatic colorectal metastases: expert consensus statement. Ann Surg Oncol. Oct. 2006;13(10):1271-1280
7. Sayagues JM, Corchete LA, Gutierrez ML. et al. Genomic characterization of liver metastases from colorectal cancer patients. Oncotarget. Nov 8. 2016;7(45):72908-72922
8. Roessler S, Lin G, Forgues M. et al. Integrative genomic and transcriptomic characterization of matched primary and metastatic liver and colorectal carcinoma. Int J Biol Sci. 2015;11(1):88-98
9. Tan IB, Malik S, Ramnarayanan K. et al. High-depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver-limited metastatic colorectal cancer. Genome Biol. Feb 12. 2015;16:32
10. Brannon AR, Vakiani E, Sylvester BE. et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. Aug 28. 2014;15(8):454
11. Kovaleva V, Geissler AL, Lutz L. et al. Spatio-temporal mutation profiles of case-matched colorectal carcinomas and their metastases reveal unique de novo mutations in metachronous lung metastases by targeted next generation sequencing. Mol Cancer. Oct 18. 2016;15(1):63
12. Kleivi K, Lind GE, Diep CB. et al. Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer. Jan 3. 2007;6:2
13. Atreya CE, Sangale Z, Xu N. et al. PTEN expression is consistent in colorectal cancer primaries and metastases and associates with patient survival. Cancer Med. Aug. 2013;2(4):496-506
14. Goos JA, Coupe VM, Diosdado B. et al. Aurora kinase A (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer. Oct 29. 2013;109(9):2445-2452
15. De Groote K, Prenen H. Intrahepatic therapy for liver-dominant metastatic colorectal cancer. World J Gastrointest Oncol. Sep 15. 2015;7(9):148-152
16. Rothbarth J, van de Velde CJ. Treatment of liver metastases of colorectal cancer. Ann Oncol. 2005;16(Suppl 2):ii144-149
17. Virdee PS, Moschandreas J, Gebski V. et al. Protocol for Combined Analysis of FOXFIRE, SIRFLOX, and FOXFIRE-Global Randomized Phase III Trials of Chemotherapy +/- Selective Internal Radiation Therapy as First-Line Treatment for Patients With Metastatic Colorectal Cancer. JMIR Res Protoc. Mar 28. 2017;6(3):e43
18. Hynes RO. Metastatic potential: generic predisposition of the primary tumor or rare, metastatic variants-or both? Cell. Jun 27. 2003;113(7):821-823
19. Sethi N, Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat Rev Cancer. Sep 23. 2011;11(10):735-748
20. Auguste P, Fallavollita L, Wang N, Burnier J, Bikfalvi A, Brodt P. The host inflammatory response promotes liver metastasis by increasing tumor cell arrest and extravasation. Am J Pathol. May. 2007;170(5):1781-1792
21. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. Oct 14. 2011;147(2):275-292
22. Vermaat JS, Nijman IJ, Koudijs MJ. et al. Primary colorectal cancers and their subsequent hepatic metastases are genetically different: implications for selection of patients for targeted treatment. Clin Cancer Res. Feb 01. 2012;18(3):688-699
23. Amaro A, Chiara S, Pfeffer U. Molecular evolution of colorectal cancer: from multistep carcinogenesis to the big bang. Cancer Metastasis Rev. Mar. 2016;35(1):63-74
24. Barrett T, Wilhite SE, Ledoux P. et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. Jan. 2013;41(Database issue):D991-995
25. Irizarry RA, Hobbs B, Collin F. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. Apr. 2003;4(2):249-264
26. Qi L, Li T, Shi G. et al. An individualized gene expression signature for prediction of lung adenocarcinoma metastases. Mol Oncol. Nov. 2017;11(11):1630-1645
27. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. Jan. 2012;40(Database issue):D109-114
28. Belanger BF, Williams WJ, Yin TC. A flexible renewal process simulator for neural spike trains. IEEE Trans Biomed Eng. May. 1976;23(3):262-266
29. Wang D, Wang C, Zhang L. et al. Evaluation of cDNA microarray data by multiple clones mapping to the same transcript. OMICS. Dec. 2009;13(6):493-499
30. Shi L, Reid LH, Jones WD. et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. Sep. 2006;24(9):1151-1161
31. Almazroo OA, Miah MK, Venkataramanan R. Drug Metabolism in the Liver. Clin Liver Dis. Feb. 2017;21(1):1-20
32. Shi YX, Huang CJ, Yang ZG. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J Gastroenterol. Sep 28. 2016;22(36):8161-8167
33. Nguyen P, Leray V, Diez M. et al. Liver lipid metabolism. J Anim Physiol Anim Nutr (Berl). Jun. 2008;92(3):272-283
34. Camenisch G, Pisabarro MT, Sherman D. et al. ANGPTL3 stimulates endothelial cell adhesion and migration via integrin alpha vbeta 3 and induces blood vessel formation in vivo. J Biol Chem. May 10. 2002;277(19):17281-17290
35. Koyama T, Ogawara K, Kasamatsu A. et al. ANGPTL3 is a novel biomarker as it activates ERK/MAPK pathway in oral cancer. Cancer Med. May. 2015;4(5):759-769
36. Zhu L, Jiang L, Wang W. et al. Angiopoietin-like protein 3 is an indicator of prognosis in esophageal cancer patients. Int J Clin Exp Med. 2015;8(9):16101-16106
37. Broxmeyer HE, Srour EF, Cooper S, Wallace CT, Hangoc G, Youn BS. Angiopoietin-like-2 and -3 act through their coiled-coil domains to enhance survival and replating capacity of human cord blood hematopoietic progenitors. Blood Cells Mol Dis. Jan 15. 2012;48(1):25-29
38. Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. May 14. 2007;26(22):3227-3239
39. McCubrey JA, Steelman LS, Franklin RA. et al. Targeting the RAF/MEK/ERK, PI3K/AKT and p53 pathways in hematopoietic drug resistance. Adv Enzyme Regul. 2007;47:64-103
40. Conti A, Fredolini C, Tamburro D. et al. Identification of novel candidate circulating biomarkers for malignant soft tissue sarcomas: Correlation with metastatic progression. Proteomics. Feb. 2016;16(4):689-697
41. Wishart DS, Knox C, Guo AC. et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. Jan 01. 2006;34(Database issue):D668-672
42. Rojas A, Zhang P, Wang Y. et al. A Positive TGF-beta/c-KIT Feedback Loop Drives Tumor Progression in Advanced Primary Liver Cancer. Neoplasia. Jun. 2016;18(6):371-386
43. Xiao X, Liu Z, Wang R. et al. Genistein suppresses FLT4 and inhibits human colorectal cancer metastasis. Oncotarget. Feb 20. 2015;6(5):3225-3239
44. Kim KT, Lee HW, Lee HO. et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. Apr 29. 2016;17:80
Author contact
![]() Corresponding author: Xianlong Wang, Department of Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350108, China; Phone: +86-133 9816 5173; E-mail: wang.xianlongcom and Zheng Guo, Department of Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350108, China; Phone: +86-451-8661-5933; Fax: +86-451-8666-9617; E-mail: guozhrbmu.edu.cn
Corresponding author: Xianlong Wang, Department of Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350108, China; Phone: +86-133 9816 5173; E-mail: wang.xianlongcom and Zheng Guo, Department of Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, School of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350108, China; Phone: +86-451-8661-5933; Fax: +86-451-8666-9617; E-mail: guozhrbmu.edu.cn

 Global reach, higher impact
Global reach, higher impact