Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(8):1506-1517. doi:10.7150/jca.22797 This issue Cite
Research Paper
The Interaction of Smoking with Gene Polymorphisms on Four Digestive Cancers: A Systematic Review and Meta-Analysis
1. Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Science, Northwest University, Xi'an 710069, China
2. Institute of Preventive Genomic Medicine, Xi'an 710069, China
Received 2017-9-12; Accepted 2018-1-22; Published 2018-4-6
Abstract
The main purpose of this study was to perform a meta-analysis to assess the interaction between smoking and nine genes (GSTM1, GSTT1, GSTP1, CYP1A1, NAT2, SULT1A1, hOGG1, XRCC1 and p53) on colorectal cancer, gastric cancer, liver cancer and oesophageal cancer. Published articles from the PubMed, ISI and EMBASE databases were retrieved. A total of 67 case-control studies or nested case-control studies were identified for the analysis. The pooled jodds ratio (OR) with 95% confidence interval (CI) was calculated using the random effect model. The overall study showed that the GSTM1 polymorphism was associated with the risk of the four digestive cancers among Asian population (OR 1.284, 95% CI: 1.122-1.470, p: 0). Subgroup analyses by cancer site showed that GSTM1 null genotype increased the gastric cancer risk in total population (OR 1.335, 95% CI: 1.145-1.556, p: 0). However, the association of GSTM1 null genotype with the oesophageal cancer risk was found in smokers (OR 1.382, 95% CI: 1.009-1.894, p:0.044), but not in non-smokers (OR 1.250, 95% CI: 0.826-1.891, p:0.290). Moreover, smokers with the CYP1A1 IIe462Val polymorphism were at an increased cancer risk in Asian population (OR=1.585, 95% CI 1.029-2.442, p: 0.037). None of the other gene-smoking interactions was observed in the above cancers. This meta-analysis reveals two potential gene-smoking interactions, one is between smoking and GSTM1 on oesophageal cancer, and the other is between smoking and CYP1A1 IIe462Val on the four cancers in Asian population. Future studies need to be conducted to verify the conclusions.
Keywords: gene polymorphisms, gene-smoking interaction, digestive cancer, meta-analysis
Introduction
Cancer was the second leading cause of non-communicable diseases deaths worldwide in 2015. Most cancer patients die from digestive cancers between 2005 and 2015, of which the death toll increased to 832,000 for colorectal cancer (CRC), 818.9,000 for gastric cancer (GC), 810.5,000 for liver cancer (LC) and 439,000 for oesophageal cancer (OC)[1]. Moreover, the incidence of these four cancers ranks in the top ten over the world, mainly in developing countries [2]. Especially, these cancers are generally recognized as tobacco-related cancers (TRCs) by the International Association of Research in Cancer (IARC) [3]. However, not all individuals exposed to tobacco develop these cancers. Because the etiology of cancer is multifactorial and complicated [4], cigarette smoking, as a prevalent environment factor, may interact with multiple genetic factors, leading to a higher susceptibility to cancer.
The research on the gene-smoking interaction in cancer risk has been popular [5]. Previously published studies clarified the molecular mechanism of the gene-smoking interaction. Most tobacco carcinogens first form DNA adducts via metabolic activation; persistent DNA adducts induce mutations in some critical genes and initiate carcinogenesis [6]. The elimination of DNA adducts requires DNA repair, implying that variations of the DNA repair genes may be related to different repair efficiencies of DNA damage [7]. Moreover, various detoxification pathways are competitive and different individuals have distinct balances between metabolic activation and detoxification, influencing the cancer risk [8]. Increasing epidemiologic studies and meta-analyses have indicated the interaction between smoking and gene polymorphisms in various cancer types [9-11]. However, most meta-analyses only assessed the interaction between single gene polymorphism and smoking on one or several cancers. Furthermore, the results were inconsistent or even conflicting. Hence, we performed a comprehensive meta-analysis on the interaction of smoking with ten gene polymorphisms in four digestive cancers. The aim was to develop a more powerful evaluation of gene-smoking interaction on major digestive cancers risk.
Materials and methods
Search strategy
PubMed, ISI and EMBASE databases were searched until Dec. 2017 with combinations of the following keywords: “smoke, cigarette, tobacco, smoking”, “gene, polymorphism”, “colorectal, colon, rectum, colorectum, liver, hepatocellular, oesophageal, oesophagus, gastric, stomach”, and “cancer, carcinoma, adenomas”. No restrictions were placed on language. References of the retrieved and review articles were also screened by hand.
Inclusion and exclusion criteria
Studies that were included in our analysis had to meet all of the following criteria: (1) evaluated the gene-smoking interaction on the risk of digestive cancers; (2) only case-control studies or cohort studies were considered; (3) provided case and control or cohort size by gene-smoking interaction; (4) showed the gene polymorphisms that were evaluated in at least five independent studies on the four digestive cancers and (5) when an author had several studies on the same patient population, only the most recent or largest sample article was included..The following exclusion criteria were used: (1) the full text was not obtained; (2) only case population; and (3) duplicated study.
Data extraction and quality assessment
All data were independently extracted by two investigators according to the above selection criteria. The information collected from each study are as follows: the first author's last name; year of publication; country of origin; ethnicity; study design; total number of cases and controls or cohort; cancer type; gene names; number of cases and controls or cohort by gene polymorphisms; number of cases and controls or cohort by gene-smoking interaction. Smoking habits were categorized as non-smoker and smoker. The number of cases and controls or cohort by gene-smoking interaction was extracted according to four combinations: non-smoker + “no risk” polymorphism; non-smoker + “at risk” polymorphism; smoker + “no risk” polymorphism; and smoker + “at risk” polymorphism. For each gene polymorphism, the “at risk” phenotype was identified based on known biological mechanisms and the classification conducted by most included articles. “At risk” polymorphism for GSTM1/GSTT1 was the null (-/-); for GSTP1, the IIe105Val substitution (Ile/Val+Val/Val); for CYP1A1, the 3801T>C substitution (MspI) (T/C+C/C) and Ile462Val substitution (Ile/Val+Val/Val), for NAT2, the fast + intermediate (at least one *4 or *12) acetylator; for SULT1A1, the slow+intermediate (at least one *2) sulphation, for hOGG1, the Ser326Cys substitution (Ser/Cys+Cys/Cys); for XRCC1, the Arg399Gln substitution (Arg/Gln+Gln/Gln); and for p53, the Arg72Pro substitution (Arg/Pro +Pro/Pro).
The quality of each study was evaluated by the Newcastle-Ottawa Scale (NOS), which is a 9-star system containing the following three dimensions: selection; comparability; and outcome (cohort studies) or exposure (case-control studies) [12]. A study with 7-9 scores was classified as a high-quality study, while those with scores of 4-6 and 0-3 are moderate- and low-quality studies, respectively [13].
Statistical methods
The reference group was identified as “no risk” polymorphism, and the odds ratios (OR) with 95% confidence intervals (CI) were calculated to determine a risk of the association between gene polymorphisms and digestive cancers. To be conservative, the random effects model was applied to calculate the summary risk. In addition, the subgroup analyses were conducted based on the cancer site and ethnicity. Heterogeneity was evaluated among studies by calculating the Q-statistic and I2 value [14]. Publication bias was assessed by constructing the funnel plots (there was no publication bias if the funnel plot was symmetric) and quantified using Begg's test and Egger's test [15, 16], in which a p-value<0.05 indicated the presence of potential publication bias. All statistical analyses were performed using Comprehensive Meta-Analysis Software, version v. 2.0 (CMA, Biostat, Englewood, NJ, USA). For the positive findings, the false-positive report probability and statistical power were calculated by G*Power software [17, 18].
Results
Literature search
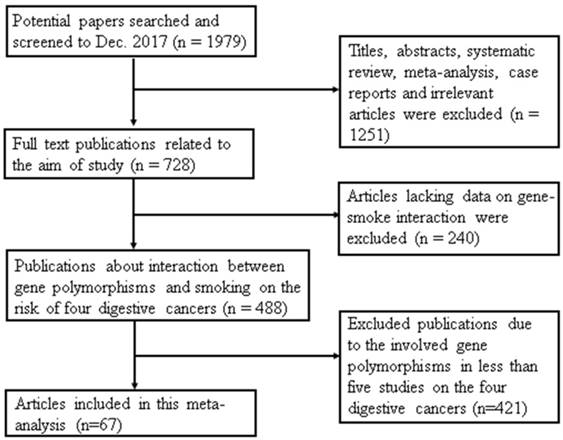
A total of 1979 articles were collected from the 3 databases. As shown in Figure 1, 1491 publications were excluded; 1251 articles were titles, abstracts, systematic reviews, meta-analyses, case reports and irrelevant articles and another 240 papers lacked data on gene-smoking interactions. Finally, a total of 67 studies were included in this meta-analysis. The reason for removing 421 studies from the remaining articles was that they evaluated the gene polymorphisms in less than five independent studies on the four digestive cancers.
Study characteristics and quality assessment
Study characteristics are summarized in Table 1. These studies were case-control or nested case-control studies, including 21,954 cases and 30,341 controls. Forty-three studies were performed in Asia, 11 studies were performed in Europe, 10 studies were performed in the Americas, and 3 studies were performed in Africa. Among all identified articles, 30 evaluated GSTM1 polymorphism [19-48], 18 evaluated GSTT1 polymorphism [20-24, 30-32, 34, 35, 40, 42-48], 12 evaluated GSTP1 polymorphism [11, 22, 30, 32, 34, 35, 42, 49-53], 8 evaluated CYP1A1 IIe462Val polymorphism [9, 27, 28, 54-58], 7 evaluated CYP1A1 MspI polymorphism [26, 28, 45, 54, 57, 58], 8 evaluated NAT2 polymorphism [24, 28, 36, 38, 46, 59-61], 6 evaluated SULT1A1 polymorphism [24, 45, 62-65], 8 evaluated hOGG1 polymorphism [66-73], 7 evaluated XRCC1 polymorphism [52, 67, 69, 74-77], and 6 evaluated p53 polymorphism [78-83].
Flow diagram of study selection in this meta-analysis. This flowchart indicates that the process of screening relevant studies based on the inclusion/exclusion criteria. A total of 67 studies were included in this meta-analysis.
As shown in Table 1, the quality scores of studies ranged from 6 to 9. Therefore, 91% of the studies (n=61) were high-quality studies (studies with a score≥7).
Tobacco metabolizing related genes
GST genes
Among 30 studies on the GSTM1 polymorphism in Table 2, the results showed the GSTM1 null genotype increased the four digestive cancers risk (OR=1.118, 95% CI 1.022-1.222). No significant publication bias was found using Begg's test (p=0.10), while there was publication bias by Egger's test (p=0.045). According to the trim and fill analysis, the adjusted estimated effect was OR 1.054 (95% CI: 0.954-1.163) based on the random-effects model. Substantial heterogeneity was observed in this analysis (Q=70.248, p=0.000, I2= 53.024 %), which suggested that GSTM1 polymorphisms have different effects on the risk of four cancers, depending on the cancer type and ethnicity. Subgroup analysis based on ethnicity revealed that such an association was observed among both African (OR=1.614, 95% CI 1.038-2.51; I2=0%, p for heterogeneity=1) and Asian (OR=1.284, 95% CI 1.122-1.47; I2=57.181%, p for heterogeneity=0.001) populations; further subgroup analysis based on the cancer type showed that the GSTM1 null genotype were associated with an increased risk of oesophageal cancer (OR=1.406, 95% CI 1.124-1.759; I2=63.644%, p for heterogeneity=0.027) and gastric cancer (OR=1.335, 95% CI 1.145-1.556; I2=52.921%, p for heterogeneity=0.019). Stratified analysis by smoking status showed the association of the GSTM1 null genotype with the four cancers risk was significant among smokers (OR=1.179, 95% CI 1.030-1.349; I2=57.328%, p for heterogeneity=0). In subgroup analyses among smokers, there was publication bias (p Begg =0.004; p Egger =0.029). According to the trim and fill analysis, the adjusted estimated effect was OR 1.012 (95%CI: 0.867-1.181) based on the random-effects model. However, the effect size was only found in Asian population (OR=1.355, 95% CI 1.089-1.686; I2=39.566%, p for heterogeneity=0.044). Smokers with the GSTM1 null genotype had an increased risk of oesophageal cancer (OR=1.382, 95% CI 1.009-1.894, I2=55.082, p for heterogeneity=0.064) and gastric cancer (OR=1.690, 95% CI 1.298-2.201, I2=69.955%, p for heterogeneity=0). Moreover, subgroup analyses in non-smokers showed that the GSTM1 null genotype also increased the gastric cancer risk (OR=1.344, 95% CI 1.054-1.715; I2=51.576%, p for heterogeneity=0.024). The GSTM1 null genotype was associated with the four cancers risk in Asian population (OR=1.237, 95% CI 1.020-1.500; I2=44.307%, p for heterogeneity=0.023), no publication bias was observed (p>0.05).
Characteristics of included case-control studies
| First author, year | NOS | Country/Ethnicity | Cancer site | Genes | Genotype distribution (cases/controls) | Genotype distribution by smoking status (cases/controls) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No risk* | At risk$ | Non-smoker | Smoker | |||||||
| No risk* | At risk$ | No risk* | At risk$ | |||||||
| Wang,2004 | 7 | China/Asia | Oesophagus | GSTM1 | 53/57^ | 74/44 | 24/37 | 33/26 | 29/20 | 41/18 |
| Rudolph,2012 | 8 | German/Europe | Colorectum | GSTM1 | 822/844 | 932/923 | 368/424 | 425/466 | 404/382 | 458/417 |
| GSTT1 | 1,433/1,459 | 313/308 | 644/722 | 142/170 | 715/672 | 147/123 | ||||
| Lilla,2007 | 9 | Germany/Europe | Colorectum | SULT1A1 | 212/263 | 292/340 | 106/132 | 132/157 | 106/131 | 160/183 |
| Gao,2002 | 9 | China/Asia | Oesophagus | GSTM1 | 35/90 | 106/133 | 13/38 | 36/58 | 22/52 | 70/75 |
| GSTT1 | 67/104 | 74/119 | 20/44 | 29/52 | 47/60 | 45/67 | ||||
| Stomach | GSTM1 | 63/90 | 90/133 | 10/38 | 20/58 | 53/52 | 70/75 | |||
| GSTT1 | 82/104 | 71/119 | 20/44 | 10/52 | 62/60 | 61/67 | ||||
| Dandara,2006 | 7 | South Africa/Africa | Oesophagus | SULT1A1 | 115/132 | 121/134 | 27/41 | 28/47 | 88/91 | 93/87 |
| Li,2010 | 7 | South Africa/Africa | Oesophagus | GSTM1 | 206/200 | 133/80 | 55/75 | 8/20 | 151/125 | 125/60 |
| GSTT1 | 127/178 | 113/102 | 27/66 | 36/29 | 100/112 | 77/73 | ||||
| GSTP1 | 92/107 | 148/173 | 21/30 | 42/65 | 71/77 | 106/108 | ||||
| Gertig,1998 | 7 | America/Americas | Colorectum | GSTM1 | 97/104 | 114/117 | 36/40 | 41/40 | 61/64 | 73/77 |
| GSTT1 | 173/169 | 36/51 | 61/60 | 16/19 | 112/109 | 20/32 | ||||
| Tiemersma,2004 | 7 | Netherlands/Europe | Colorectum | GSTM1 | 203/206 | 228/226 | 81/102 | 85/118 | 119/103 | 143/108 |
| GSTT1 | 370/363 | 61/69 | 139/177 | 27/43 | 228/185 | 34/26 | ||||
| NAT2 | 262/254 | 169/178 | 89/132 | 66/67 | 169/121 | 79/77 | ||||
| SULT1A1 | 149/169 | 282/263 | 66/97 | 72/97 | 83/62 | 128/106 | ||||
| Abo-Hashem,2016 | 7 | Egypt/Africa | Liver | GSTP1 | 23/31 | 17/9 | 11/27 | 9/3 | 12/4 | 8/6 |
| Li,2005 | 6 | China/Asia | Stomach | GSTM1 | 33/36 | 67/26 | 16/23 | 30/19 | 17/13 | 37/7 |
| Tsukino,2004 | 7 | Japan/Asia | Stomach | hOGG1 | 32/74 | 110/197 | 11/38 | 39/99 | 21/36 | 71/98 |
| Inoue,2000 | 7 | Japan/Asia | Colorectum | GSTM1 | 97/97 | 108/123 | 19/37 | 17/36 | 78/60 | 91/87 |
| CYP1A1& | 86/87 | 119/133 | 14/20 | 22/53 | 72/67 | 97/80 | ||||
| Lee,2000 | 7 | China/Asia | Oesophagus | GSTP1 | 65/160 | 25/94 | 11/98 | 11/50 | 54/55 | 14/40 |
| Shen,2005 | 7 | China/Asia | Stomach | GSTM1 | 41/314 | 71/361 | 31/302 | 54/345 | 10/12 | 17/16 |
| CYP1A1# | 70/412 | 42/264 | 57/391 | 29/254 | 13/21 | 13/10 | ||||
| Yoshida,2007 | 7 | Japan/Asia | Colorectum | GSTM1 | 30/59 | 36/62 | 20/26 | 15/29 | 8/29 | 18/32 |
| CYP1A1# | 34/79 | 32/42 | 20/36 | 15/19 | 14/40 | 12/21 | ||||
| CYP1A1& | 20/49 | 46/72 | 8/24 | 27/31 | 12/23 | 14/38 | ||||
| NAT2 | 2/9 | 64/112 | 0/5 | 35/50 | 1/4 | 25/57 | ||||
| Zendehdel,2009 | 9 | Sweden/Europe | Oesophagus | GSTM1, | 52/230 | 43/239 | 17/112 | 13/87 | 35/127 | 30/143 |
| GSTT1 | 80/394 | 15/76 | 24/173 | 6/26 | 56/221 | 9/49 | ||||
| GSTP1 | 44/208 | 50/245 | 13/82 | 16/110 | 31/126 | 34/135 | ||||
| Oesophagus | GSTM1, | 35/230 | 42/239 | 4/112 | 4/87 | 30/127 | 38/143 | |||
| GSTT1 | 70/394 | 7/76 | 8/173 | 1/26 | 62/221 | 6/49 | ||||
| GSTP1 | 26/208 | 52/245 | 5/82 | 5/110 | 21/126 | 47/135 | ||||
| Stomach | GSTM1, | 54/230 | 70/239 | 6/112 | 8/87 | 4/127 | 62/143 | |||
| GSTT1 | 111/394 | 13/76 | 12/173 | 2/26 | 99/221 | 11/49 | ||||
| GSTP1 | 47/208 | 75/245 | 6/82 | 8/110 | 41/126 | 67/135 | ||||
| Lee,2006 | 7 | Chile/Americas | Stomach | GSTM1, | 60/207 | 13/56 | 29/128 | 2/33 | 31/79 | 11/23 |
| CYP1A1& | 38/153 | 35/110 | 16/90 | 15/71 | 22/63 | 20/39 | ||||
| Huang,2006 | 9 | America/Americas | Colon | GSTM1 | 297/503 | 257/371 | 111/211 | 97/151 | 184/292 | 158/219 |
| GSTT1 | 428/603 | 130/271 | 162/247 | 46/115 | 259/356 | 83/155 | ||||
| Moore,2005 | 7 | U.S./Americas | Colorectum | GSTM1 | 311/313 | 352/376 | 105/122 | 115/150 | 190/173 | 217/205 |
| GSTT1 | 561/584 | 129/118 | 182/230 | 44/50 | 350/325 | 77/56 | ||||
| GSTP1 | 282/317 | 399/381 | 97/132 | 123/140 | 173/171 | 251/251 | ||||
| Cai,2001 | 8 | China/Asia | Stomach | GSTM1 | 35/51 | 60/43 | 12/28 | 22/32 | 23/23 | 38/11 |
| Tamer,2005 | 7 | Turkey/Asia | Stomach | GSTM1 | 30/116 | 40/88 | 17/75 | 19/45 | 13/41 | 21/43 |
| GSTT1 | 49/151 | 21/53 | 25/85 | 11/35 | 24/66 | 10/18 | ||||
| GSTP1 | 38/90 | 32/114 | 20/49 | 16/71 | 18/41 | 16/43 | ||||
| Slattery,2002 | 7 | USA/Americas | Colon | GSTM1 | 761/892 | 816/1012 | 332/413 | 326/486 | 429/479 | 490/526 |
| NAT2 | 920/1154 | 688/804 | 366/540 | 298/380 | 554/614 | 390/424 | ||||
| García-González,2012 | 8 | Spain/Europe | Stomach | GSTM1 | 274/290 | 283/267 | 125/151 | 120/147 | 51/40 | 71/35 |
| GSTT1 | 437/440 | 120/117 | 188/228 | 57/70 | 97/56 | 25/19 | ||||
| GSTP1 | 255/251 | 302/306 | 119/138 | 126/160 | 50/36 | 72/39 | ||||
| Malik,2010 | 8 | India/Aisa | Stomach | hOGG1 | 50/94 | 58/101 | 15/68 | 17/79 | 35/21 | 40/17 |
| Malik,2009 | 7 | India/Asia | Stomach | GSTM1 | 44/116 | 64/79 | 12/85 | 20/62 | 32/26 | 43/12 |
| Slattery,2003 | 9 | US/Americas | Rectum | GSTM1 | 230/279 | 243/295 | 84/123 | 88/124 | 145/156 | 153/171 |
| NAT2 | 247/306 | 204/255 | 90/143 | 74/105 | 156/163 | 128/150 | ||||
| Kasahara,2008 | 7 | Japan/Asia | Colorectum | hOGG1 | 17/39 | 51/82 | 8/14 | 28/41 | 7/23 | 19/37 |
| XRCC1 | 42/62 | 26/59 | 20/29 | 16/26 | 18/30 | 8/30 | ||||
| Wu,2003 | 6 | China/Asia | Oesophagus | SULT1A1 | 135/274 | 52/34 | 44/153 | 25/19 | 91/121 | 27/15 |
| Yu,1995 | 7 | China/Asia | Liver | GSTM1 | 14/55 | 16/95 | 7/34 | 10/61 | 7/21 | 6/34 |
| Yu,1999 a | 7 | China/ Asia | Liver | GSTM1 | 38/151 | 42/177 | 25/94 | 22/104 | 13/57 | 20/73 |
| Yu,1999 b | 7 | China/Asia | Liver | GSTM1 | 42/159 | 42/216 | 26/91 | 23/132 | 16/67 | 19/84 |
| GSTT1 | 42/194 | 41/181 | 25/110 | 24/113 | 17/84 | 17/67 | ||||
| Moslehi,2006 | 6 | USA/Americas | Colorectum | NAT2 | 413/376 | 272/317 | 140/158 | 92/124 | 249/195 | 168/168 |
| Malakar,2012 | 9 | India/Asia | Stomach | GSTM1 | 45/107 | 57/97 | 7/52 | 14/30 | 38/55 | 43/67 |
| GSTT1 | 65/111 | 37/93 | 11/45 | 10/37 | 54/66 | 27/56 | ||||
| Yu,2000 | 7 | China/Asia | Liver | NAT2 | 27/55 | 124/156 | 16/30 | 59/100 | 11/25 | 65/56 |
| Songserm,2014 | 8 | Thailand/Asia | Liver | hOGG1 | 34/95 | 111/234 | 14/55 | 50/123 | 20/40 | 61/111 |
| XRCC1 | 4/21 | 156/318 | 2/11 | 70/170 | 2/10 | 86/148 | ||||
| Ates,2005 | 7 | Turkey/Asia | Colorectum | GSTM1 | 83/116 | 98/88 | 44/75 | 46/45 | 39/41 | 52/43 |
| GSTT1 | 118/151 | 63/53 | 56/85 | 34/35 | 62/66 | 29/18 | ||||
| GSTP1 | 73/90 | 108/114 | 30/49 | 60/71 | 43/41 | 47/43 | ||||
| Van der Hel,2003 a | 8 | Netherlands/Europe | Colorectum | GSTM1 | 124/396 | 88/369 | 73/271 | 65/257 | 51/125 | 23/112 |
| GSTT1 | 154/541 | 58/224 | 104/385 | 34/143 | 50/156 | 24/81 | ||||
| Van der Hel,2003 b | 7 | Netherlands/Europe | Colorectum | NAT2 | 146/495 | 112/362 | 99/341 | 63/249 | 42/153 | 45/113 |
| Moaven,2010 | 7 | Iran/Asia | Oesophagus | GSTP1 | 84/74 | 64/62 | 51/65 | 50/46 | 33/10 | 14/16 |
| Zhang,2014 | 8 | China/Asia | Colorectum | hOGG1 | 44/48 | 203/252 | 30/32 | 129/158 | 14/16 | 74/94 |
| Ghosh,2016 | 9 | India/Asia | Stomach | GSTP1 | 41/61 | 29/21 | 10/38 | 9/16 | 31/23 | 20/5 |
| XRCC1 | 28/48 | 42/34 | 8/33 | 11/21 | 20/15 | 31/13 | ||||
| Boccia,2005 | 7 | Italy/Europe | Stomach | SULT1A1 | 40/160 | 36/100 | 33/126 | 24/83 | 7/31 | 10/15 |
| Boccia,2015 | 7 | Italy/Europe | Liver | GSTM1 | 96/139 | 105/150 | 31/91 | 48/81 | 62/48 | 57/69 |
| GSTT1 | 141/220 | 60/69 | 59/129 | 20/43 | 81/91 | 38/26 | ||||
| CYP1A1& | 165/226 | 56/64 | 65/136 | 20/37 | 98/90 | 35/27 | ||||
| SULT1A1 | 132/180 | 89/110 | 52/103 | 33/70 | 78/77 | 55/40 | ||||
| Yuan,2012 | 8 | China/Asia | Liver | hOGG1 | 67/144 | 283/256 | 30/48 | 83/84 | 37/96 | 200/172 |
| Sakamoto,2006 | 7 | Japan/Asia | Liver | hOGG1 | 56/73 | 153/202 | 35/56 | 105/152 | 21/17 | 48/50 |
| Hanaoka,2001 | 8 | Brazil/Americas | Stomach | hOGG1 | 133/123 | 75/82 | 72/85 | 48/55 | 61/38 | 27/25 |
| Gelatti,2005 | 8 | Italy/Europe | Liver | GSTM1 | 101/185 | 99/215 | 41/60 | 34/80 | 60/125 | 65/135 |
| GSTT1 | 168/328 | 32/72 | 67/124 | 8/16 | 101/204 | 24/56 | ||||
| NAT2 | 105/201 | 95/199 | 40/65 | 35/75 | 65/136 | 60/124 | ||||
| Setiawan,2000 | 9 | China/Asia | Stomach | GSTM1 | 45/207 | 42/212 | 26/131 | 18/143 | 19/76 | 24/69 |
| GSTT1 | 37/228 | 44/190 | 18/146 | 21/127 | 19/82 | 23/63 | ||||
| Setiawan,2001 | 9 | China/Asia | Stomach | GSTP1 | 61/296 | 20/123 | 30/199 | 10/75 | 31/97 | 10/48 |
| Zhang,2012 | 7 | China/Asia | Stomach | GSTP1 | 331/343 | 219/207 | 69/136 | 37/77 | 82/100 | 59/71 |
| Chen,2004 | 8 | China/Asia | Colon | GSTM1 | 23/151 | 30/188 | 17/92 | 16/108 | 6/57 | 14/79 |
| GSTT1 | 41/270 | 12/69 | 26/153 | 7/47 | 15/116 | 5/20 | ||||
| Rectum | GSTM1 | 33/151 | 39/188 | 23/92 | 26/108 | 10/57 | 13/79 | |||
| GSTT1 | 61/270 | 11/69 | 43/153 | 6/47 | 18/116 | 5/20 | ||||
| Bhat, 2014 | 7 | India/Asia | Oesophagus | CYP1A1# | 253/300 | 273/226 | 101/134 | 99/122 | 152/166 | 174/104 |
| Chen, 2011 | 7 | China/Asia | Stomach | XRCC1 | 177/132 | 157/202 | 83/88 | 67/124 | 94/44 | 90/78 |
| Fernandes, 2016 | 8 | Brazil/Americas | Colorectum | CYP1A1# | 193/312 | 34/88 | 107/190 | 24/53 | 86/122 | 10/35 |
| CYP1A1& | 165/246 | 62/154 | 96/156 | 35/87 | 69/90 | 27/67 | ||||
| Hou, 2005 | 7 | USA/Americas | Colorectum | CYP1A1# | 633/643 | 42/36 | 219/258 | 9/19 | 387/344 | 29/15 |
| Li, 2009 | 7 | China/Asia | Liver | CYP1A1# | 560/598 | 410/402 | 313/320 | 223/212 | 247/278 | 187/190 |
| Little, 2006 | 8 | Northeast Scotland/Europe | Colorectum | CYP1A1# | 235/372 | 16/24 | 75/128 | 5/5 | 84/142 | 7/10 |
| CYP1A1& | 190/310 | 42/68 | 63/107 | 12/19 | 68/122 | 16/27 | ||||
| Malakar,2014 | 7 | India/Asia | Stomach | p53 | 11/36 | 94/174 | 1/14 | 20/71 | 10/22 | 74/103 |
| Qiu, 2016 | 6 | China/Asia | Liver | p53 | 221/244 | 764/748 | 137/207 | 488/645 | 84/37 | 276/103 |
| Shao, 2008 | 6 | China/Asia | Oesophagus | p53 | 163/195 | 510/499 | 61/90 | 229/219 | 102/105 | 281/280 |
| Shen,2004 | 7 | China/Asia | Stomach | p53 | 96/94 | 228/223 | 36/46 | 97/76 | 60/48 | 131/147 |
| Yan,2009 | 6 | China/Asia | Stomach | XRCC1 | 241/345 | 214/305 | 121/186 | 91/163 | 106/155 | 111/136 |
| Yang, 2008 | 7 | China/Asia | Oesophagus | p53 | 373/273 | 62/277 | 222/200 | 43/200 | 151/73 | 19/77 |
| Yu, 1999 c | 9 | China/Asia | Liver | CYP1A1# | 46/239 | 35/170 | 33/147 | 15/97 | 13/92 | 20/73 |
| CYP1A1& | 25/152 | 56/257 | 19/86 | 29/158 | 6/66 | 27/99 | ||||
| Yu,2004 | 7 | China/Asia | Oesophagus | XRCC1 | 65/88 | 70/64 | 33/50 | 28/35 | 32/38 | 42/29 |
| Cai, 2017 | 7 | China/Asia | Liver | p53 | 63/65 | 279/282 | 33/55 | 146/171 | 30/10 | 133/111 |
| Putthanachote, 2017 | 7 | Putthanachote/ Asia | Stomach | XRCC1 | 12/8 | 89/194 | 8/3 | 41/105 | 4/5 | 48/89 |
Aberrations: NOS, the Newcastle-Ottawa-Scale.
^Number of cases and controls.
*The wild type of each gene.
$The mutant type of each gene.
# For CYP1A1, the IIe462Val substitution (IIe/Val+Val/Val).
& For CYP1A1, the 3801T>C substitution (MspI) (T/C+C/C).
Meta-analysis of the association between GSTM1, GSTT1 polymorphisms and the four digestive cancers risk
| Stratified analysis | Subgroup analysis | No. of studies | OR (95% CI) | Heterogeneity test | Publication bias | False-positive report probability | Statistical power | ||
|---|---|---|---|---|---|---|---|---|---|
| Q | P | I2 (%) | p | ||||||
| GSTM1 total population | Overall cancer | 30 | 1.118(1.022-1.222) | 70.248 | 0 | 53.024 | 0.100* | 0.050 | 0.659 |
| Cancer type | 0.045$ | ||||||||
| Colorectum | 12 | 1.010(0.911-1.121) | 11.808 | 0.461 | 0 | ||||
| Oesophagus | 4 | 1.406(1.124-1.759) | 11.002 | 0.027 | 63.644 | 0.047 | 0.337 | ||
| Stomach | 11 | 1.335(1.145-1.556) | 21.241 | 0.019 | 52.921 | 0.048 | 0.991 | ||
| Liver | 5 | 0.866(0.691-1.086) | 1.763 | 0.779 | 0 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 1.614(1.038-2.51) | 0 | 1 | 0 | 0.042 | 0.782 | ||
| Americas | 6 | 1(0.853-1.172) | 3.552 | 0.616 | 0 | ||||
| Asia | 17 | 1.284(1.122-1.47) | 39.702 | 0.001 | 57.181 | 0.048 | 0.976 | ||
| Europe | 7 | 0.991(0.862-1.141) | 7.724 | 0.461 | 0 | ||||
| GSTM1 non-smokers | Overall cancer | 30 | 1.071(0.948-1.210) | 54.333 | 0.011 | 39.263 | 0.486* | ||
| Cancer type | 0.186$ | ||||||||
| Colorectum | 12 | 0.993(0.847-1.163) | 10.507 | 0.572 | 0 | ||||
| Oesophagus | 4 | 1.250(0.826-1.891) | 6.091 | 0.192 | 34.331 | ||||
| Stomach | 11 | 1.344(1.054-1.715) | 20.651 | 0.024 | 51.576 | 0.047 | 0.716 | ||
| Liver | 5 | 0.866(0.622-1.206) | 8.996 | 0.061 | 55.538 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 0.545(0.207-1.435) | 0 | 1 | 0 | ||||
| Americas | 6 | 0.956(0.759-1.205) | 7.012 | 0.220 | 28.698 | ||||
| Asia | 17 | 1.237(1.020-1.500) | 30.524 | 0.023 | 44.307 | 0.048 | 0.542 | ||
| Europe | 7 | 1.018(0.828-1.253) | 8.301 | 0.405 | 3.625 | ||||
| GSTM1 smokers | Overall cancer | 30 | 1.179(1.030-1.349) | 77.335 | 0 | 57.328 | 0.004* | 0.050 | 0.728 |
| Cancer type | 0.029$ | ||||||||
| Colorectum | 12 | 1.014(0.855-1.203) | 12.204 | 0.429 | 1.673 | ||||
| Oesophagus | 4 | 1.382(1.009-1.894) | 8.905 | 0.064 | 55.082 | 0.046 | 0.301 | ||
| Stomach | 11 | 1.690(1.298-2.201) | 33.284 | 0 | 69.955 | 0.047 | 0.999 | ||
| Liver | 5 | 0.862(0.606-1.227) | 3.146 | 0.534 | 0 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 1.725(0.891-3.339) | 0 | 1 | 0 | ||||
| Americas | 6 | 1.035(0.794-1.349) | 1.146 | 0.950 | 0 | ||||
| Asia | 17 | 1.355(1.089-1.686) | 28.106 | 0.044 | 39.566 | 0.048 | 0.755 | ||
| Europe | 7 | 1.054(0.826-1.343) | 37.431 | 0 | 78.628 | ||||
| GSTT1 total population | Overall cancer | 18 | 0.970(0.863-1.092) | 38.800 | 0.010 | 45.876 | 0.150* | ||
| Cancer type | 0.628$ | ||||||||
| Colorectum | 8 | 0.935(0.782-1.119) | 17.558 | 0.025 | 54.438 | ||||
| Oesophagus | 3 | 1.068(0.778-1.466) | 7.426 | 0.060 | 59.599 | ||||
| Stomach | 6 | 0.923(0.722-1.180) | 8.715 | 0.121 | 42.626 | ||||
| Liver | 3 | 1.084(0.772-1.521) | 2.108 | 0.348 | 5.136 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 1.553(0.978-2.465) | 0 | 1 | 0 | ||||
| Americas | 3 | 0.827(0.643-1.063) | 8.377 | 0.015 | 76.124 | ||||
| Asia | 8 | 1.017(0.837-1.237) | 11.380 | 0.181 | 29.703 | ||||
| Europe | 8 | 0.950(0.805-1.122) | 8.447 | 0.391 | 5.297 | ||||
| GSTT1 non-smokers | Overall cancer | 18 | 0.979(0.838-1.143) | 28.943 | 0.115 | 27.443 | 0.554* | ||
| Cancer type | 0.610$ | ||||||||
| Colorectum | 8 | 0.881(0.752-1.031) | 9.792 | 0.280 | 18.297 | ||||
| Oesophagus | 3 | 1.845(1.204-2.829) | 4.065 | 0.255 | 26.196 | 0.043 | 0.999 | ||
| Stomach | 6 | 0.973(0.732-1.293) | 4.639 | 0.462 | 0 | ||||
| Liver | 3 | 0.965(0.649-1.436) | 0.047 | 0.977 | 0 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 3.034(1.564-5.889) | 0 | 1 | 0 | 0.040 | 0.909 | ||
| Americas | 3 | 0.797(0.605-1.051) | 3.882 | 0.144 | 48.487 | ||||
| Asia | 8 | 0.999(0.779-1.280) | 9.695 | 0.287 | 17.481 | ||||
| Europe | 8 | 0.944(0.801-1.112) | 1.915 | 0.984 | 0 | ||||
| GSTT1 smokers | Overall cancer | 18 | 0.977(0.843-1.132) | 31.747 | 0.062 | 33.852 | 0.888* | ||
| Cancer type | 0.996$ | ||||||||
| Colorectum | 8 | 1.043(0.834-1.305) | 13.100 | 0.108 | 38.930 | ||||
| Oesophagus | 3 | 0.858(0.593-1.240) | 4.475 | 0.215 | 32.963 | ||||
| Stomach | 6 | 0.844(0.615-1.159) | 8.526 | 0.130 | 41.354 | ||||
| Liver | 3 | 1.192(0.778-1.825) | 2.556 | 0.279 | 21.741 | ||||
| Ethnicity | |||||||||
| Africa | 1 | 1.181(0.638-2.186) | 0 | 1 | 0 | ||||
| Americas | 3 | 0.864(0.606-1.232) | 6.401 | 0.041 | 68.754 | ||||
| Asia | 8 | 1.117(0.844-1.478) | 10.680 | 0.221 | 25.093 | ||||
| Europe | 8 | 0.907(0.710-1.158) | 12.237 | 0.141 | 34.627 | ||||
The bold letters show statistically significant results.
* Begg's test for publication bias.
$ Egger's test for publication bias.
Among 18 studies on the GSTT1 polymorphism in Table 2, we found that the GSTT1 null genotype could increase the oesophageal cancer risk in non-smokers (OR=1.845, 95% CI 1.204-2.829; I2=26.196%, p for heterogeneity=0.255). By subgroup analysis in non-smokers, Only one study showed the GSTT1 polymorphisms were related to the risk of four cancers in African population (OR=3.034, 95% CI 1.564-5.889)[22]. No publication bias was detected in this analysis (p>0.05).
Among 12 studies on the GSTP1 polymorphism in Supplementary Table S1, no significant correlations were found except one study on liver cancer in non-smokers (OR=7.364, 95% CI 1.671-32.440)[49]. There was no publication bias (p>0.05).
CYP1A1 gene
Eight papers provided data on the CYP1A1 IIe462Val polymorphism in Table 3. The results indicated that smokers with the CYP1A1 Ile462Val polymorphisms were at an increased risk of four cancers in Asian population (OR=1.585, 95%CI 1.029-2.442; I2=41.870%, p for heterogeneity=0.142). Seven articles were about CYP1A1 MspI polymorphism in Supplementary Table S1. The CYP1A1 MspI polymorphisms were not associated with the risk of four cancers in stratified analysis and subgroup analysis.
SULT1A1 gene
In Table 3, the SULT1A1 slow/intermediate phenotypes were associated with a 31.5% increase in the risk of four cancers (OR=1.315, 95% CI 1.009-1.715) from 6 studies. However, such an association was not observed in stratified analysis and subgroup analysis. Only one paper showed the association was significant in Asian population (OR=3.104, 95% CI 1.923-5.011)[64].
NAT2 gene
Eight papers provided data on the NAT2 polymorphism, as shown in Table 4. Two studies indicated that the NAT2 polymorphism was associated with the risk of four cancers in Asian population (OR=1.701, 95% CI 1.019-2.838) [28, 60]. Moreover, the association was also observed in smokers (OR=2.513, 95% CI 1.156-5.462).
DNA repair genes
Neither hOGG1 gene nor XRCC1 gene polymorphism was not associated with the risk of four cancers, as shown in Supplementary Table S1.
Tumour suppressor gene
We also found no significant association of p53 polymorphism with the risk of four cancers (Supplementary Table S1).
Discussion
A total of 67 case-control studies on the interaction of gene-smoking on the risk of four digestive cancers were identified in this review. This study included six tobacco metabolizing genes (GSTM1, GSTT1, GSTP1, CYP1A1, SULT1A1, and NAT2), two DNA repair genes (hOGG1 and XRCC1) and one tumour suppressor gene (p53). To the best of our knowledge, this is the first meta-analysis that investigated the joint effect of the most gene polymorphisms and smoking on four digestive cancers. Our data indicated the GSTM1 polymorphism was associated with the risk of four digestive cancers among Asian population (OR 1.284, 95% CI: 1.122-1.470). The GSTM1 null genotype could increase the gastric cancer risk (OR 1.335, 95% CI: 1.145-1.556) in total population. However, the association of the GSTM1 null genotype with the oesophageal cancer risk was found in smokers (OR 1.382, 95% CI: 1.009-1.894), not in non-smokers (OR 1.250, 95% CI: 0.826-1.891). Interestingly, we found the GSTT1 null genotype could increase the oesophageal cancer risk among non-smokers in only 3 studies (OR 1.845, 95% CI: 1.204-2.829). The SULT1A1 polymorphism was related to the risk of four digestive cancers (OR 1.315, 95% CI: 1.009-1.715), but such an association was not observed in stratified analysis and subgroup analysis except one study in Asian population (OR=3.104, 95% CI 1.923-5.011). Two studies indicated that the NAT2 polymorphism was associated with the risk of four cancers in Asian population (OR=1.701, 95% CI 1.019-2.838), and the association was also observed in smokers (OR=2.513, 95% CI 1.156-5.462). Moreover, smokers with the CYP1A1 Ile462Val polymorphism were at an increased cancer risk in Asian population (OR=1.585, 95% CI 1.029-2.442). None of the other gene-smoking interactions was observed in the above cancers.
Increasing studies investigated the gene-smoking interaction on the risk of cancer during these years. Two previously published studies indicated smokers with GSTM1 null genotype were at an increased oesophageal cancer risk [19, 21]. Moreover, the significant association was found between CYP1A1 IIe462Val and liver cancer risk among the cigarette smoking subjects in a meta-analysis (OR = 1.40, 95% CI 1.06-1.85) [84]. These results were similar to our findings. Zhang et al indicated the NAT2 polymorphisms were correlated to an increased liver cancer risk in smokers [11]. Whereas our study only provided two studies to support this conclusion. The SULT1A1 Arg213His polymorphism was associated with an increased oesophageal cancer risk [85], but such an association was not founded in our subgroup analysis. We also found no interaction of smoking with other genetic polymorphisms on four digestive cancers. Several reasons account for the null results.
First, the association between gene polymorphism and cancer risk could be modified by various smoking habits, including the age of initiating smoking, duration of smoking, pack-years of smoking, the method of tobacco use and cigarette categories. One study showed that lifetime exposure to tobacco increased the risk of upper aero-digestive tract (UADT) cancers. Furthermore, chewing tobacco was more likely to increase the risk of UADT cancers (OR=7.61; 95% CI 4.65-12.45) compared to smoking [86]. The categories of cigarette also play a role in cancer progression and affect the association of gene polymorphisms with cancer susceptibility [87]. Remarkably, Liang et al reported on the significant interactions of smoking pack years with HEL308 genotypes (Pinteraction=0.026) and ADH1B genotypes (Pinteraction=0.0016) in the head and neck squamous cell carcinoma (HNSCC) risk, respectively [88]. Most of the included studies only provided data to evaluate the smoking status and we could not verify the findings in our study. Moreover, the age of initiating smoking is rarely measured in published studies, but this factor could be related to genetic polymorphisms in subgroups. Second, many other genes could be relevant to the metabolism of harmful compounds in tobacco except for the included genes, and the gene-gene interaction also existed in cancer susceptibility [89, 90]. It is probable that combinations of multiple gene polymorphisms are more significant as risk factors than a single gene polymorphism.
Interestingly, we found the GSTT1 null genotype could increase oesophageal cancer risk among non-smokers, but not among smokers. It was conflictive with the recognized conclusion on tobacco use increasing the cancer risk. However, this result also suggested not all the smokers with high-risk genetic variants were at an increased cancer risk. Because other benefical environmental factors, such as dietary habits, play an important role in cancer prevention [91]. A previous study indicated that regular tea consumption decreased the OC (OR: 0.38, 95% CI: 0.17-0.87) and GC (OR: 0.30, 95% CI: 0.14-0.66) risk among those with GSTT1 null genotype [21]. Ko et al also showed soy product consumption was associated with lower breast cancer risk in BRCA mutation carriers (HR: 0.39; 95% CI: 0.19-0.79) [92]. It was resonalble to assume that a protective factor also interacted with the GSTT1 null genotype among smokers. Moreover, our finding was based on only 3 papers, and needed to be further verified by more studies.
Regarding the interaction between smoking and GSTM1 and CYP1A1 IIe462Val on digestive cancers risk, evidence regarding the molecular mechanism also supported the results of this meta-analysis. Tobacco smoke contains various carcinogens, for example, polycyclic aromatic hydrocarbons (PAHs) and tobacco specific nitrosamines (TSNA) [93]. These carcinogens are first metabolically activated by phase I enzymes, e.g., cytochrome P4501A1 (CYP1A1), into their final forms and then combine with DNA, forming aromatic-DNA adducts that are considered as an early stage in carcinogenesis. Moreover, these activated forms are detoxified by phase II enzymes, especially glutathione S-transferases (GSTs)[94]. Thus, the susceptibility to cancer determined by genetic factors may depend on the metabolic balance between phase I and phase II enzymes[8]. Because the CYP and GST genetic polymorphisms regulate the metabolism of xenobiotics, they are thought to affect individual's sensitivity to environmental factors and susceptibility to cancer. Although this meta-analysis suggested that there was no significant interaction between smoking and other gene polymorphisms, several related molecular mechanisms remain biologically plausible. Except for the CYP and GST family genes, the carcinogens in tobacco smoke can be activated by SULT1A1 and NAT2 [95, 96]. DNA repair genes, e.g, hOGG1 and XRCC, are involved in the elimination of DNA adducts, which suggests that the DNA repair genes polymorphisms may be associated with different repair efficiencies of DNA damage [69]. Moreover, the p53 is a tumour suppressor gene and plays a key role in regulating the cell cycle and maintaining genomic integrity [79]. Thus, it may modify individual's susceptibility to various carcinogens.
Compared with a single study that investigated the role of some metabolic gene polymorphisms in cancer risk, we evaluated the interaction between ten gene polymorphisms and smoking for four digestive cancers, and this is the first such report to date. Therefore, we could provide more comprehensive information on the gene-smoking interaction in main digestive cancers. However, there are several limitations in this meta-analysis. First, there is strong heterogeneity in the risk estimates for most gene polymorphisms and stratified analyses. Second, the ORs were only adjusted for the cancer type and ethnicity. A more precise analysis should be performed based on the data adjusted for confounding factors including the age, sex, family history, environmental factors, cancer stage, and lifestyle. In addition, we were not able to evaluate the interaction of genes with genes or other environmental factors, which should be assessed in future studies.
Meta-analysis of the association between CYP1A1, SULT1A1 polymorphisms and the four digestive cancers risk
| Stratified analysis | Subgroup analysis | No. of studies | OR (95% CI) | Heterogeneity test | False-positive report probability | Statistical power | ||
|---|---|---|---|---|---|---|---|---|
| Q | P | I2 (%) | ||||||
| CYP1A1 IIe462Val total population | Overall cancer | 8 | 1.102(0.911-1.332) | 13.969 | 0.052 | 49.888 | ||
| Cancer type | ||||||||
| Colorectum | 4 | 1.039(0.724-1.490) | 8.418 | 0.038 | 64.360 | |||
| Oesophagus | 1 | 1.432(0.829-2.474) | 0 | 1 | 0 | |||
| Stomach | 1 | 0.936(0.494-1.776) | 0 | 1 | 0 | |||
| Liver | 2 | 1.082(0.714-1.639) | 0.005 | 0.945 | 0 | |||
| Ethnicity | ||||||||
| Americas | 2 | 0.851(0.572-1.267) | 3.949 | 0.047 | 74.676 | |||
| Asia | 5 | 1.197(0.961-1.492) | 6.270 | 0.180 | 36.201 | |||
| Europe | 1 | 1.055(0.505-2.207) | 0 | 1 | 0 | |||
| CYP1A1 IIe462Val non-smokers | Overall cancer | 8 | 0.973(0.827-1.145) | 6.539 | 0.478 | 0 | ||
| Cancer type | ||||||||
| Colorectum | 4 | 0.901(0.586-1.384) | 3.519 | 0.318 | 14.759 | |||
| Oesophagus | 1 | 1.077(0.647-1.792) | 0 | 1 | 0 | |||
| Stomach | 1 | 0.783(0.434-1.412) | 0 | 1 | 0 | |||
| Liver | 2 | 0.964(0.665-1.397) | 1.531 | 0.216 | 34.681 | |||
| Ethnicity | ||||||||
| Americas | 2 | 0.720(0.460-1.127) | 0.539 | 0.463 | 0 | |||
| Asia | 5 | 1.009(0.846-1.204) | 3.349 | 0.501 | 0 | |||
| Europe | 1 | 1.707(0.478-6.089) | 0 | 1 | 0 | |||
| Overall cancer | 8 | 1.341(0.959-1.876) | 17.436 | 0.015 | 59.853 | |||
| Cancer type | ||||||||
| CYP1A1 IIe462Val smokers | Colorectum | 4 | 1.067(0.565-2.015) | 9.278 | 0.026 | 67.667 | ||
| Oesophagus | 1 | 1.827(0.658-5.075) | 0 | 1 | 0 | |||
| Stomach | 1 | 2.100(0.494-8.926) | 0 | 1 | 0 | |||
| Liver | 2 | 1.385(0.636-3.014) | 1.846 | 0.174 | 45.832 | |||
| Ethnicity | ||||||||
| Americas | 2 | 0.883(0.431-1.807) | 8.182 | 0.004 | 87.778 | |||
| Asia | 5 | 1.585(1.029-2.442) | 6.881 | 0.142 | 41.870 | 0.046 | 0.932 | |
| Europe | 1 | 1.183(0.341-4.108) | 0 | 1 | 0 | |||
| SULT1A1 total population | Overall cancer | 6 | 1.315(1.009-1.715) | 17.371 | 0.004 | 71.216 | 0.048 | 0.993 |
| Cancer type | ||||||||
| Colorectum | 2 | 1.137(0.649-1.994) | 0.502 | 0.478 | 0 | |||
| Oesophagus | 2 | 1.724(0.940-3.163) | 13.122 | 0 | 92.379 | |||
| Stomach | 1 | 1.440(0.579-3.579) | 0 | 1 | 0 | |||
| Liver | 1 | 1.103(0.480-2.536) | 0 | 1 | 0 | |||
| Ethnicity | ||||||||
| Africa | 1 | 1.036(0.730-1.472) | 0 | 1 | 0 | |||
| Asia | 1 | 3.104(1.923-5.011) | 0 | 1 | 0 | 0.044 | 0.997 | |
| Europe | 4 | 1.148(0.984-1.339) | 1.332 | 0.722 | 0 | |||
| SULT1A1 non-smokers | Overall cancer | 6 | 1.257(0.849-1.861) | 17.039 | 0.004 | 70.656 | ||
| Cancer type | ||||||||
| Colorectum | 2 | 1.068(0.494-2.311) | 0.021 | 0.885 | 0 | |||
| Oesophagus | 2 | 2.027(0.853-4.819) | 10.933 | 0.001 | 90.853 | |||
| Stomach | 1 | 1.104(0.339-3.591) | 0 | 1 | 0 | |||
| Liver | 1 | 0.934(0.296-2.947) | 0 | 1 | 0 | |||
| Ethnicity | ||||||||
| Africa | 1 | 0.905(0.461-1.776) | 0 | 1 | 0 | |||
| Asia | 1 | 4.575(2.308-9.070) | 0 | 1 | 0 | 0.041 | 0.991 | |
| Europe | 4 | 1.045(0.836-1.307) | 0.242 | 0.970 | 0 | |||
| SULT1A1 smokers | Overall cancer | 6 | 1.248(0.952-1.637) | 8.766 | 0.119 | 42.964 | ||
| Cancer type | ||||||||
| Colorectum | 2 | 0.996(0.660-1.501) | 0.439 | 0.507 | 0 | |||
| Oesophagus | 2 | 1.454(0.893-2.369) | 3.562 | 0.059 | 71.922 | |||
| Stomach | 1 | 2.952(0.864-10.091) | 0 | 1 | 0 | |||
| Liver | 1 | 1.357(0.688-2.680) | 0 | 1 | 0 | |||
| Ethnicity | ||||||||
| Africa | 1 | 1.105(0.652-1.875) | 0 | 1 | 0 | |||
| Asia | 1 | 2.393(1.117-5.126) | 0 | 1 | 0 | 0.040 | 0.694 | |
| Europe | 4 | 1.146(0.854-1.539) | 4.358 | 0.225 | 31.169 | |||
The bold letters show statistically significant results.
Meta-analysis of the association between NAT2 polymorphism and the four digestive cancers risk
| Stratified analysis | Subgroup analysis | No. of studies | OR (95% CI) | Heterogeneity test | False-positive report probability | Statistical power | ||
|---|---|---|---|---|---|---|---|---|
| Q | P | I2 (%) | ||||||
| total population | Overall cancer | 8 | 0.990(0.872-1.125) | 11.662 | 0.112 | 39.978 | ||
| Cancer type | ||||||||
| Colorectum | 6 | 0.970(0.837-1.123) | 7.981 | 0.157 | 37.351 | |||
| Liver | 2 | 1.115(0.796-1.561) | 3.280 | 0.070 | 69.515 | |||
| Ethnicity | ||||||||
| Americas | 3 | 0.961(0.832-1.109) | 6.101 | 0.047 | 67.219 | |||
| Asia | 2 | 1.701(1.019-2.838) | 0.303 | 0.582 | 0 | 0.044 | 0.426 | |
| Europe | 3 | 0.963(0.793-1.168) | 0.553 | 0.759 | 0 | |||
| non-smokers | Overall cancer | 8 | 1.047(0.889-1.232) | 8.934 | 0.257 | 21.649 | ||
| Cancer type | ||||||||
| Colorectum | 6 | 1.072(0.894-1.285) | 7.483 | 0.187 | 33.179 | |||
| Liver | 2 | 0.886(0.556-1.414) | 0.695 | 0.404 | 0 | |||
| Ethnicity | ||||||||
| Americas | 3 | 1.044(0.802-1.359) | 2.478 | 0.290 | 19.295 | |||
| Asia | 2 | 1.251(0.597-2.622) | 1.607 | 0.205 | 37.779 | |||
| Europe | 3 | 1.004(0.730-1.382) | 4.444 | 0.108 | 54.996 | |||
| smokers | Overall cancer | 8 | 0.993(0.817-1.205) | 13.717 | 0.056 | 48.968 | ||
| Cancer type | ||||||||
| Colorectum | 6 | 0.933(0.755-1.152) | 7.364 | 0.195 | 32.101 | |||
| Liver | 2 | 1.334(0.837-2.125) | 4.334 | 0.037 | 76.927 | |||
| Ethnicity | ||||||||
| Americas | 3 | 0.913(0.749-1.113) | 2.470 | 0.291 | 19.037 | |||
| Asia | 2 | 2.513(1.156-5.462) | 0.113 | 0.737 | 0 | 0.040 | 0.379 | |
| Europe | 3 | 0.988(0.745-1.310) | 4.613 | 0.100 | 56.648 | |||
The bold letters show statistically significant results.
In summary, our meta-analysis provides the evidence of two potential gene-smoking interactions, one is between smoking and GSTM1 on oesophageal cancer, and the other is between smoking and CYP1A1 Ile462Val on the four cancers in Asian populations. None of the other gene-smoking interactions was observed in the above cancer. Future studies need to be conducted to verify the conclusions.
Supplementary Material
Supplementary table S1.
Acknowledgements
We would like to thank our laboratory members Ziqing Zhu, Qiqi Li and Ying Yang for discussion. Le Du thanks Jingchuan Li for consistent support during this research. This work was not supported by any grant.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459-1544
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:9
3. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1-1438
4. Bhowmik A, Das S, Bhattacharjee A, Choudhury B, Naiding M, Deka S. et al. MDM2 and TP53 Polymorphisms as Predictive Markers for Head and Neck Cancer in Northeast Indian Population: Effect of Gene-Gene and Gene-Environment Interactions. Asian Pacific Journal of Cancer Prevention. 2015;16:5767-5772
5. Simonds NI, Ghazarian AA, Pimentel CB, Schully SD, Ellison GL, Gillanders EM. et al. Review of the Gene-Environment Interaction Literature in Cancer: What Do We Know? Genet Epidemiol. 2016;40:356-65
6. Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473-86
7. Shen H, Spitz MR, Qiao Y, Guo Z, Wang LE, Bosken CH. et al. Smoking, DNA repair capacity and risk of nonsmall cell lung cancer. Int J Cancer. 2003;107:84-8
8. Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733-44
9. Bhat GA, Shah IA, Makhdoomi MA, Iqbal B, Rafiq R, Nabi S. et al. CYP1A1 and CYP2E1 genotypes and risk of esophageal squamous cell carcinoma in a high-incidence region, Kashmir. Tumor Biology. 2014;35:5323-5330
10. Wang Q, Chen Y, Zhang Y, Xu W, He H, Li X. et al. Quantitative assessment of the influence of glutathione S-transferase T1 null variant on gastric cancer risk. Tumour Biol. 2014;35:849-58
11. Zhang J, Xu F, Ouyang C. Joint effect of polymorphism in the N-acetyltransferase 2 gene and smoking on hepatocellular carcinoma. Tumour Biol. 2012;33:1059-63
12. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25:603-605
13. Yang L, Liu B, Huang B, Deng J, Li H, Yu B. et al. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Human Molecular Genetics. 2013;22:1886-1894
14. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557-60
15. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-101
16. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629-34
17. Jia Z, Chen C, Wu Y, Ding F, Tian X, Li W. et al. No difference in clinical outcomes after total knee arthroplasty between patellar eversion and non-eversion. Knee Surg Sports Traumatol Arthrosc. 2016;24:141-7
18. Mizuguchi T, Kawamoto M, Meguro M, Shibata T, Nakamura Y, Kimura Y. et al. Laparoscopic hepatectomy: a systematic review, meta-analysis, and power analysis. Surg Today. 2011;41:39-47
19. Wang AH, Sun CS, Li LS, Huang JY, Chen QS, Xu DZ. Genetic susceptibility and environmental factors of esophageal cancer in Xi'an. World J Gastroenterol. 2004;10:940-4
20. Rudolph A, Hein R, Hoffmeister M, Försti A, Hemminki K, Risch A. et al. Copy number variations of GSTT1 and GSTM1, colorectal cancer risk and possible effect modification of cigarette smoking and menopausal hormone therapy. International Journal of Cancer. 2012;131:E841-E848
21. Gao CM, Takezaki T, Wu JZ, Li ZY, Liu YT, Li SP. et al. Glutathione-S-transferases M1 (GSTM1) and GSTT1 genotype, smoking, consumption of alcohol and tea and risk of esophageal and stomach cancers: a case-control study of a high-incidence area in Jiangsu Province, China. Cancer Lett. 2002;188:95-102
22. Li D, Dandara C, Parker MI. The 341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genetics. 2010;11:47
23. Gertig DM, Stampfer M, Haiman C, Hennekens CH, Kelsey K, Hunter DJ. Glutathione S-transferase GSTM1 and GSTT1 polymorphisms and colorectal cancer risk: a prospective study. Cancer Epidemiol Biomarkers Prev. 1998;7:1001-5
24. Tiemersma EW, Bunschoten A, Kok FJ, Glatt H, de Boer SY, Kampman E. Effect ofSULT1A1 andNAT2 genetic polymorphism on the association between cigarette smoking and colorectal adenomas. International Journal of Cancer. 2004;108:97-103
25. Li H, Chen XL, Li HQ. Polymorphism of CYPIA1 and GSTM1 genes associated with susceptibility of gastric cancer in Shandong Province of China. World J Gastroenterol. 2005;11:5757-62
26. Inoue H, Kiyohara C, Marugame T, Shinomiya S, Tsuji E, Handa K. et al. Cigarette smoking, CYP1A1 MspI and GSTM1 genotypes, and colorectal adenomas. Cancer Res. 2000;60:3749-52
27. Shen J, Wang RT, Xu YC, Wang LW, Wang XR. Interaction models of CYP1A1, GSTM1 polymorphisms and tobacco smoking in intestinal gastric cancer. World J Gastroenterol. 2005;11:6056-60
28. Yoshida K, Osawa K, Kasahara M, Miyaishi A, Nakanishi K, Hayamizu S. et al. Association of CYP1A1, CYP1A2, GSTM1 and NAT2 gene polymorphisms with colorectal cancer and smoking. Asian Pac J Cancer Prev. 2007;8:438-44
29. Lee K, Caceres D, Varela N, Csendes DA, Rios RH, Quinones SL. [Allelic variants of cytochrome P4501A1 (CYP1A1), glutathione S transferase M1 (GSTM1) polymorphisms and their association with smoking and alcohol consumption as gastric cancer susceptibility biomarkers]. Rev Med Chil. 2006;134:1107-15
30. Zendehdel K, Bahmanyar S, McCarthy S, Nyren O, Andersson B, Ye W. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer Causes & Control. 2009;20:2031-2038
31. Huang K, Sandler RS, Millikan RC, Schroeder JC, North KE, Hu J. GSTM1 and GSTT1 Polymorphisms, Cigarette Smoking, and Risk of Colon Cancer: A Population-based Case-control Study in North Carolina (United States). Cancer Causes & Control. 2006;17:385-394
32. Moore LE, Huang WY, Chatterjee N, Gunter M, Chanock S, Yeager M. et al. GSTM1, GSTT1, and GSTP1 polymorphisms and risk of advanced colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2005;14:1823-7
33. Cai L, Yu SZ, Zhang ZF. Glutathione S-transferases M1, T1 genotypes and the risk of gastric cancer: a case-control study. World J Gastroenterol. 2001;7:506-9
34. Tamer L, Ates NA, Ates C, Ercan B, Elipek T, Yildirim H. et al. Glutathione S-transferase M1, T1 and P1 genetic polymorphisms, cigarette smoking and gastric cancer risk. Cell Biochem Funct. 2005;23:267-72
35. Garcia-Gonzalez MA, Quintero E, Bujanda L, Nicolas D, Benito R, Strunk M. et al. Relevance of GSTM1, GSTT1, and GSTP1 gene polymorphisms to gastric cancer susceptibility and phenotype. Mutagenesis. 2012;27:771-777
36. Slattery ML, Curtin K, Ma K, Schaffer D, Potter J, Samowitz W. GSTM-1 and NAT2 and genetic alterations in colon tumors. Cancer Causes Control. 2002;13:527-34
37. Malik MA, Upadhyay R, Mittal RD, Zargar SA, Modi DR, Mittal B. Role of Xenobiotic-Metabolizing Enzyme Gene Polymorphisms and Interactions with Environmental Factors in Susceptibility to Gastric Cancer in Kashmir Valley. Journal of Gastrointestinal Cancer. 2009;40:26-32
38. Slattery ML, Edwards S, Curtin K, Schaffer D, Neuhausen S. Associations between smoking, passive smoking, GSTM-1, NAT2, and rectal cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:882-9
39. Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, Chen CJ. Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology. 1995;109:1266-73
40. Yu MW, Chiu YH, Chiang YC, Chen CH, Lee TH, Santella RM. et al. Plasma carotenoids, glutathione S-transferase M1 and T1 genetic polymorphisms, and risk of hepatocellular carcinoma: independent and interactive effects. Am J Epidemiol. 1999;149:621-9
41. Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697-702
42. Ateş NA, Tamer L, Ateş C, Ercan B, Elipek T, Öcal K. et al. Glutathione S-Transferase M1, T1, P1 Genotypes and Risk for Development of Colorectal Cancer. Biochemical Genetics. 2005;43:149-163
43. Malakar M, Devi KR, Phukan RK, Kaur T, Deka M, Puia L. et al. Genetic Polymorphism of Glutathione S-transferases M1 and T1, Tobacco Habits and Risk of Stomach Cancer in Mizoram, India. Asian Pacific Journal of Cancer Prevention. 2012;13:4725-4732
44. van der Hel OL, Bueno de Mesquita HB, Roest M, Slothouber B, van Gils C, van Noord PA. et al. No modifying effect of NAT1, GSTM1, and GSTT1 on the relation between smoking and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:681-2
45. Boccia S, Miele L, Panic N, Turati F, Arzani D, Cefalo C. et al. The Effect of CYP, GST,andSULTPolymorphisms and Their Interaction with Smoking on the Risk of Hepatocellular Carcinoma. BioMed Research International. 2015;2015:1-7
46. Gelatti U, Covolo L, Talamini R, Tagger A, Barbone F, Martelli C. et al. N-Acetyltransferase-2, glutathione S-transferase M1 andT1 genetic polymorphisms, cigarette smoking and hepatocellular carcinoma: A case-control study. International Journal of Cancer. 2005;115:301-306
47. Setiawan VW, Zhang ZF, Yu GP, Li YL, Lu ML, Tsai CJ. et al. GSTT1 and GSTM1 null genotypes and the risk of gastric cancer: a case-control study in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2000;9:73-80
48. Chen K, Jiang QT, Ma XY, Yao KY, Leng SG, Yu WP. et al. [Associations between genetic polymorphisms of glutathione S-transferase M1 and T1, smoking and susceptibility to colorectal cancer: a case-control study]. Zhonghua Zhong Liu Za Zhi. 2004;26:645-8
49. Abo-Hashem EM, El-Emshaty WM, Farag RES, Zakaria S, Abd El-Aziz M, Ghonaim A. Genetic Polymorphisms of Cytochrome P4501A1 (CYP1A1) and Glutathione S-Transferase P1 (GSTP1) and Risk of Hepatocellular Carcinoma Among Chronic Hepatitis C Patients in Egypt. Biochemical Genetics. 2016;54:696-713
50. Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP. et al. Genetic polymorphisms of p53 and GSTP1,but not NAT2,are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458-64
51. Moaven O, Raziee HR, Sima HR, Ganji A, Malekzadeh R, A'rabi A. et al. Interactions between Glutathione-S-Transferase M1, T1 and P1 polymorphisms and smoking, and increased susceptibility to esophageal squamous cell carcinoma. Cancer Epidemiology. 2010;34:285-290
52. Ghosh S, Ghosh S, Bankura B, Saha ML, Maji S, Ghatak S. et al. Association of DNA repair and xenobiotic pathway gene polymorphisms with genetic susceptibility to gastric cancer patients in West Bengal, India. Tumor Biology. 2016;37:9139-9149
53. Setiawan VW, Zhang ZF, Yu GP, Lu QY, Li YL, Lu ML. et al. GSTP1 polymorphisms and gastric cancer in a high-risk Chinese population. Cancer Causes Control. 2001;12:673-81
54. Fernandes GMM, Russo A, Proença MA, Gazola NF, Rodrigues GH, Biselli-Chicote PM. et al. CYP1A1, CYP2E1 and EPHX1 polymorphisms in sporadic colorectal neoplasms. World Journal of Gastroenterology. 2016;22:9974
55. Hou L. CYP1A1 Val462 and NQO1 Ser187 polymorphisms, cigarette use, and risk for colorectal adenoma. Carcinogenesis. 2005;26:1122-1128
56. Li R, Shugart YY, Zhou W, An Y, Yang Y, Zhou Y. et al. Common genetic variations of the cytochrome P450 1A1 gene and risk of hepatocellular carcinoma in a Chinese population. European Journal of Cancer. 2009;45:1239-1247
57. Little J, Sharp L, Masson LF, Brockton NT, Cotton SC, Haites NE. et al. Colorectal cancer and genetic polymorphisms ofCYP1A1,GSTM1andGSTT1: A case-control study in the Grampian region of Scotland. International Journal of Cancer. 2006;119:2155-2164
58. Yu MW, Chiu YH, Yang SY, Santella RM, Chern HD, Liaw YF. et al. Cytochrome P450 1A1 genetic polymorphisms and risk of hepatocellular carcinoma among chronic hepatitis B carriers. Br J Cancer. 1999;80:598-603
59. Moslehi R, Chatterjee N, Church TR, Chen J, Yeager M, Weissfeld J. et al. Cigarette smoking, N-acetyltransferase genes and the risk of advanced colorectal adenoma. Pharmacogenomics. 2006;7:819-29
60. Yu MW, Pai CI, Yang SY, Hsiao TJ, Chang HC, Lin SM. et al. Role of N-acetyltransferase polymorphisms in hepatitis B related hepatocellular carcinoma: impact of smoking on risk. Gut. 2000;47:703-9
61. van der Hel OL, Bueno de Mesquita HB, Sandkuijl L, van Noord PA, Pearson PL, Grobbee DE. et al. Rapid N-acetyltransferase 2 imputed phenotype and smoking may increase risk of colorectal cancer in women (Netherlands). Cancer Causes Control. 2003;14:293-8
62. Lilla C, Risch A, Verla-Tebit E, Hoffmeister M, Brenner H, Chang-Claude J. SULT1A1 genotype and susceptibility to colorectal cancer. International Journal of Cancer. 2007;120:201-206
63. Dandara C. Gene-environment interaction: the role of SULT1A1 and CYP3A5 polymorphisms as risk modifiers for squamous cell carcinoma of the oesophagus. Carcinogenesis. 2005;27:791-797
64. Wu M-T, Wang Y-T, Ho C-K, Wu D-C, Lee Y-C, Hsu H-K. et al. SULT1A1 polymorphism and esophageal cancer in males. International Journal of Cancer. 2003;103:101-104
65. Boccia S, Persiani R, La Torre G, Rausei S, Arzani D, Gianfagna F. et al. Sulfotransferase 1A1 polymorphism and gastric cancer risk: a pilot case-control study. Cancer Letters. 2005;229:235-243
66. Tsukino H, Hanaoka T, Otani T, Iwasaki M, Kobayashi M, Hara M. et al. hOGG1 Ser326Cys polymorphism, interaction with environmental exposures, and gastric cancer risk in Japanese populations. Cancer Sci. 2004;95:977-83
67. Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N. et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. Journal of Experimental & Clinical Cancer Research. 2008;27:49
68. Malik MA, Zargar SA, Mittal B. Lack of influence of DNA repair gene OGG1 codon 326 polymorphisms of gastric cancer risk in the Kashmir valley. Asian Pac J Cancer Prev. 2010;11:165-8
69. Songserm N, Promthet S, Pientong C, Ekalaksananan T, Chopjitt P, Wiangnon S. Gene-environment interaction involved in cholangiocarcinoma in the Thai population: polymorphisms of DNA repair genes, smoking and use of alcohol. BMJ Open. 2014;4:e005447
70. Zhang SH. APE1polymorphisms are associated with colorectal cancer susceptibility in Chinese Hans. World Journal of Gastroenterology. 2014;20:8700
71. Yuan T, Wei J, Luo J, Liu M, Deng S, Chen P. Polymorphisms of Base-Excision Repair Genes hOGG1 326cys and XRCC1 280His Increase Hepatocellular Carcinoma Risk. Digestive Diseases and Sciences. 2012;57:2451-2457
72. Sakamoto T, Higaki Y, Hara M, Ichiba M, Horita M, Mizuta T. et al. hOGG1 Ser326Cys polymorphism and risk of hepatocellular carcinoma among Japanese. J Epidemiol. 2006;16:233-9
73. Hanaoka T, Sugimura H, Nagura K, Ihara M, Li XJ, Hamada GS. et al. hOGG1 exon7 polymorphism and gastric cancer in case-control studies of Japanese Brazilians and non-Japanese Brazilians. Cancer Lett. 2001;170:53-61
74. Chen DJ, Ding R, Cao W, Ye DQ. [Interaction between polymorphisms in NQO1(C609T) and XRCC1(G28152A) and their correlation with smoking on gastric cancer]. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:5-8
75. Yan L, Yanan D, Donglan S, Na W, Rongmiao Z, Zhifeng C. Polymorphisms of XRCC1 gene and risk of gastric cardiac adenocarcinoma. Diseases of the Esophagus. 2009;22:396-401
76. Yu HP, Zhang XY, Wang XL, Shi LY, Li YY, Li F. et al. DNA repair gene XRCC1 polymorphisms, smoking, and esophageal cancer risk. Cancer Detection and Prevention. 2004;28:194-199
77. Putthanachote. TheXRCC 1DNA repair gene modifies the environmental risk of stomach cancer: a hospital-based matched case-control study. BMC Cancer. 2017;17:680
78. Malakar M, Devi KR, Phukan RK, Kaur T, Deka M, Puia L. et al. p53 Codon 72 Polymorphism Interactions with Dietary and Tobacco Related Habits and Risk of Stomach Cancer in Mizoram, India. Asian Pacific Journal of Cancer Prevention. 2014;15:717-723
79. Qiu M, Liu Y, Yu X, Qin L, Bei C, Zeng X. et al. Interaction between p53 codon 72 and MDM2 309T>G polymorphisms and the risk of hepatocellular carcinoma. Tumor Biology. 2015;37:3863-3870
80. Shao Y, Tan W, Zhang S. P53 gene codon 72 polymorphism and risk of esophageal squamous cell carcinoma: a case/control study in a Chinese population. Diseases of the Esophagus. 2008;21:139-143
81. Shen H, Solari A, Wang X, Zhang Z, Xu Y, Wang L. et al. P53 codon 72 polymorphism and risk of gastric cancer in a Chinese population. Oncol Rep. 2004;11:1115-20
82. Yang W, Zhang Y, Tian X, Ning T, Ke Y. p53 Codon 72 polymorphism and the risk of esophageal squamous cell carcinoma. Molecular Carcinogenesis. 2008;47:100-104
83. Cai J, Cai Y, Ma Q, Chang F, Xu L, Zhang G. et al. Association of p53 codon 72 polymorphism with susceptibility to hepatocellular carcinoma in a Chinese population from northeast Sichuan. Biomedical Reports. 2017;6:217-222
84. Yu L, Sun L, Jiang Y-F, Lu B-L, Sun D-R, Zhu L-Y. Interactions between CYP1A1 polymorphisms and cigarette smoking are associated with the risk of hepatocellular carcinoma: evidence from epidemiological studies. Molecular Biology Reports. 2012;39:6641-6646
85. Li K, Ren YW, Wan Y, Yin ZH, Wu W, Zhou BS. SULT1A1 Arg213His polymorphism and susceptibility of environment-related cancers: a meta analysis of 5,915 cases and 7,900 controls. Mol Biol Rep. 2012;39:2597-605
86. Gupta B, Kumar N, Johnson NW. Relationship of Lifetime Exposure to Tobacco, Alcohol and Second Hand Tobacco Smoke with Upper aero-digestive tract cancers in India: a Case-Control Study with a Life-Course Perspective. Asian Pac J Cancer Prev. 2017;18:347-356
87. Ramôa CP, Eissenberg T, Sahingur SE. Increasing popularity of waterpipe tobacco smoking and electronic cigarette use: Implications for oral healthcare. Journal of Periodontal Research. 2017;52:813-823
88. Liang C, Marsit CJ, Houseman EA, Butler R, Nelson HH, McClean MD. et al. Gene-environment interactions of novel variants associated with head and neck cancer. Head & Neck. 2012;34:1111-1118
89. Hamachi T. CYP1A1,GSTM1,GSTT1andNQO1polymorphisms and colorectal adenomas in Japanese men. World Journal of Gastroenterology. 2013;19:4023
90. Lao X, Peng Q, Lu Y, Li S, Qin X, Chen Z. et al. Glutathione S-transferase gene GSTM1, gene-gene interaction, and gastric cancer susceptibility: evidence from an updated meta-analysis. Cancer Cell Int. 2014;14:014-0127
91. Koumbi L. Dietary factors can protect against liver cancer development. World Journal of Hepatology. 2017;9:119
92. Ko KP, Kim SW, Ma SH, Park B, Ahn Y, Lee JW. et al. Dietary intake and breast cancer among carriers and noncarriers of BRCA mutations in the Korean Hereditary Breast Cancer Study. American Journal of Clinical Nutrition. 2013;98:1493-1501
93. Molina E, Perez-Morales R, Rubio J, Petrosyan P, Cadena LH, Arlt VM. et al. The GSTM1null (deletion) and MGMT84 rs12917 (Phe/Phe) haplotype are associated with bulky DNA adduct levels in human leukocytes. Mutat Res. 2013;758:62-8
94. Kiyohara C, Wakai K, Mikami H, Sido K, Ando M, Ohno Y. Risk modification byCYP1A1 andGSTM1 polymorphisms in the association of environmental tobacco smoke and lung cancer: A case-control study in Japanese nonsmoking women. International Journal of Cancer. 2003;107:139-144
95. Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129:141-70
96. Hirvonen A. Polymorphic NATs and cancer predisposition. IARC Sci Publ. 1999;148:251-70
Author contact
![]() Corresponding author: Jin Yang, email: yangjinedu.cn; telephone: 86-13572177146
Corresponding author: Jin Yang, email: yangjinedu.cn; telephone: 86-13572177146

 Global reach, higher impact
Global reach, higher impact